MAGI1, a New Potential Tumor Suppressor Gene in Estrogen Receptor Positive Breast Cancer
Abstract
1. Introduction
2. Results
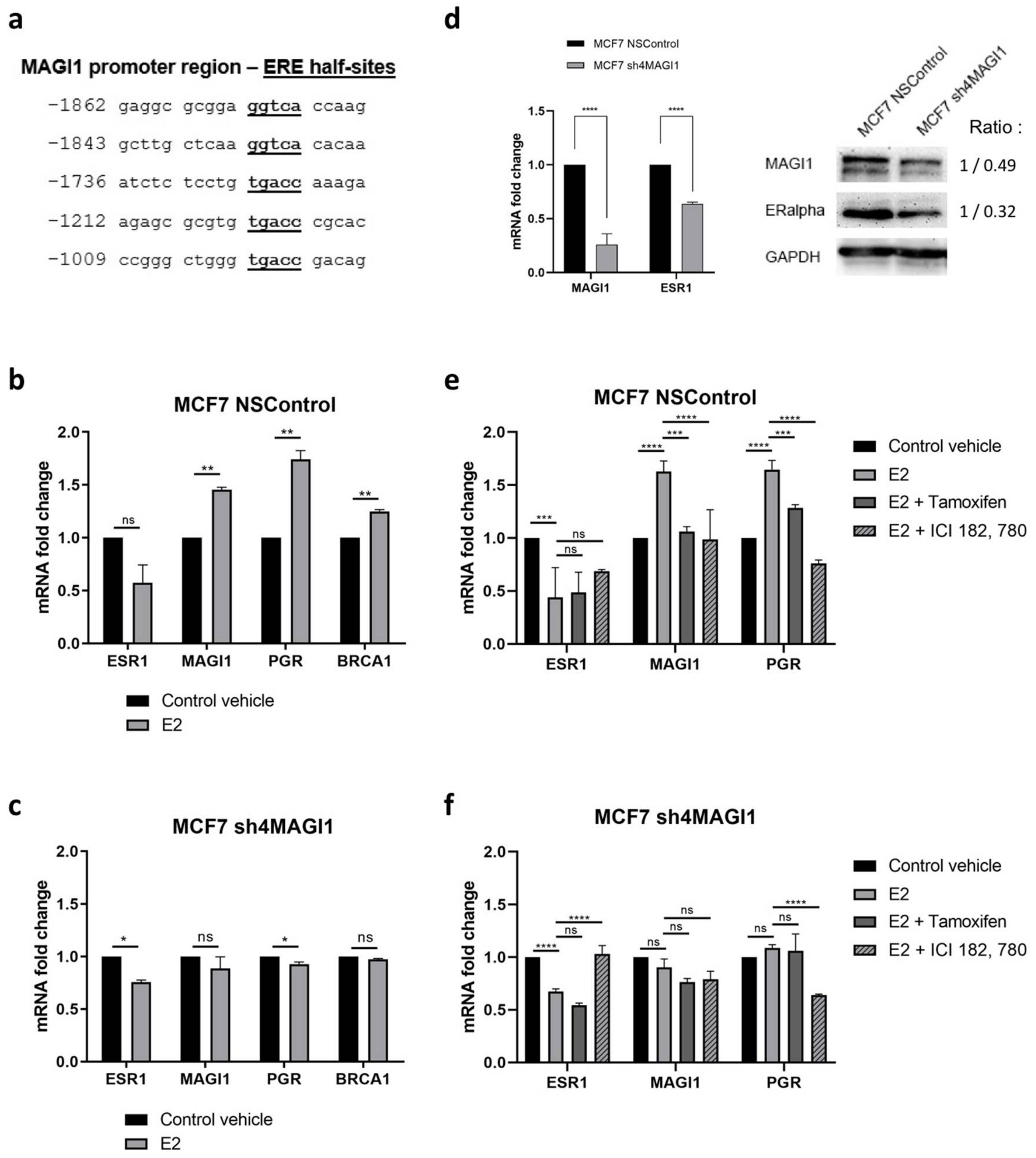
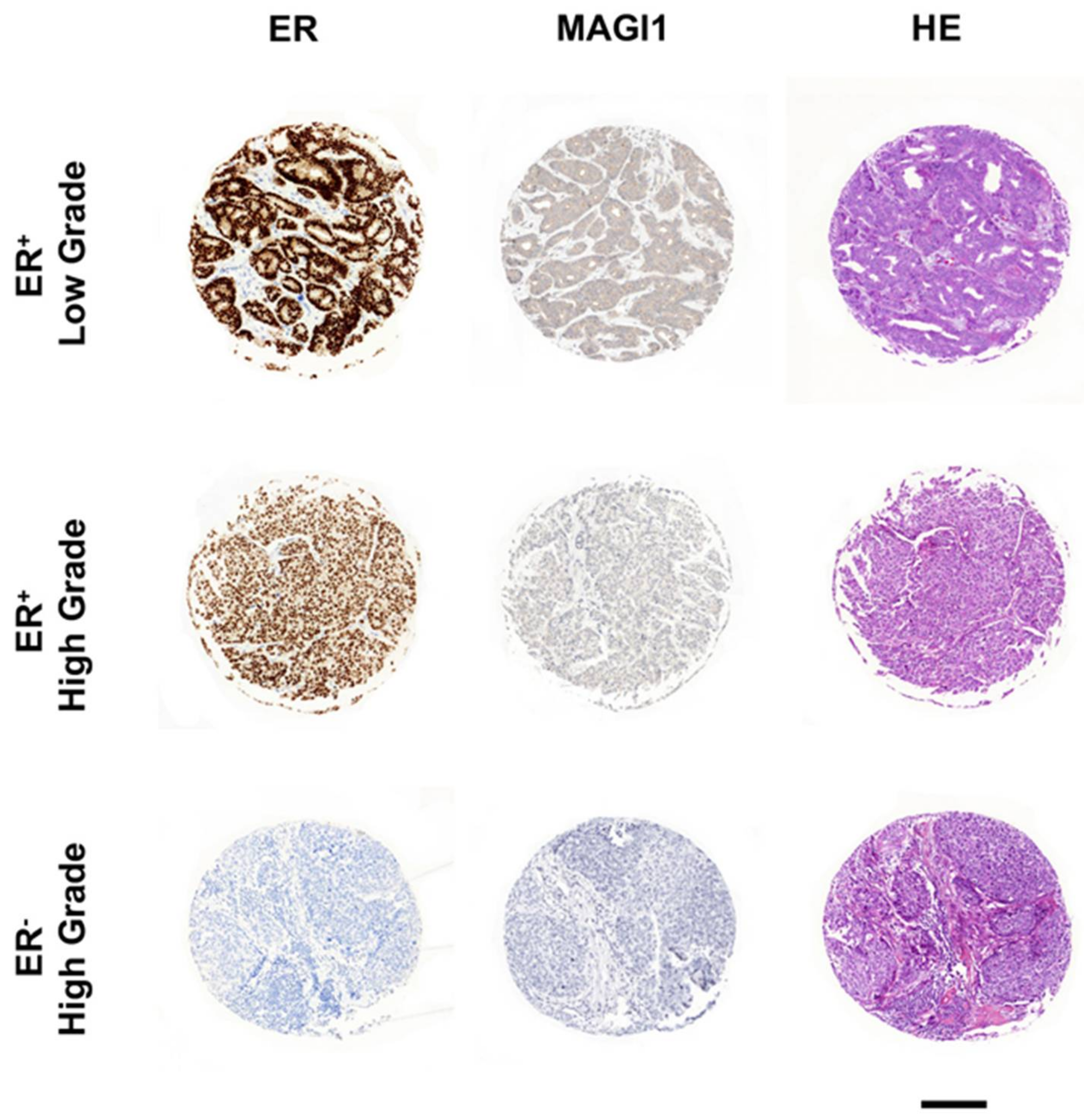
2.1. MAGI1 Is Highly Expressed in ER+ Breast Cancer
2.2. MAGI1 Is Upregulated by Estrogen Receptor Alpha (erα) and Contributes to ER Signaling
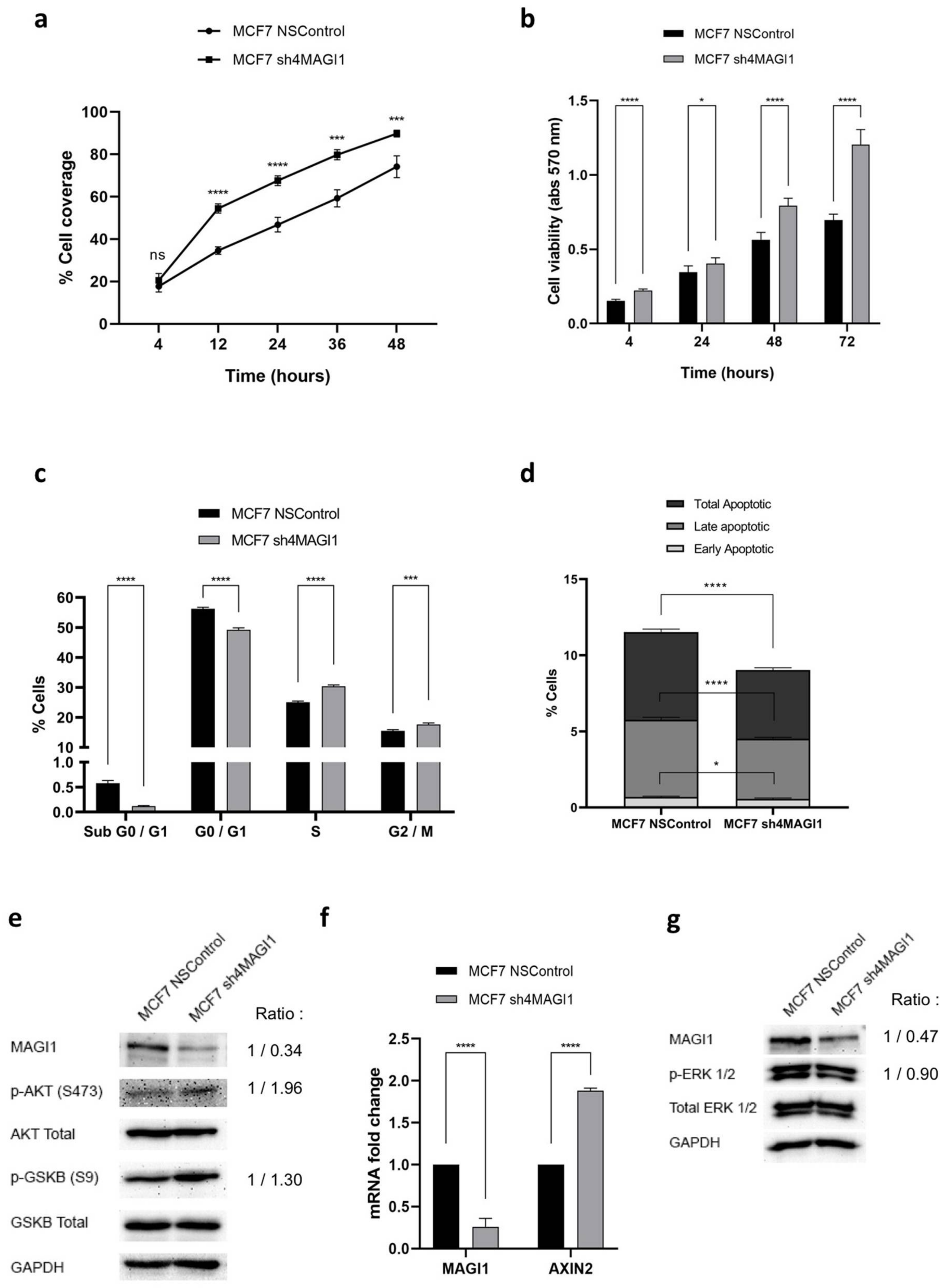
2.3. MAGI1 Downregulation Increases Proliferation, Reduces Apoptosis and Activates PI3K/Wnt Signaling in MCF7 Cells
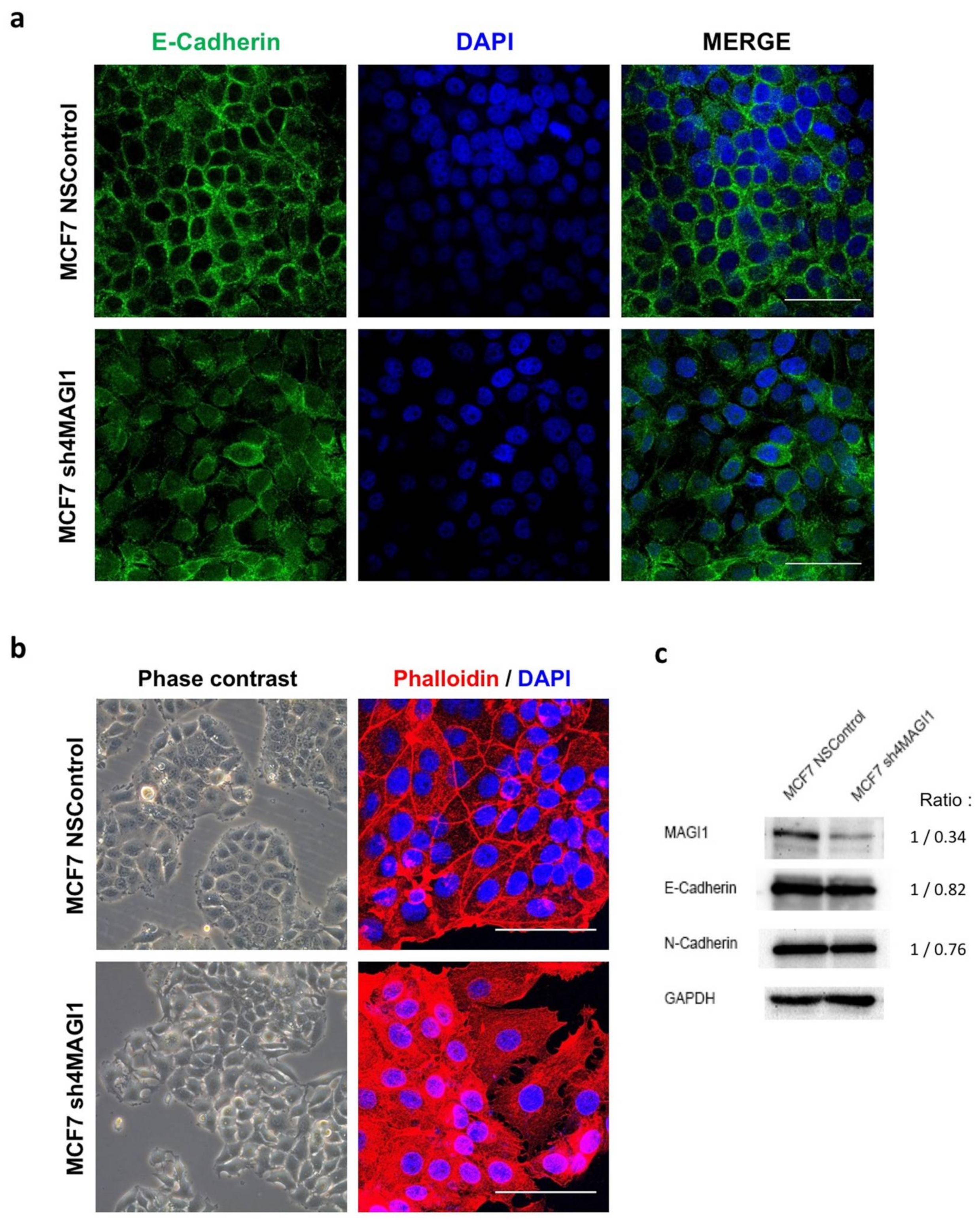
2.4. MAGI1 Downregulation in MCF7 Breast Cancer Cells Reduces Epithelial Differentiation
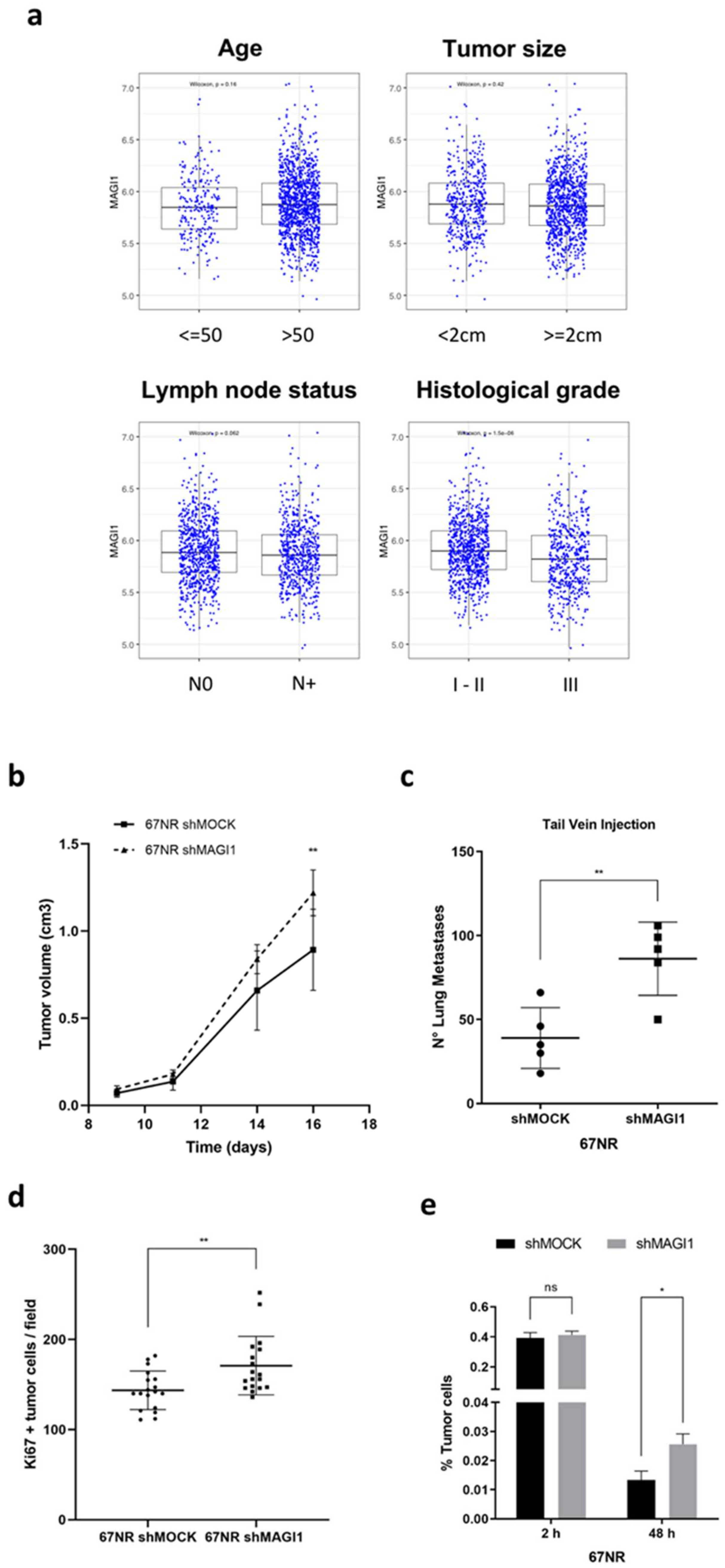
2.5. MAGI1 Downregulation Promotes Metastasis of Disseminated ER+ Cancer Cells
2.6. Low MAGI1 Expression Predicts Poor Prognosis in ER+ Breast Cancer
2.7. MAGI1 Expression Negatively Correlates with Inflammation in ER+ Breast Cancer and Is Downregulated by the Prostaglandin E2 (PGE2)/Cyclooxygenase-2 (COX-2)axis
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Cell Treatments
4.3. Animal Procedures Authorization
4.4. Spontaneous Tumor Models
4.5. Orthotopic Tumor Models
4.6. Experimental Lung Metastasis Model
4.7. Cytoimmunofluorescence Staining
4.8. Tissue Staining
4.9. Bioinformatic Analyses—Gene Expression and Survival Analyses
4.10. Bioinformatic Analyses—Gene Onthology Analyses
4.11. Statistical Analysis (Except Bioinformatics Analysis)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Patrie, K.M.; Drescher, A.J.; Welihinda, A.; Mundel, P.; Margolis, B. Interaction of two actin-binding proteins, synaptopodin and alpha-actinin-4, with the tight junction protein MAGI-1. J. Biol. Chem. 2002, 277, 30183–30190. [Google Scholar] [CrossRef] [PubMed]
- Ide, N.; Hata, Y.; Nishioka, H.; Hirao, K.; Yao, I.; Deguchi, M.; Mizoguchi, A.; Nishimori, H.; Tokino, T.; Nakamura, Y.; et al. Localization of membrane-associated guanylate kinase (MAGI)-1/BAI-associated protein (BAP) 1 at tight junctions of epithelial cells. Oncogene 1999, 18, 7810–7815. [Google Scholar] [CrossRef] [PubMed]
- Laura, R.P.; Ross, S.; Koeppen, H.; Lasky, L.A. MAGI-1: A widely expressed, alternatively spliced tight junction protein. Exp. Cell Res. 2002, 275, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Zaric, J.; Joseph, J.M.; Tercier, S.; Sengstag, T.; Ponsonnet, L.; Delorenzi, M.; Ruegg, C. Identification of MAGI1 as a tumor-suppressor protein induced by cyclooxygenase-2 inhibitors in colorectal cancer cells. Oncogene 2012, 31, 48–59. [Google Scholar] [CrossRef]
- Zhang, G.; Liu, T.; Wang, Z. Downregulation of MAGI1 associates with poor prognosis of hepatocellular carcinoma. J. Investig. Surg. 2012, 25, 93–99. [Google Scholar] [CrossRef]
- Jia, S.; Lu, J.; Qu, T.; Feng, Y.; Wang, X.; Liu, C.; Ji, J. MAGI1 inhibits migration and invasion via blocking MAPK/ERK signaling pathway in gastric cancer. Chin. J. Cancer Res. 2017, 29, 25–35. [Google Scholar] [CrossRef]
