TYK2 in Tumor Immunosurveillance
Abstract
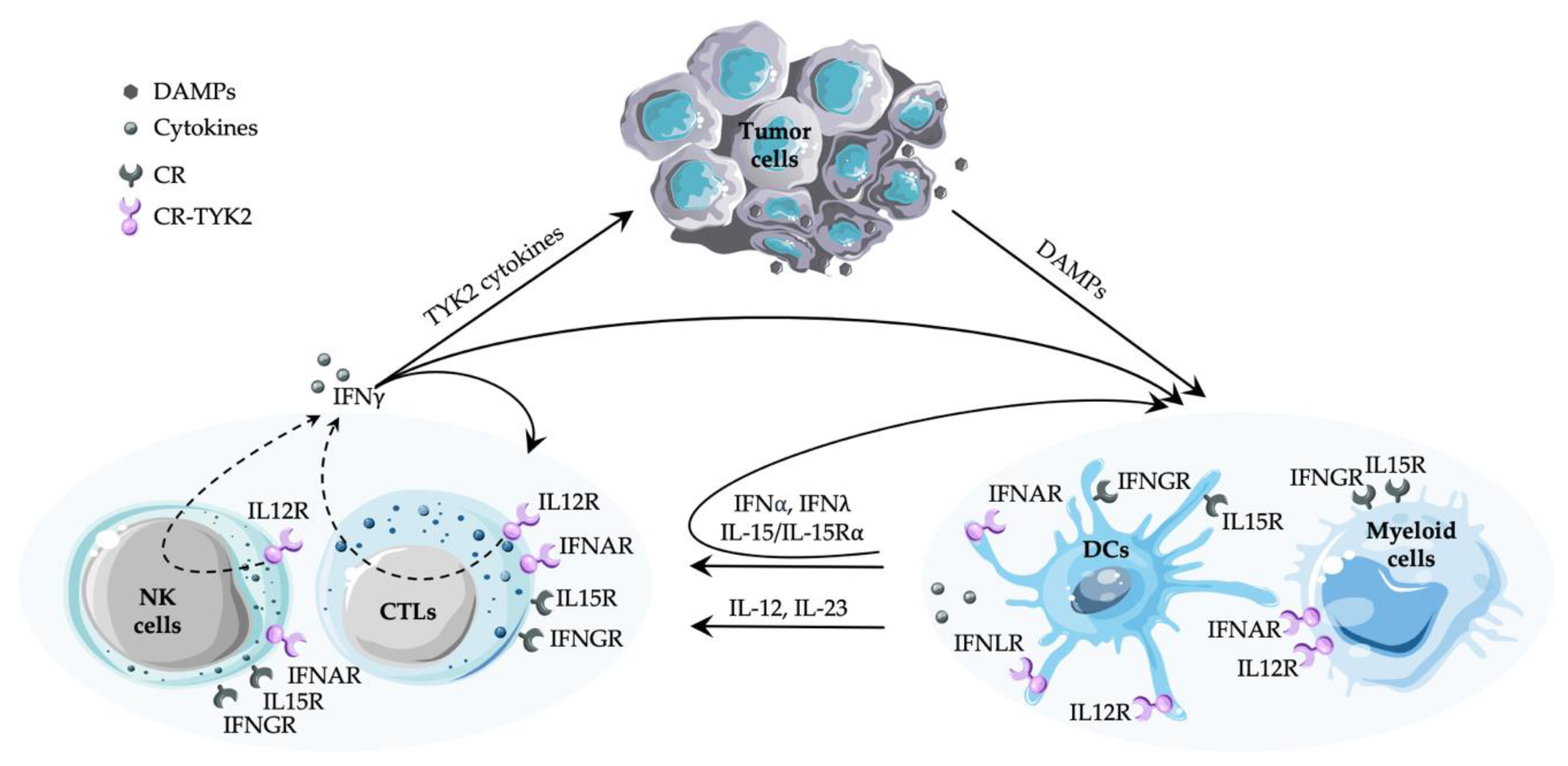
1. Tumor Immunosurveillance and Tumor Microenvironment
2. Identification and Structure-Function Relations of TYK2
3. Loss-of-Function Mutations of TYK2 in Men and Mice
4. TYK2-Dependent Cytokine Responses and Their Involvement in Immunity to Cancer
4.1. Type I IFNs
4.2. IL-12 and IL-23
4.2.1. IL-12
4.2.2. IL-23
4.3. IL-10 Family Cytokines
4.3.1. IL-10
4.3.2. IL-22
4.3.3. IL-26
4.3.4. Type III IFNs
5. Cytokines Produced in Major Dependence of TYK2
5.1. Type I IFN
5.2. IL-15
5.3. Type II IFN
5.4. IL-17
6. TYK2 and Immunity to Cancer
7. Conclusions and Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Finn, O.J. A Believer’s overview of cancer immunosurveillance and immunotherapy. J. Immunol. 2018, 200, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Fridman, W.H. From cancer immune surveillance to cancer immunoediting: Birth of modern immuno-oncology. J. Immunol. 2018, 201, 825–826. [Google Scholar] [CrossRef] [PubMed]
- Ribatti, D. The concept of immune surveillance against tumors. The first theories. Oncotarget 2017, 8, 7175–7180. [Google Scholar] [CrossRef] [PubMed]
- Dunn, G.P.; Bruce, A.T.; Ikeda, H.; Old, L.J.; Schreiber, R.D. Cancer immunoediting: from immunosurveillance to tumor escape. Nat. Immunol. 2002, 3, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Dunn, G.P.; Old, L.J.; Schreiber, R.D. The immunobiology of cancer immunosurveillance and immunoediting. Immunity 2004, 21, 137–148. [Google Scholar] [CrossRef]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer immunoediting: Integrating immunity’s roles in cancer suppression and promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef]
- Mittal, D.; Gubin, M.M.; Schreiber, R.D.; Smyth, M.J. New insights into cancer immunoediting and its three component phases--elimination, equilibrium and escape. Curr. Opin. Immunol. 2014, 27, 16–25. [Google Scholar] [CrossRef]
- Ostrand-Rosenberg, S. Immune surveillance: a balance between protumor and antitumor immunity. Curr. Opin. Genet. Dev. 2008, 18, 11–18. [Google Scholar] [CrossRef]
- Vesely, M.D.; Schreiber, R.D. Cancer immunoediting: antigens, mechanisms, and implications to cancer immunotherapy. Ann. N. Y. Acad. Sci. 2013, 1284, 1–5. [Google Scholar] [CrossRef]
- Alspach, E.; Lussier, D.M.; Miceli, A.P.; Kizhvatov, I.; DuPage, M.; Luoma, A.M.; Meng, W.; Lichti, C.F.; Esaulova, E.; Vomund, A.N.; et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 2019, 574, 696–701. [Google Scholar] [CrossRef]
- Chen, D.S.; Mellman, I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Pio, R.; Ajona, D.; Ortiz-Espinosa, S.; Mantovani, A.; Lambris, J.D. Complementing the cancer-immunity cycle. Front. Immunol. 2019, 10, 774. [Google Scholar] [CrossRef] [PubMed]
- Ritter, B.; Greten, F.R. Modulating inflammation for cancer therapy. J. Exp. Med. 2019, 216, 1234–1243. [Google Scholar] [CrossRef] [PubMed]
- Demaria, O.; Cornen, S.; Daeron, M.; Morel, Y.; Medzhitov, R.; Vivier, E. Harnessing innate immunity in cancer therapy. Nature 2019, 574, 45–56. [Google Scholar] [CrossRef]
- Gonzalez, H.; Hagerling, C.; Werb, Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev. 2018, 32, 1267–1284. [Google Scholar] [CrossRef]
- Hopken, U.E.; Rehm, A. Targeting the tumor microenvironment of leukemia and lymphoma. Trends Cancer 2019, 5, 351–364. [Google Scholar] [CrossRef]
- Salmon, H.; Remark, R.; Gnjatic, S.; Merad, M. Host tissue determinants of tumour immunity. Nat. Rev. Cancer 2019, 19, 215–227. [Google Scholar] [CrossRef]
- Scott, D.W.; Gascoyne, R.D. The tumour microenvironment in B cell lymphomas. Nat. Rev. Cancer 2014, 14, 517–534. [Google Scholar] [CrossRef]
- Valkenburg, K.C.; de Groot, A.E.; Pienta, K.J. Targeting the tumour stroma to improve cancer therapy. Nat. Rev. Clin. Oncol. 2018, 15, 366–381. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef]
- Hagerling, C.; Casbon, A.J.; Werb, Z. Balancing the innate immune system in tumor development. Trends Cell Biol. 2015, 25, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Dunn, G.P.; Bruce, A.T.; Sheehan, K.C.; Shankaran, V.; Uppaluri, R.; Bui, J.D.; Diamond, M.S.; Koebel, C.M.; Arthur, C.; White, J.M.; et al. A critical function for type I interferons in cancer immunoediting. Nat. Immunol. 2005, 6, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Shankaran, V.; Ikeda, H.; Bruce, A.T.; White, J.M.; Swanson, P.E.; Old, L.J.; Schreiber, R.D. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 2001, 410, 1107–1111. [Google Scholar] [CrossRef] [PubMed]