- Kranjec, C.; Massimi, P.; Banks, L. Restoration of MAGI-1 expression in human papillomavirus-positive tumor cells induces cell growth arrest and apoptosis. J. Virol. 2014, 88, 7155–7169. [Google Scholar] [CrossRef]
- Kitamura, K.; Seike, M.; Okano, T.; Matsuda, K.; Miyanaga, A.; Mizutani, H.; Noro, R.; Minegishi, Y.; Kubota, K.; Gemma, A. MiR-134/487b/655 cluster regulates TGF-beta-induced epithelial-mesenchymal transition and drug resistance to gefitinib by targeting MAGI2 in lung adenocarcinoma cells. Mol. Cancer Ther. 2014, 13, 444–453. [Google Scholar] [CrossRef]
- Kotelevets, L.; van Hengel, J.; Bruyneel, E.; Mareel, M.; van Roy, F.; Chastre, E. Implication of the MAGI-1b/PTEN signalosome in stabilization of adherens junctions and suppression of invasiveness. FASEB J. 2005, 19, 115–117. [Google Scholar] [CrossRef]
- Dobrosotskaya, I.Y.; James, G.L. MAGI-1 interacts with beta-catenin and is associated with cell-cell adhesion structures. Biochem. Biophys. Res. Commun. 2000, 270, 903–909. [Google Scholar] [CrossRef]
- Ma, Q.; Zhang, Y.; Meng, R.; Xie, K.M.; Xiong, Y.; Lin, S.; He, Z.L.; Tao, T.; Yang, Y.; Zhao, J.Z.; et al. MAGI3 Suppresses Glioma Cell Proliferation via Upregulation of PTEN Expression. Biomed. Environ. Sci. 2015, 28, 502–509. [Google Scholar] [CrossRef] [PubMed]
- Turashvili, G.; Brogi, E. Tumor Heterogeneity in Breast Cancer. Front. Med. 2017, 4, 227. [Google Scholar] [CrossRef] [PubMed]
- Yersal, O.; Barutca, S. Biological subtypes of breast cancer: Prognostic and therapeutic implications. World J. Clin. Oncol. 2014, 5, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Colozza, M.; de Azambuja, E.; Personeni, N.; Lebrun, F.; Piccart, M.J.; Cardoso, F. Achievements in systemic therapies in the pregenomic era in metastatic breast cancer. Oncologist 2007, 12, 253–270. [Google Scholar] [CrossRef]
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef]
- Lyons, T.G. Targeted Therapies for Triple-Negative Breast Cancer. Curr. Treat. Options Oncol. 2019, 20, 82. [Google Scholar] [CrossRef]
- Savard, M.F.; Khan, O.; Hunt, K.K.; Verma, S. Redrawing the Lines: The Next Generation of Treatment in Metastatic Breast Cancer. Am. Soc. Clin. Oncol. Educ. Book 2019, 39, e8–e21. [Google Scholar] [CrossRef]
- Andre, F.; Ciruelos, E.; Rubovszky, G.; Campone, M.; Loibl, S.; Rugo, H.S.; Iwata, H.; Conte, P.; Mayer, I.A.; Kaufman, B.; et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N. Engl. J. Med. 2019, 380, 1929–1940. [Google Scholar] [CrossRef]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef]
- Sorlie, T.; Tibshirani, R.; Parker, J.; Hastie, T.; Marron, J.S.; Nobel, A.; Deng, S.; Johnsen, H.; Pesich, R.; Geisler, S.; et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. USA 2003, 100, 8418–8423. [Google Scholar] [CrossRef] [PubMed]
- Sotiriou, C.; Neo, S.Y.; McShane, L.M.; Korn, E.L.; Long, P.M.; Jazaeri, A.; Martiat, P.; Fox, S.B.; Harris, A.L.; Liu, E.T. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc. Natl. Acad. Sci. USA 2003, 100, 10393–10398. [Google Scholar] [CrossRef] [PubMed]
- Tran, B.; Bedard, P.L. Luminal-B breast cancer and novel therapeutic targets. Breast Cancer Res. 2011, 13, 221. [Google Scholar] [CrossRef] [PubMed]
- Paik, S.; Shak, S.; Tang, G.; Kim, C.; Baker, J.; Cronin, M.; Baehner, F.L.; Walker, M.G.; Watson, D.; Park, T.; et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004, 351, 2817–2826. [Google Scholar] [CrossRef] [PubMed]
- Sparano, J.A.; Gray, R.J.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E., Jr.; Dees, E.C.; Goetz, M.P.; Olson, J.A., Jr.; et al. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N. Engl. J. Med. 2018, 379, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Sparano, J.A.; Gray, R.J.; Ravdin, P.M.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E., Jr.; Dees, E.C.; Goetz, M.P.; et al. Clinical and Genomic Risk to Guide the Use of Adjuvant Therapy for Breast Cancer. N. Engl. J. Med. 2019, 380, 2395–2405. [Google Scholar] [CrossRef]
- Olsen, S.N.; Wronski, A.; Castano, Z.; Dake, B.; Malone, C.; De Raedt, T.; Enos, M.; DeRose, Y.S.; Zhou, W.; Guerra, S.; et al. Loss of RasGAP Tumor Suppressors Underlies the Aggressive Nature of Luminal B Breast Cancers. Cancer Discov. 2017, 7, 202–217. [Google Scholar] [CrossRef]
- Pietras, R.J.; Marquez-Garban, D.C. Membrane-associated estrogen receptor signaling pathways in human cancers. Clin. Cancer Res. 2007, 13, 4672–4676. [Google Scholar] [CrossRef]
- Creighton, C.J. The molecular profile of luminal B breast cancer. Biologics 2012, 6, 289–297. [Google Scholar] [CrossRef]
- Dai, X.; Li, T.; Bai, Z.; Yang, Y.; Liu, X.; Zhan, J.; Shi, B. Breast cancer intrinsic subtype classification, clinical use and future trends. Am. J. Cancer Res. 2015, 5, 2929–2943. [Google Scholar]
- Sotiriou, C.; Pusztai, L. Gene-expression signatures in breast cancer. N. Engl. J. Med. 2009, 360, 790–800. [Google Scholar] [CrossRef] [PubMed]
- van ’t Veer, L.J.; Dai, H.; van de Vijver, M.J.; He, Y.D.; Hart, A.A.; Mao, M.; Peterse, H.L.; van der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, F.; van’t Veer, L.J.; Bogaerts, J.; Slaets, L.; Viale, G.; Delaloge, S.; Pierga, J.Y.; Brain, E.; Causeret, S.; DeLorenzi, M.; et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N. Engl. J. Med. 2016, 375, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Pereira, B.; Chin, S.F.; Rueda, O.M.; Vollan, H.K.; Provenzano, E.; Bardwell, H.A.; Pugh, M.; Jones, L.; Russell, R.; Sammut, S.J.; et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 2016, 7, 11479. [Google Scholar] [CrossRef] [PubMed]
- Ciriello, G.; Gatza, M.L.; Beck, A.H.; Wilkerson, M.D.; Rhie, S.K.; Pastore, A.; Zhang, H.; McLellan, M.; Yau, C.; Kandoth, C.; et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015, 163, 506–519. [Google Scholar] [CrossRef]
- Rangel, N.; Villegas, V.E.; Rondon-Lagos, M. Profiling of gene expression regulated by 17beta-estradiol and tamoxifen in estrogen receptor-positive and estrogen receptor-negative human breast cancer cell lines. Breast Cancer 2017, 9, 537–550. [Google Scholar] [CrossRef]