- Firmbach-Kraft, I.; Byers, M.; Shows, T.; Dalla-Favera, R.; Krolewski, J.J. tyk2, prototype of a novel class of non-receptor tyrosine kinase genes. Oncogene 1990, 5, 1329–1336. [Google Scholar] [PubMed]
- Krolewski, J.J.; Lee, R.; Eddy, R.; Shows, T.B.; Dalla-Favera, R. Identification and chromosomal mapping of new human tyrosine kinase genes. Oncogene 1990, 5, 277–282. [Google Scholar] [PubMed]
- Wilks, A.F. The JAK kinases: not just another kinase drug discovery target. Semin. Cell Dev. Biol. 2008, 19, 319–328. [Google Scholar] [CrossRef]
- Velazquez, L.; Fellous, M.; Stark, G.R.; Pellegrini, S. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell 1992, 70, 313–322. [Google Scholar] [CrossRef]
- Darnell, J.E., Jr.; Kerr, I.M.; Stark, G.R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994, 264, 1415–1421. [Google Scholar] [CrossRef]
- Ihle, J.N.; Witthuhn, B.A.; Quelle, F.W.; Yamamoto, K.; Silvennoinen, O. Signaling through the hematopoietic cytokine receptors. Annu. Rev. Immunol. 1995, 13, 369–398. [Google Scholar] [CrossRef]
- O’Shea, J.J.; Schwartz, D.M.; Villarino, A.V.; Gadina, M.; McInnes, I.B.; Laurence, A. The JAK-STAT pathway: Impact on human disease and therapeutic intervention. Annu. Rev. Med. 2015, 66, 311–328. [Google Scholar] [CrossRef]
- Picard, C.; Bobby Gaspar, H.; Al-Herz, W.; Bousfiha, A.; Casanova, J.L.; Chatila, T.; Crow, Y.J.; Cunningham-Rundles, C.; Etzioni, A.; Franco, J.L.; et al. International union of immunological societies: 2017 primary immunodeficiency diseases committee report on inborn errors of immunity. J. Clin. Immunol. 2018, 38, 96–128. [Google Scholar] [CrossRef] [PubMed]
- Stark, G.R.; Darnell, J.E. The JAK-STAT pathway at twenty. Immunity 2012, 36, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Babon, J.J.; Lucet, I.S.; Murphy, J.M.; Nicola, N.A.; Varghese, L.N. The molecular regulation of janus kinase (JAK) activation. Biochem. J. 2014, 462, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ferrao, R.; Lupardus, P.J. The janus kinase (JAK) FERM and SH2 domains: Bringing specificity to JAK-receptor interactions. Front. Endocrinol. 2017, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Hammaren, H.M.; Virtanen, A.T.; Raivola, J.; Silvennoinen, O. The regulation of JAKs in cytokine signaling and its breakdown in disease. Cytokine 2019, 118, 48–63. [Google Scholar] [CrossRef]
- Hubbard, S.R. Mechanistic insights into regulation of JAK2 tyrosine kinase. Front. Endocrinol. 2017, 8, 361. [Google Scholar] [CrossRef]
- Morris, R.; Kershaw, N.J.; Babon, J.J. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. 2018, 27, 1984–2009. [Google Scholar] [CrossRef]
- Cendrowski, J.; Maminska, A.; Miaczynska, M. Endocytic regulation of cytokine receptor signaling. Cytokine Growth Factor Rev. 2016, 32, 63–73. [Google Scholar] [CrossRef]
- Moraga, I.; Spangler, J.; Mendoza, J.L.; Garcia, K.C. Multifarious determinants of cytokine receptor signaling specificity. Adv. Immunol 2014, 121, 1–39. [Google Scholar] [CrossRef]
- Uze, G.; Schreiber, G.; Piehler, J.; Pellegrini, S. The receptor of the type I interferon family. Curr. Top. Microbiol. Immunol. 2007, 316, 71–95. [Google Scholar] [CrossRef]
- Zheng, H.; Qian, J.; Baker, D.P.; Fuchs, S.Y. Tyrosine phosphorylation of protein kinase D2 mediates ligand-inducible elimination of the Type 1 interferon receptor. J. Biol. Chem. 2011, 286, 35733–35741. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.; Kaiser-Labusch, P.; Bank, J.; Ammann, S.; Kolb-Kokocinski, A.; Edelbusch, C.; Omran, H.; Ehl, S. Tyrosine kinase 2 is not limiting human antiviral type III interferon responses. Eur. J. Immunol. 2016, 46, 2639–2649. [Google Scholar] [CrossRef] [PubMed]
- Kreins, A.Y.; Ciancanelli, M.J.; Okada, S.; Kong, X.F.; Ramirez-Alejo, N.; Kilic, S.S.; El Baghdadi, J.; Nonoyama, S.; Mahdaviani, S.A.; Ailal, F.; et al. Human TYK2 deficiency: Mycobacterial and viral infections without hyper-IgE syndrome. J. Exp. Med. 2015, 212, 1641–1662. [Google Scholar] [CrossRef] [PubMed]
- Lupardus, P.J.; Ultsch, M.; Wallweber, H.; Bir Kohli, P.; Johnson, A.R.; Eigenbrot, C. Structure of the pseudokinase-kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 8025–8030. [Google Scholar] [CrossRef] [PubMed]
- Min, X.; Ungureanu, D.; Maxwell, S.; Hammaren, H.; Thibault, S.; Hillert, E.K.; Ayres, M.; Greenfield, B.; Eksterowicz, J.; Gabel, C.; et al. Structural and functional characterization of the JH2 pseudokinase domain of JAK family tyrosine kinase 2 (TYK2). J. Biol. Chem. 2015, 290, 27261–27270. [Google Scholar] [CrossRef] [PubMed]