- Stoica, G.E.; Franke, T.F.; Moroni, M.; Mueller, S.; Morgan, E.; Iann, M.C.; Winder, A.D.; Reiter, R.; Wellstein, A.; Martin, M.B.; et al. Effect of estradiol on estrogen receptor-alpha gene expression and activity can be modulated by the ErbB2/PI 3-K/Akt pathway. Oncogene 2003, 22, 7998–8011. [Google Scholar] [CrossRef]
- Improta-Brears, T.; Whorton, A.R.; Codazzi, F.; York, J.D.; Meyer, T.; McDonnell, D.P. Estrogen-induced activation of mitogen-activated protein kinase requires mobilization of intracellular calcium. Proc. Natl. Acad. Sci. USA 1999, 96, 4686–4691. [Google Scholar] [CrossRef]
- Ejlertsen, B. Adjuvant chemotherapy in early breast cancer. Dan. Med. J. 2016, 63, B5222. [Google Scholar]
- Miller, T.W.; Rexer, B.N.; Garrett, J.T.; Arteaga, C.L. Mutations in the phosphatidylinositol 3-kinase pathway: Role in tumor progression and therapeutic implications in breast cancer. Breast Cancer Res. 2011, 13, 224. [Google Scholar] [CrossRef]
- Bosch, A.; Li, Z.; Bergamaschi, A.; Ellis, H.; Toska, E.; Prat, A.; Tao, J.J.; Spratt, D.E.; Viola-Villegas, N.T.; Castel, P.; et al. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor-positive breast cancer. Sci. Transl. Med. 2015, 7, 283ra51. [Google Scholar] [CrossRef] [PubMed]
- Beaver, J.A.; Gustin, J.P.; Yi, K.H.; Rajpurohit, A.; Thomas, M.; Gilbert, S.F.; Rosen, D.M.; Ho Park, B.; Lauring, J. PIK3CA and AKT1 mutations have distinct effects on sensitivity to targeted pathway inhibitors in an isogenic luminal breast cancer model system. Clin. Cancer Res. 2013, 19, 5413–5422. [Google Scholar] [CrossRef] [PubMed]
- Usary, J.; Llaca, V.; Karaca, G.; Presswala, S.; Karaca, M.; He, X.; Langerod, A.; Karesen, R.; Oh, D.S.; Dressler, L.G.; et al. Mutation of GATA3 in human breast tumors. Oncogene 2004, 23, 7669–7678. [Google Scholar] [CrossRef] [PubMed]
- Cross, D.A.; Alessi, D.R.; Cohen, P.; Andjelkovich, M.; Hemmings, B.A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 1995, 378, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Jho, E.H.; Zhang, T.; Domon, C.; Joo, C.K.; Freund, J.N.; Costantini, F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell Biol. 2002, 22, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Aslakson, C.J.; Miller, F.R. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992, 52, 1399–1405. [Google Scholar]
- Yang, J.; Mani, S.A.; Donaher, J.L.; Ramaswamy, S.; Itzykson, R.A.; Come, C.; Savagner, P.; Gitelman, I.; Richardson, A.; Weinberg, R.A. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004, 117, 927–939. [Google Scholar] [CrossRef]
- Johnston, S.R.; Saccani-Jotti, G.; Smith, I.E.; Salter, J.; Newby, J.; Coppen, M.; Ebbs, S.R.; Dowsett, M. Changes in estrogen receptor, progesterone receptor, and pS2 expression in tamoxifen-resistant human breast cancer. Cancer Res. 1995, 55, 3331–3338. [Google Scholar]
- Lorusso, G.; Ruegg, C. New insights into the mechanisms of organ-specific breast cancer metastasis. Semin. Cancer Biol. 2012, 22, 226–233. [Google Scholar] [CrossRef]
- Haibe-Kains, B.; Desmedt, C.; Loi, S.; Culhane, A.C.; Bontempi, G.; Quackenbush, J.; Sotiriou, C. A three-gene model to robustly identify breast cancer molecular subtypes. J. Natl. Cancer Inst. 2012, 104, 311–325. [Google Scholar] [CrossRef]
- Jasem, J.; Amini, A.; Rabinovitch, R.; Borges, V.F.; Elias, A.; Fisher, C.M.; Kabos, P. 21-Gene Recurrence Score Assay As a Predictor of Adjuvant Chemotherapy Administration for Early-Stage Breast Cancer: An Analysis of Use, Therapeutic Implications, and Disparity Profile. J. Clin. Oncol. 2016, 34, 1995–2002. [Google Scholar] [CrossRef] [PubMed]
- Vieira, A.F.; Schmitt, F. An Update on Breast Cancer Multigene Prognostic Tests-Emergent Clinical Biomarkers. Front. Med. 2018, 5, 248. [Google Scholar] [CrossRef] [PubMed]
- Multhoff, G.; Molls, M.; Radons, J. Chronic inflammation in cancer development. Front. Immunol. 2011, 2, 98. [Google Scholar] [CrossRef]
- Nakanishi, M.; Rosenberg, D.W. Multifaceted roles of PGE2 in inflammation and cancer. Semin. Immunopathol. 2013, 35, 123–137. [Google Scholar] [CrossRef]
- Fischer, S.M.; Hawk, E.T.; Lubet, R.A. Coxibs and other nonsteroidal anti-inflammatory drugs in animal models of cancer chemoprevention. Cancer Prev. Res. 2011, 4, 1728–1735. [Google Scholar] [CrossRef]
- Liu, C.H.; Chang, S.H.; Narko, K.; Trifan, O.C.; Wu, M.T.; Smith, E.; Haudenschild, C.; Lane, T.F.; Hla, T. Overexpression of cyclooxygenase-2 is sufficient to induce tumorigenesis in transgenic mice. J. Biol. Chem. 2001, 276, 18563–18569. [Google Scholar] [CrossRef]
- Kuukasjarvi, T.; Kononen, J.; Helin, H.; Holli, K.; Isola, J. Loss of estrogen receptor in recurrent breast cancer is associated with poor response to endocrine therapy. J. Clin. Oncol. 1996, 14, 2584–2589. [Google Scholar] [CrossRef]
- Elzawahry, H.M.; Saber, M.M.; Mokhtar, N.M.; Zeeneldin, A.A.; Ismail, Y.M.; Alieldin, N.H. Role of Ki67 in predicting resistance to adjuvant tamoxifen in postmenopausal breast cancer patients. J. Egypt. Natl. Cancer Inst. 2013, 25, 181–191. [Google Scholar] [CrossRef]
- Cianfrocca, M.; Goldstein, L.J. Prognostic and predictive factors in early-stage breast cancer. Oncologist 2004, 9, 606–616. [Google Scholar] [CrossRef]
- Paik, S.; Tang, G.; Shak, S.; Kim, C.; Baker, J.; Kim, W.; Cronin, M.; Baehner, F.L.; Watson, D.; Bryant, J.; et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J. Clin. Oncol. 2006, 24, 3726–3734. [Google Scholar] [CrossRef]
- Rosen, P.P.; Groshen, S.; Saigo, P.E.; Kinne, D.W.; Hellman, S. Pathological prognostic factors in stage I (T1N0M0) and stage II (T1N1M0) breast carcinoma: A study of 644 patients with median follow-up of 18 years. J. Clin. Oncol. 1989, 7, 1239–1251. [Google Scholar] [CrossRef] [PubMed]
- Labelle, M.; Begum, S.; Hynes, R.O. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell 2011, 20, 576–590. [Google Scholar] [CrossRef] [PubMed]