- Wallweber, H.J.; Tam, C.; Franke, Y.; Starovasnik, M.A.; Lupardus, P.J. Structural basis of recognition of interferon-alpha receptor by tyrosine kinase 2. Nat. Struct. Mol. Biol. 2014, 21, 443–448. [Google Scholar] [CrossRef]
- Levy, D.E.; Darnell, J.E., Jr. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002, 3, 651–662. [Google Scholar] [CrossRef]
- Schindler, C.; Plumlee, C. Inteferons pen the JAK-STAT pathway. Semin. Cell Dev. Biol. 2008, 19, 311–318. [Google Scholar] [CrossRef]
- Majoros, A.; Platanitis, E.; Kernbauer-Holzl, E.; Rosebrock, F.; Muller, M.; Decker, T. Canonical and non-canonical aspects of JAK-STAT signaling: Lessons from interferons for cytokine responses. Front. Immunol. 2017, 8, 29. [Google Scholar] [CrossRef]
- Yang, J.; Stark, G.R. Roles of unphosphorylated STATs in signaling. Cell Res. 2008, 18, 443–451. [Google Scholar] [CrossRef]
- Bohmer, F.D.; Friedrich, K. Protein tyrosine phosphatases as wardens of STAT signaling. JAKSTAT 2014, 3, e28087. [Google Scholar] [CrossRef] [PubMed]
- Leitner, N.R.; Witalisz-Siepracka, A.; Strobl, B.; Muller, M. Tyrosine kinase 2-Surveillant of tumours and bona fide oncogene. Cytokine 2017, 89, 209–218. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kershaw, N.J.; Murphy, J.M.; Lucet, I.S.; Nicola, N.A.; Babon, J.J. Regulation of janus kinases by SOCS proteins. Biochem. Soc. Trans. 2013, 41, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Bocchini, C.E.; Kasembeli, M.M.; Roh, S.H.; Tweardy, D.J. Contribution of chaperones to STAT pathway signaling. JAKSTAT 2014, 3, e970459. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y.; Yang, L.; Liu, X.J.; Wang, X.Z.; Pan, Y.X.; Luo, J.M. The long noncoding RNA MEG3 and its target miR-147 regulate JAK/STAT pathway in advanced chronic myeloid leukemia. EBioMedicine 2018, 34, 61–75. [Google Scholar] [CrossRef]
- Witte, S.; Muljo, S.A. Integrating non-coding RNAs in JAK-STAT regulatory networks. JAKSTAT 2014, 3, e28055. [Google Scholar] [CrossRef]
- Stephen-Victor, E.; Fickenscher, H.; Bayry, J. IL-26: An emerging proinflammatory member of the IL-10 cytokine family with multifaceted actions in antiviral, antimicrobial, and autoimmune responses. PLoS Pathog. 2016, 12, e1005624. [Google Scholar] [CrossRef]
- Tengvall, S.; Che, K.F.; Linden, A. Interleukin-26: An emerging player in host defense and inflammation. J. Innate Immun. 2016, 8, 15–22. [Google Scholar] [CrossRef]
- Hemann, E.A.; Gale, M., Jr.; Savan, R. Interferon lambda genetics and biology in regulation of viral control. Front. Immunol. 2017, 8, 1707. [Google Scholar] [CrossRef]
- Ye, L.; Schnepf, D.; Staeheli, P. Interferon-lambda orchestrates innate and adaptive mucosal immune responses. Nat. Rev. Immunol. 2019, 19, 614–625. [Google Scholar] [CrossRef]
- Strobl, B.; Stoiber, D.; Sexl, V.; Mueller, M. Tyrosine kinase 2 (TYK2) in cytokine signalling and host immunity. Front. Biosci. 2011, 16, 3214–3232. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, A.; Shukla, M.; Yakubenko, V.P.; Mulya, A.; Kundu, S.; Cathcart, M.K. IL-4 and IL-13 employ discrete signaling pathways for target gene expression in alternatively activated monocytes/macrophages. Free Radic. Biol. Med. 2013, 54, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Junttila, I.S. Tuning the cytokine responses: An update on interleukin (IL)-4 and IL-13 receptor complexes. Front. Immunol. 2018, 9, 888. [Google Scholar] [CrossRef]
- Karaghiosoff, M.; Neubauer, H.; Lassnig, C.; Kovarik, P.; Schindler, H.; Pircher, H.; McCoy, B.; Bogdan, C.; Decker, T.; Brem, G.; et al. Partial impairment of cytokine responses in Tyk2-deficient mice. Immunity 2000, 13, 549–560. [Google Scholar] [CrossRef]
- Sheehan, K.C.; Lai, K.S.; Dunn, G.P.; Bruce, A.T.; Diamond, M.S.; Heutel, J.D.; Dungo-Arthur, C.; Carrero, J.A.; White, J.M.; Hertzog, P.J.; et al. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J. Interferon Cytokine Res. 2006, 26, 804–819. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, K.; Kato, K.; Aoki, K.; Matsuda, T.; Miyamoto, A.; Shibamori, M.; Yamashita, M.; Numata, A.; Takase, K.; Kobayashi, S.; et al. Tyk2 plays a restricted role in IFN alpha signaling, although it is required for IL-12-mediated T cell function. Immunity 2000, 13, 561–571. [Google Scholar] [CrossRef]
- Vielnascher, R.M.; Hainzl, E.; Leitner, N.R.; Rammerstorfer, M.; Popp, D.; Witalisz, A.; Rom, R.; Karaghiosoff, M.; Kolbe, T.; Muller, S.; et al. Conditional ablation of TYK2 in immunity to viral infection and tumor surveillance. Transgenic Res. 2014, 23, 519–529. [Google Scholar] [CrossRef]