- Labelle, M.; Begum, S.; Hynes, R.O. Platelets guide the formation of early metastatic niches. Proc. Natl. Acad. Sci. USA 2014, 111, E3053–E3061. [Google Scholar] [CrossRef] [PubMed]
- Labelle, M.; Hynes, R.O. The initial hours of metastasis: The importance of cooperative host-tumor cell interactions during hematogenous dissemination. Cancer Discov. 2012, 2, 1091–1099. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hao, Q.; Cao, W.; Vadgama, J.V.; Wu, Y. Celecoxib in breast cancer prevention and therapy. Cancer Manag. Res. 2018, 10, 4653–4667. [Google Scholar] [CrossRef]
- Markosyan, N.; Chen, E.P.; Ndong, V.N.; Yao, Y.; Sterner, C.J.; Chodosh, L.A.; Lawson, J.A.; Fitzgerald, G.A.; Smyth, E.M. Deletion of cyclooxygenase 2 in mouse mammary epithelial cells delays breast cancer onset through augmentation of type 1 immune responses in tumors. Carcinogenesis 2011, 32, 1441–1449. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Dubois, R.N. Prostaglandins and cancer. Gut 2006, 55, 115–122. [Google Scholar] [CrossRef]
- Moris, D.; Kontos, M.; Spartalis, E.; Fentiman, I.S. The Role of NSAIDs in Breast Cancer Prevention and Relapse: Current Evidence and Future Perspectives. Breast Care 2016, 11, 339–344. [Google Scholar] [CrossRef]
- Bardia, A.; Olson, J.E.; Vachon, C.M.; Lazovich, D.; Vierkant, R.A.; Wang, A.H.; Limburg, P.J.; Anderson, K.E.; Cerhan, J.R. Effect of aspirin and other NSAIDs on postmenopausal breast cancer incidence by hormone receptor status: Results from a prospective cohort study. Breast Cancer Res. Treat. 2011, 126, 149–155. [Google Scholar] [CrossRef]
- Brasky, T.M.; Bonner, M.R.; Moysich, K.B.; Ambrosone, C.B.; Nie, J.; Tao, M.H.; Edge, S.B.; Kallakury, B.V.; Marian, C.; Goerlitz, D.S.; et al. Non-steroidal anti-inflammatory drugs (NSAIDs) and breast cancer risk: Differences by molecular subtype. Cancer Causes Control 2011, 22, 965–975. [Google Scholar] [CrossRef]
- Desmedt, C.; Demicheli, R.; Fornili, M.; Bachir, I.; Duca, M.; Viglietti, G.; Berliere, M.; Piccart, M.; Sotiriou, C.; Sosnowski, M.; et al. Potential Benefit of Intra-operative Administration of Ketorolac on Breast Cancer Recurrence According to the Patient’s Body Mass Index. J. Natl. Cancer Inst. 2018, 110, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- Fabi, A.; Metro, G.; Papaldo, P.; Mottolese, M.; Melucci, E.; Carlini, P.; Sperduti, I.; Russillo, M.; Gelibter, A.; Ferretti, G.; et al. Impact of celecoxib on capecitabine tolerability and activity in pretreated metastatic breast cancer: Results of a phase II study with biomarker evaluation. Cancer Chemother. Pharmacol. 2008, 62, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Chow, L.W.; Loo, W.T.; Yip, A.Y.; Ng, E.L. Acceptable cardiac safety profile of neoadjuvant 5-fluorouracil, epirubicin, cyclophosphamide and celecoxib (FEC-C) for breast cancer: A subanalysis of biomarkers for cardiac injury. Int. J. Biol. Markers 2013, 28, E92–E99. [Google Scholar] [CrossRef] [PubMed]