- Shaw, M.H.; Boyartchuk, V.; Wong, S.; Karaghiosoff, M.; Ragimbeau, J.; Pellegrini, S.; Muller, M.; Dietrich, W.F.; Yap, G.S. A natural mutation in the Tyk2 pseudokinase domain underlies altered susceptibility of B10.Q/J mice to infection and autoimmunity. Proc. Natl. Acad. Sci. USA 2003, 100, 11594–11599. [Google Scholar] [CrossRef]
- Izumi, K.; Mine, K.; Inoue, Y.; Teshima, M.; Ogawa, S.; Kai, Y.; Kurafuji, T.; Hirakawa, K.; Miyakawa, D.; Ikeda, H.; et al. Reduced Tyk2 gene expression in beta-cells due to natural mutation determines susceptibility to virus-induced diabetes. Nat. Commun. 2015, 6, 6748. [Google Scholar] [CrossRef]
- Mine, K.; Hirakawa, K.; Kondo, S.; Minami, M.; Okada, A.; Tsutsu, N.; Yokogawa, Y.; Hibio, Y.; Kojima, F.; Fujimoto, S.; et al. Subtyping of type 1 diabetes as classified by anti-GAD antibody, IgE levels, and tyrosine kinase 2 (TYK2) promoter variant in the japanese. EBioMedicine 2017, 23, 46–51. [Google Scholar] [CrossRef][Green Version]
- Nagafuchi, S.; Kamada-Hibio, Y.; Hirakawa, K.; Tsutsu, N.; Minami, M.; Okada, A.; Kai, K.; Teshima, M.; Moroishi, A.; Murakami, Y.; et al. TYK2 promoter variant and diabetes mellitus in the japanese. EBioMedicine 2015, 2, 744–749. [Google Scholar] [CrossRef] [PubMed]
- Minegishi, Y.; Saito, M.; Morio, T.; Watanabe, K.; Agematsu, K.; Tsuchiya, S.; Takada, H.; Hara, T.; Kawamura, N.; Ariga, T.; et al. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity 2006, 25, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Sarrafzadeh, S.A.; Mahloojirad, M.; Casanova, J.L.; Badalzadeh, M.; Bustamante, J.; Boisson-Dupuis, S.; Pourpak, Z.; Nourizadeh, M.; Moin, M. A new patient with inherited TYK2 deficiency. J. Clin. Immunol. 2019. [Google Scholar] [CrossRef]
- Prchal-Murphy, M.; Semper, C.; Lassnig, C.; Wallner, B.; Gausterer, C.; Teppner-Klymiuk, I.; Kobolak, J.; Muller, S.; Kolbe, T.; Karaghiosoff, M.; et al. TYK2 kinase activity is required for functional type I interferon responses in vivo. PLoS ONE 2012, 7, e39141. [Google Scholar] [CrossRef] [PubMed]
- Raje, V.; Derecka, M.; Cantwell, M.; Meier, J.; Szczepanek, K.; Sisler, J.D.; Strobl, B.; Gamero, A.; Harris, T.E.; Larner, A.C. Kinase inactive tyrosine kinase (Tyk2) supports differentiation of brown fat cells. Endocrinology 2017, 158, 148–157. [Google Scholar] [CrossRef]
- Prchal-Murphy, M.; Witalisz-Siepracka, A.; Bednarik, K.T.; Putz, E.M.; Gotthardt, D.; Meissl, K.; Sexl, V.; Müller, M.; Strobl, B. In vivo tumor surveillance by NK cells requires TYK2 but not TYK2 kinase activity. Oncoimmunology 2015, 4, e1047579. [Google Scholar] [CrossRef]
- Boisson-Dupuis, S.; Ramirez-Alejo, N.; Li, Z.; Patin, E.; Rao, G.; Kerner, G.; Lim, C.K.; Krementsov, D.N.; Hernandez, N.; Ma, C.S.; et al. Tuberculosis and impaired IL-23-dependent IFN-gamma immunity in humans homozygous for a common TYK2 missense variant. Sci. Immunol. 2018, 3, eaau8714. [Google Scholar] [CrossRef]
- Couturier, N.; Bucciarelli, F.; Nurtdinov, R.N.; Debouverie, M.; Lebrun-Frenay, C.; Defer, G.; Moreau, T.; Confavreux, C.; Vukusic, S.; Cournu-Rebeix, I.; et al. Tyrosine kinase 2 variant influences T lymphocyte polarization and multiple sclerosis susceptibility. Brain 2011, 134, 693–703. [Google Scholar] [CrossRef]
- Dendrou, C.A.; Cortes, A.; Shipman, L.; Evans, H.G.; Attfield, K.E.; Jostins, L.; Barber, T.; Kaur, G.; Kuttikkatte, S.B.; Leach, O.A.; et al. Resolving TYK2 locus genotype-to-phenotype differences in autoimmunity. Sci. Transl. Med. 2016, 8, 363ra149. [Google Scholar] [CrossRef]
- Diogo, D.; Bastarache, L.; Liao, K.P.; Graham, R.R.; Fulton, R.S.; Greenberg, J.D.; Eyre, S.; Bowes, J.; Cui, J.; Lee, A.; et al. TYK2 protein-coding variants protect against rheumatoid arthritis and autoimmunity, with no evidence of major pleiotropic effects on non-autoimmune complex traits. PLoS ONE 2015, 10, e0122271. [Google Scholar] [CrossRef]
- Enerback, C.; Sandin, C.; Lambert, S.; Zawistowski, M.; Stuart, P.E.; Verma, D.; Tsoi, L.C.; Nair, R.P.; Johnston, A.; Elder, J.T. The psoriasis-protective TYK2 I684S variant impairs IL-12 stimulated pSTAT4 response in skin-homing CD4+ and CD8+ memory T-cells. Sci. Rep. 2018, 8, 7043. [Google Scholar] [CrossRef] [PubMed]
- Gorman, J.A.; Hundhausen, C.; Kinsman, M.; Arkatkar, T.; Allenspach, E.J.; Clough, C.; West, S.E.; Thomas, K.; Eken, A.; Khim, S.; et al. The TYK2-P1104A Autoimmune Protective Variant Limits Coordinate Signals Required to Generate Specialized T Cell Subsets. Front. Immunol. 2019, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- Kerner, G.; Ramirez-Alejo, N.; Seeleuthner, Y.; Yang, R.; Ogishi, M.; Cobat, A.; Patin, E.; Quintana-Murci, L.; Boisson-Dupuis, S.; Casanova, J.L.; et al. Homozygosity for TYK2 P1104A underlies tuberculosis in about 1% of patients in a cohort of European ancestry. Proc. Natl. Acad. Sci. USA 2019, 116, 10430–10434. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Gakovic, M.; Ragimbeau, J.; Eloranta, M.L.; Ronnblom, L.; Michel, F.; Pellegrini, S. Two rare disease-associated Tyk2 variants are catalytically impaired but signaling competent. J. Immunol. 2013, 190, 2335–2344. [Google Scholar] [CrossRef]
- Shaw, M.H.; Freeman, G.J.; Scott, M.F.; Fox, B.A.; Bzik, D.J.; Belkaid, Y.; Yap, G.S. Tyk2 negatively regulates adaptive Th1 immunity by mediating IL-10 signaling and promoting IFN-gamma-dependent IL-10 reactivation. J. Immunol. 2006, 176, 7263–7271. [Google Scholar] [CrossRef]
- Stoiber, D.; Kovacic, B.; Schuster, C.; Schellack, C.; Karaghiosoff, M.; Kreibich, R.; Weisz, E.; Artwohl, M.; Kleine, O.C.; Muller, M.; et al. TYK2 is a key regulator of the surveillance of B lymphoid tumors. J. Clin. Investig. 2004, 114, 1650–1658. [Google Scholar] [CrossRef]
- Zhang, Q.; Sturgill, J.L.; Kmieciak, M.; Szczepanek, K.; Derecka, M.; Koebel, C.; Graham, L.J.; Dai, Y.; Chen, S.; Grant, S.; et al. The role of Tyk2 in regulation of breast cancer growth. J. Interferon Cytokine Res. 2011, 31, 671–677. [Google Scholar] [CrossRef]
- Aizu, K.; Li, W.; Yajima, T.; Arai, T.; Shimoda, K.; Nimura, Y.; Yoshikai, Y. An important role of Tyk2 in APC function of dendritic cells for priming CD8+ T cells producing IFN-gamma. Eur. J. Immunol. 2006, 36, 3060–3070. [Google Scholar] [CrossRef]
- Hashiguchi, T.; Oyamada, A.; Sakuraba, K.; Shimoda, K.; Nakayama, K.I.; Iwamoto, Y.; Yoshikai, Y.; Yamada, H. Tyk2-dependent bystander activation of conventional and nonconventional Th1 cell subsets contributes to innate host defense against Listeria monocytogenes infection. J. Immunol. 2014, 192, 4739–4747. [Google Scholar] [CrossRef]
- Hosogi, M.; Tonogaito, H.; Aioi, A.; Hamada, K.; Shimoda, K.; Muromoto, R.; Matsuda, T.; Miyachi, Y. Hapten-induced contact hypersensitivity is enhanced in Tyk2-deficient mice. J. Dermatol. Sci. 2004, 36, 51–56. [Google Scholar] [CrossRef]
- Oyamada, A.; Ikebe, H.; Itsumi, M.; Saiwai, H.; Okada, S.; Shimoda, K.; Iwakura, Y.; Nakayama, K.I.; Iwamoto, Y.; Yoshikai, Y.; et al. Tyrosine kinase 2 plays critical roles in the pathogenic CD4 T cell responses for the development of experimental autoimmune encephalomyelitis. J. Immunol. 2009, 183, 7539–7546. [Google Scholar] [CrossRef] [PubMed]
- Schleicher, U.; Mattner, J.; Blos, M.; Schindler, H.; Rollinghoff, M.; Karaghiosoff, M.; Muller, M.; Werner-Felmayer, G.; Bogdan, C. Control of Leishmania major in the absence of Tyk2 kinase. Eur. J. Immunol. 2004, 34, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Simonovic, N.; Witalisz-Siepracka, A.; Meissl, K.; Lassnig, C.; Reichart, U.; Kolbe, T.; Farlik, M.; Bock, C.; Sexl, V.; Müller, M.; et al. NK Cells Require Cell-Extrinsic and -Intrinsic TYK2 for Full Functionality in Tumor Surveillance and Antibacterial Immunity. J. Immunol. 2019, 202, 1724–1734. [Google Scholar] [CrossRef] [PubMed]
- Tokumasa, N.; Suto, A.; Kagami, S.; Furuta, S.; Hirose, K.; Watanabe, N.; Saito, Y.; Shimoda, K.; Iwamoto, I.; Nakajima, H. Expression of Tyk2 in dendritic cells is required for IL-12, IL-23, and IFN-gamma production and the induction of Th1 cell differentiation. Blood 2007, 110, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Hainzl, E.; Stockinger, S.; Rauch, I.; Heider, S.; Berry, D.; Lassnig, C.; Schwab, C.; Rosebrock, F.; Milinovich, G.; Schlederer, M.; et al. Intestinal epithelial cell tyrosine kinase 2 transduces IL-22 signals to protect from acute colitis. J. Immunol. 2015, 195, 5011–5024. [Google Scholar] [CrossRef]
- Ishizaki, M.; Muromoto, R.; Akimoto, T.; Sekine, Y.; Kon, S.; Diwan, M.; Maeda, H.; Togi, S.; Shimoda, K.; Oritani, K.; et al. Tyk2 is a therapeutic target for psoriasis-like skin inflammation. Int. Immunol. 2014, 26, 257–267. [Google Scholar] [CrossRef]
- Nakamura, R.; Shibata, K.; Yamada, H.; Shimoda, K.; Nakayama, K.; Yoshikai, Y. Tyk2-signaling plays an important role in host defense against Escherichia coli through IL-23-induced IL-17 production by gammadelta T cells. J. Immunol. 2008, 181, 2071–2075. [Google Scholar] [CrossRef]