- Blair, C.K.; Sweeney, C.; Anderson, K.E.; Folsom, A.R. NSAID use and survival after breast cancer diagnosis in post-menopausal women. Breast Cancer Res. Treat. 2007, 101, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Fraser, D.M.; Sullivan, F.M.; Thompson, A.M.; McCowan, C. Aspirin use and survival after the diagnosis of breast cancer: A population-based cohort study. Br. J. Cancer 2014, 111, 623–627. [Google Scholar] [CrossRef] [PubMed]
- Holmes, M.D.; Chen, W.Y.; Li, L.; Hertzmark, E.; Spiegelman, D.; Hankinson, S.E. Aspirin intake and survival after breast cancer. J. Clin. Oncol. 2010, 28, 1467–1472. [Google Scholar] [CrossRef] [PubMed]
- Kwan, M.L.; Habel, L.A.; Slattery, M.L.; Caan, B. NSAIDs and breast cancer recurrence in a prospective cohort study. Cancer Causes Control. 2007, 18, 613–620. [Google Scholar] [CrossRef]
- Hamy, A.S.; Tury, S.; Wang, X.; Gao, J.; Pierga, J.Y.; Giacchetti, S.; Brain, E.; Pistilli, B.; Marty, M.; Espie, M.; et al. Celecoxib With Neoadjuvant Chemotherapy for Breast Cancer Might Worsen Outcomes Differentially by COX-2 Expression and ER Status: Exploratory Analysis of the REMAGUS02 Trial. J. Clin. Oncol. 2019, 37, 624–635. [Google Scholar] [CrossRef]
- Frisk, G.; Ekberg, S.; Lidbrink, E.; Eloranta, S.; Sund, M.; Fredriksson, I.; Lambe, M.; Smedby, K.E. No association between low-dose aspirin use and breast cancer outcomes overall: A Swedish population-based study. Breast Cancer Res. 2018, 20, 142. [Google Scholar] [CrossRef]
- Baumgarten, S.C.; Frasor, J. Minireview: Inflammation: An instigator of more aggressive estrogen receptor (ER) positive breast cancers. Mol. Endocrinol. 2012, 26, 360–371. [Google Scholar] [CrossRef]
- Murray, J.I.; West, N.R.; Murphy, L.C.; Watson, P.H. Intratumoural inflammation and endocrine resistance in breast cancer. Endocr. Relat. Cancer 2015, 22, R51–R67. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.; Jordan, V.C. New insights into acquired endocrine resistance of breast cancer. Cancer Drug Resist. 2019, 2, 198–209. [Google Scholar] [CrossRef] [PubMed]
- Weitzenfeld, P.; Meron, N.; Leibovich-Rivkin, T.; Meshel, T.; Ben-Baruch, A. Progression of luminal breast tumors is promoted by menage a trois between the inflammatory cytokine TNFalpha and the hormonal and growth-supporting arms of the tumor microenvironment. Mediat. Inflamm. 2013, 2013, 720536. [Google Scholar] [CrossRef] [PubMed]
- Guy, C.T.; Webster, M.A.; Schaller, M.; Parsons, T.J.; Cardiff, R.D.; Muller, W.J. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc. Natl. Acad. Sci. USA 1992, 89, 10578–10582. [Google Scholar] [CrossRef] [PubMed]
- Desmedt, C.; Haibe-Kains, B.; Wirapati, P.; Buyse, M.; Larsimont, D.; Bontempi, G.; Delorenzi, M.; Piccart, M.; Sotiriou, C. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin. Cancer Res. 2008, 14, 5158–5165. [Google Scholar] [CrossRef] [PubMed]