- Simma, O.; Zebedin, E.; Neugebauer, N.; Schellack, C.; Pilz, A.; Chang-Rodriguez, S.; Lingnau, K.; Weisz, E.; Putz, E.M.; Pickl, W.F.; et al. Identification of an indispensable role for tyrosine kinase 2 in CTL-mediated tumor surveillance. Cancer Res. 2009, 69, 203–211. [Google Scholar] [CrossRef][Green Version]
- Bosmann, M.; Strobl, B.; Kichler, N.; Rigler, D.; Grailer, J.J.; Pache, F.; Murray, P.J.; Muller, M.; Ward, P.A. Tyrosine kinase 2 promotes sepsis-associated lethality by facilitating production of interleukin-27. J. Leukoc. Biol. 2014, 96, 123–131. [Google Scholar] [CrossRef]
- Karaghiosoff, M.; Steinborn, R.; Kovarik, P.; Kriegshauser, G.; Baccarini, M.; Donabauer, B.; Reichart, U.; Kolbe, T.; Bogdan, C.; Leanderson, T.; et al. Central role for type I interferons and Tyk2 in lipopolysaccharide-induced endotoxin shock. Nat. Immunol. 2003, 4, 471–477. [Google Scholar] [CrossRef]
- Strobl, B.; Bubic, I.; Bruns, U.; Steinborn, R.; Lajko, R.; Kolbe, T.; Karaghiosoff, M.; Kalinke, U.; Jonjic, S.; Muller, M. Novel functions of tyrosine kinase 2 in the antiviral defense against murine cytomegalovirus. J. Immunol. 2005, 175, 4000–4008. [Google Scholar] [CrossRef] [PubMed]
- Ishizaki, M.; Muromoto, R.; Akimoto, T.; Ohshiro, Y.; Takahashi, M.; Sekine, Y.; Maeda, H.; Shimoda, K.; Oritani, K.; Matsuda, T. Tyk2 deficiency protects joints against destruction in anti-type II collagen antibody-induced arthritis in mice. Int. Immunol. 2011, 23, 575–582. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ma, C.S.; Wong, N.; Rao, G.; Nguyen, A.; Avery, D.T.; Payne, K.; Torpy, J.; O’Young, P.; Deenick, E.; Bustamante, J.; et al. Unique and shared signaling pathways cooperate to regulate the differentiation of human CD4+ T cells into distinct effector subsets. J. Exp. Med. 2016, 213, 1589–1608. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Biehl, A.; Gadina, M.; Hasni, S.; Schwartz, D.M. JAK-STAT signaling as a target for inflammatory and autoimmune diseases: Current and future prospects. Drugs 2017, 77, 521–546. [Google Scholar] [CrossRef] [PubMed]
- Bousoik, E.; Montazeri Aliabadi, H. “Do we know jack” about JAK? A closer look at JAK/STAT signaling pathway. Front. Oncol. 2018, 8, 287. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Janus kinase (JAK) inhibitors in the treatment of inflammatory and neoplastic diseases. Pharmacol. Res. 2016, 111, 784–803. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.; Zscheppang, K.; Fatykhova, D.; Tonnies, M.; Bauer, T.T.; Schneider, P.; Neudecker, J.; Ruckert, J.C.; Eggeling, S.; Schimek, M.; et al. Tyk2 as a target for immune regulation in human viral/bacterial pneumonia. Eur. Respir. J. 2017, 50, 1601953. [Google Scholar] [CrossRef]
- Burke, J.R.; Cheng, L.; Gillooly, K.M.; Strnad, J.; Zupa-Fernandez, A.; Catlett, I.M.; Zhang, Y.; Heimrich, E.M.; McIntyre, K.W.; Cunningham, M.D.; et al. Autoimmune pathways in mice and humans are blocked by pharmacological stabilization of the TYK2 pseudokinase domain. Sci. Transl. Med. 2019, 11, eaaw1736. [Google Scholar] [CrossRef]
- Sohn, S.J.; Barrett, K.; Van Abbema, A.; Chang, C.; Kohli, P.B.; Kanda, H.; Smith, J.; Lai, Y.; Zhou, A.; Zhang, B.; et al. A restricted role for TYK2 catalytic activity in human cytokine responses revealed by novel TYK2-selective inhibitors. J. Immunol. 2013, 191, 2205–2216. [Google Scholar] [CrossRef]
- Works, M.G.; Yin, F.; Yin, C.C.; Yiu, Y.; Shew, K.; Tran, T.T.; Dunlap, N.; Lam, J.; Mitchell, T.; Reader, J.; et al. Inhibition of TYK2 and JAK1 ameliorates imiquimod-induced psoriasis-like dermatitis by inhibiting IL-22 and the IL-23/IL-17 axis. J. Immunol. 2014, 193, 3278–3287. [Google Scholar] [CrossRef]
- Liongue, C.; Ward, A.C. Evolution of Class I cytokine receptors. BMC Evol. Biol. 2007, 7, 120. [Google Scholar] [CrossRef] [PubMed]
- Renauld, J.C. Class II cytokine receptors and their ligands: key antiviral and inflammatory modulators. Nat. Rev. Immunol. 2003, 3, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Liongue, C.; Sertori, R.; Ward, A.C. Evolution of cytokine receptor signaling. J. Immunol. 2016, 197, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Liongue, C.; Taznin, T.; Ward, A.C. Signaling via the CytoR/JAK/STAT/SOCS pathway: Emergence during evolution. Mol. Immunol. 2016, 71, 166–175. [Google Scholar] [CrossRef]
- Bu, L.; Baba, H.; Yoshida, N.; Miyake, K.; Yasuda, T.; Uchihara, T.; Tan, P.; Ishimoto, T. Biological heterogeneity and versatility of cancer-associated fibroblasts in the tumor microenvironment. Oncogene 2019, 38, 4887–4901. [Google Scholar] [CrossRef]
- Vilgelm, A.E.; Richmond, A. Chemokines modulate immune surveillance in tumorigenesis, metastasis, and response to immunotherapy. Front. Immunol. 2019, 10, 333. [Google Scholar] [CrossRef]
- Lazear, H.M.; Schoggins, J.W.; Diamond, M.S. Shared and distinct functions of type I and type III interferons. Immunity 2019, 50, 907–923. [Google Scholar] [CrossRef]
- van Pesch, V.; Lanaya, H.; Renauld, J.C.; Michiels, T. Characterization of the murine alpha interferon gene family. J. Virol. 2004, 78, 8219–8228. [Google Scholar] [CrossRef]
- Xu, L.; Yang, L.; Liu, W. Distinct evolution process among type I interferon in mammals. Protein Cell. 2013, 4, 383–392. [Google Scholar] [CrossRef]
- Gibbert, K.; Schlaak, J.F.; Yang, D.; Dittmer, U. IFN-alpha subtypes: distinct biological activities in anti-viral therapy. Br. J. Pharmacol 2013, 168, 1048–1058. [Google Scholar] [CrossRef]
- Ng, C.T.; Mendoza, J.L.; Garcia, K.C.; Oldstone, M.B. Alpha and beta type 1 interferon SIgnaling: Passage for diverse biologic outcomes. Cell 2016, 164, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, G. The molecular basis for differential type I interferon signaling. J. Biol. Chem. 2017, 292, 7285–7294. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Mann-Nuttel, R.; Schulze, A.; Richter, L.; Alferink, J.; Scheu, S. Sources of type I interferons in infectious immunity: Plasmacytoid dendritic cells not always in the driver’s seat. Front. Immunol. 2019, 10, 778. [Google Scholar] [CrossRef] [PubMed]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Swiecki, M.; Colonna, M. Type I interferons: Diversity of sources, production pathways and effects on immune responses. Curr. Opin. Virol. 2011, 1, 463–475. [Google Scholar] [CrossRef]
- Schreiber, G.; Piehler, J. The molecular basis for functional plasticity in type I interferon signaling. Trends Immunol. 2015, 36, 139–149. [Google Scholar] [CrossRef]
- Platanitis, E.; Demiroz, D.; Schneller, A.; Fischer, K.; Capelle, C.; Hartl, M.; Gossenreiter, T.; Muller, M.; Novatchkova, M.; Decker, T. A molecular switch from STAT2-IRF9 to ISGF3 underlies interferon-induced gene transcription. Nat. Commun. 2019, 10, 2921. [Google Scholar] [CrossRef]
- Abdul-Sater, A.A.; Majoros, A.; Plumlee, C.R.; Perry, S.; Gu, A.D.; Lee, C.; Shresta, S.; Decker, T.; Schindler, C. Different STAT transcription complexes drive early and delayed responses to type I IFNs. J. Immunol. 2015, 195, 210–216. [Google Scholar] [CrossRef]
- Au-Yeung, N.; Mandhana, R.; Horvath, C.M. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT 2013, 2, e23931. [Google Scholar] [CrossRef]
- Blaszczyk, K.; Nowicka, H.; Kostyrko, K.; Antonczyk, A.; Wesoly, J.; Bluyssen, H.A. The unique role of STAT2 in constitutive and IFN-induced transcription and antiviral responses. Cytokine Growth Factor Rev. 2016, 29, 71–81. [Google Scholar] [CrossRef]
- Fink, K.; Grandvaux, N. STAT2 and IRF9: Beyond ISGF3. JAKSTAT 2013, 2, e27521. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yin, Y.; Xu, L.; Su, J.; Huang, F.; Wang, Y.; Boor, P.P.C.; Chen, K.; Wang, W.; Cao, W.; et al. Unphosphorylated ISGF3 drives constitutive expression of interferon-stimulated genes to protect against viral infections. Sci. Signal. 2017, 10, eaah4248. [Google Scholar] [CrossRef] [PubMed]
- van Boxel-Dezaire, A.H.; Rani, M.R.; Stark, G.R. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity 2006, 25, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Rusinova, I.; Forster, S.; Yu, S.; Kannan, A.; Masse, M.; Cumming, H.; Chapman, R.; Hertzog, P.J. Interferome v2.0: An updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013, 41, D1040–D1046. [Google Scholar] [CrossRef]
- Barrat, F.J.; Crow, M.K.; Ivashkiv, L.B. Interferon target-gene expression and epigenomic signatures in health and disease. Nat. Immunol. 2019, 20, 1574–1583. [Google Scholar] [CrossRef]
- Crow, M.K.; Olferiev, M.; Kirou, K.A. Type I interferons in autoimmune disease. Annu. Rev. Pathol 2019, 14, 369–393. [Google Scholar] [CrossRef]
- Kretschmer, S.; Lee-Kirsch, M.A. Type I interferon-mediated autoinflammation and autoimmunity. Curr. Opin. Immunol. 2017, 49, 96–102. [Google Scholar] [CrossRef]
- McNab, F.; Mayer-Barber, K.; Sher, A.; Wack, A.; O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015, 15, 87–103. [Google Scholar] [CrossRef]
- Dunn, G.P.; Koebel, C.M.; Schreiber, R.D. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 2006, 6, 836–848. [Google Scholar] [CrossRef]
- Musella, M.; Manic, G.; De Maria, R.; Vitale, I.; Sistigu, A. Type-I-interferons in infection and cancer: Unanticipated dynamics with therapeutic implications. Oncoimmunology 2017, 6, e1314424. [Google Scholar] [CrossRef]