- Sotiriou, C.; Wirapati, P.; Loi, S.; Harris, A.; Fox, S.; Smeds, J.; Nordgren, H.; Farmer, P.; Praz, V.; Haibe-Kains, B.; et al. Gene expression profiling in breast cancer: Understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 2006, 98, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Finak, G.; Bertos, N.; Pepin, F.; Sadekova, S.; Souleimanova, M.; Zhao, H.; Chen, H.; Omeroglu, G.; Meterissian, S.; Omeroglu, A.; et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat. Med. 2008, 14, 518–527. [Google Scholar] [CrossRef]
- Perez, E.A.; Thompson, E.A.; Ballman, K.V.; Anderson, S.K.; Asmann, Y.W.; Kalari, K.R.; Eckel-Passow, J.E.; Dueck, A.C.; Tenner, K.S.; Jen, J.; et al. Genomic analysis reveals that immune function genes are strongly linked to clinical outcome in the North Central Cancer Treatment Group n9831 Adjuvant Trastuzumab Trial. J. Clin. Oncol. 2015, 33, 701–708. [Google Scholar] [CrossRef]
- Teschendorff, A.E.; Miremadi, A.; Pinder, S.E.; Ellis, I.O.; Caldas, C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007, 8, R157. [Google Scholar] [CrossRef]
- Tamborero, D.; Rubio-Perez, C.; Muinos, F.; Sabarinathan, R.; Piulats, J.M.; Muntasell, A.; Dienstmann, R.; Lopez-Bigas, N.; Gonzalez-Perez, A. A Pan-cancer Landscape of Interactions between Solid Tumors and Infiltrating Immune Cell Populations. Clin. Cancer Res. 2018, 24, 3717–3728. [Google Scholar] [CrossRef]
- Gu-Trantien, C.; Loi, S.; Garaud, S.; Equeter, C.; Libin, M.; de Wind, A.; Ravoet, M.; Le Buanec, H.; Sibille, C.; Manfouo-Foutsop, G.; et al. CD4+ follicular helper T cell infiltration predicts breast cancer survival. J. Clin. Investig. 2013, 123, 2873–2892. [Google Scholar] [CrossRef] [PubMed]
- Ignatiadis, M.; Singhal, S.K.; Desmedt, C.; Haibe-Kains, B.; Criscitiello, C.; Andre, F.; Loi, S.; Piccart, M.; Michiels, S.; Sotiriou, C. Gene modules and response to neoadjuvant chemotherapy in breast cancer subtypes: A pooled analysis. J. Clin. Oncol. 2012, 30, 1996–2004. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alday-Parejo, B.; Richard, F.; Wörthmüller, J.; Rau, T.; Galván, J.A.; Desmedt, C.; Santamaria-Martinez, A.; Rüegg, C. MAGI1, a New Potential Tumor Suppressor Gene in Estrogen Receptor Positive Breast Cancer. Cancers 2020, 12, 223. https://doi.org/10.3390/cancers12010223
Alday-Parejo B, Richard F, Wörthmüller J, Rau T, Galván JA, Desmedt C, Santamaria-Martinez A, Rüegg C. MAGI1, a New Potential Tumor Suppressor Gene in Estrogen Receptor Positive Breast Cancer. Cancers. 2020; 12(1):223. https://doi.org/10.3390/cancers12010223
Chicago/Turabian StyleAlday-Parejo, Begoña, François Richard, Janine Wörthmüller, Tilman Rau, José A. Galván, Christine Desmedt, Albert Santamaria-Martinez, and Curzio Rüegg. 2020. "MAGI1, a New Potential Tumor Suppressor Gene in Estrogen Receptor Positive Breast Cancer" Cancers 12, no. 1: 223. https://doi.org/10.3390/cancers12010223
APA StyleAlday-Parejo, B., Richard, F., Wörthmüller, J., Rau, T., Galván, J. A., Desmedt, C., Santamaria-Martinez, A., & Rüegg, C. (2020). MAGI1, a New Potential Tumor Suppressor Gene in Estrogen Receptor Positive Breast Cancer. Cancers, 12(1), 223. https://doi.org/10.3390/cancers12010223