- Owen, K.L.; Brockwell, N.K.; Parker, B.S. JAK-STAT signaling: A double-edged sword of immune regulation and cancer progression. Cancers 2019, 11, 2002. [Google Scholar] [CrossRef]
- Parker, B.S.; Rautela, J.; Hertzog, P.J. Antitumour actions of interferons: implications for cancer therapy. Nat. Rev. Cancer 2016, 16, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Gangaplara, A.; Martens, C.; Dahlstrom, E.; Metidji, A.; Gokhale, A.S.; Glass, D.D.; Lopez-Ocasio, M.; Baur, R.; Kanakabandi, K.; Porcella, S.F.; et al. Type I interferon signaling attenuates regulatory T cell function in viral infection and in the tumor microenvironment. PLoS Pathog. 2018, 14, e1006985. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, H.; Ueda, R.; Narumi, K.; Heike, Y.; Yoshida, T.; Aoki, K. Type I IFN gene delivery suppresses regulatory T cells within tumors. Cancer Gene Ther. 2014, 21, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Pace, L.; Vitale, S.; Dettori, B.; Palombi, C.; La Sorsa, V.; Belardelli, F.; Proietti, E.; Doria, G. APC activation by IFN-alpha decreases regulatory T cell and enhances Th cell functions. J. Immunol. 2010, 184, 5969–5979. [Google Scholar] [CrossRef] [PubMed]
- Metzger, P.; Kirchleitner, S.V.; Kluge, M.; Koenig, L.M.; Horth, C.; Rambuscheck, C.A.; Bohmer, D.; Ahlfeld, J.; Kobold, S.; Friedel, C.C.; et al. Immunostimulatory RNA leads to functional reprogramming of myeloid-derived suppressor cells in pancreatic cancer. J. Immunother. Cancer 2019, 7, 288. [Google Scholar] [CrossRef]
- Zhang, C.X.; Ye, S.B.; Ni, J.J.; Cai, T.T.; Liu, Y.N.; Huang, D.J.; Mai, H.Q.; Chen, Q.Y.; He, J.; Zhang, X.S.; et al. STING signaling remodels the tumor microenvironment by antagonizing myeloid-derived suppressor cell expansion. Cell. Death Differ. 2019, 26, 2314–2328. [Google Scholar] [CrossRef]
- Jacquelot, N.; Yamazaki, T.; Roberti, M.P.; Duong, C.P.M.; Andrews, M.C.; Verlingue, L.; Ferrere, G.; Becharef, S.; Vetizou, M.; Daillere, R.; et al. Sustained type I interferon signaling as a mechanism of resistance to PD-1 blockade. Cell Res. 2019, 29, 846–861. [Google Scholar] [CrossRef]
- Xiao, W.; Klement, J.D.; Lu, C.; Ibrahim, M.L.; Liu, K. IFNAR1 controls autocrine type I IFN regulation of PD-L1 expression in myeloid-derived suppressor cells. J. Immunol. 2018, 201, 264–277. [Google Scholar] [CrossRef]
- Brockwell, N.K.; Parker, B.S. Tumor inherent interferons: Impact on immune reactivity and immunotherapy. Cytokine 2019, 118, 42–47. [Google Scholar] [CrossRef]
- McLane, L.M.; Abdel-Hakeem, M.S.; Wherry, E.J. CD8 T cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol. 2019, 37, 457–495. [Google Scholar] [CrossRef] [PubMed]
- Robinson, R.T. IL12Rbeta1: the cytokine receptor that we used to know. Cytokine 2015, 71, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Tait Wojno, E.D.; Hunter, C.A.; Stumhofer, J.S. The immunobiology of the interleukin-12 family: Room for discovery. Immunity 2019, 50, 851–870. [Google Scholar] [CrossRef] [PubMed]
- Vignali, D.A.; Kuchroo, V.K. IL-12 family cytokines: Immunological playmakers. Nat. Immunol. 2012, 13, 722–728. [Google Scholar] [CrossRef]
- Chognard, G.; Bellemare, L.; Pelletier, A.N.; Dominguez-Punaro, M.C.; Beauchamp, C.; Guyon, M.J.; Charron, G.; Morin, N.; Sivanesan, D.; Kuchroo, V.; et al. The dichotomous pattern of IL-12r and IL-23R expression elucidates the role of IL-12 and IL-23 in inflammation. PLoS ONE 2014, 9, e89092. [Google Scholar] [CrossRef]
- Croxford, A.L.; Kulig, P.; Becher, B. IL-12-and IL-23 in health and disease. Cytokine Growth Factor Rev. 2014, 25, 415–421. [Google Scholar] [CrossRef]
- Teng, M.W.; Bowman, E.P.; McElwee, J.J.; Smyth, M.J.; Casanova, J.L.; Cooper, A.M.; Cua, D.J. IL-12 and IL-23 cytokines: from discovery to targeted therapies for immune-mediated inflammatory diseases. Nat. Med. 2015, 21, 719–729. [Google Scholar] [CrossRef]
- Liu, J.; Cao, S.; Kim, S.; Chung, E.Y.; Homma, Y.; Guan, X.; Jimenez, V.; Ma, X. Interleukin-12: an update on its immunological activities, signaling and regulation of gene expression. Curr. Immunol. Rev. 2005, 1, 119–137. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef]
- Trinchieri, G.; Pflanz, S.; Kastelein, R.A. The IL-12 family of heterodimeric cytokines: New players in the regulation of T cell responses. Immunity 2003, 19, 641–644. [Google Scholar] [CrossRef]
- Watford, W.T.; Hissong, B.D.; Bream, J.H.; Kanno, Y.; Muul, L.; O’Shea, J.J. Signaling by IL-12 and IL-23 and the immunoregulatory roles of STAT4. Immunol. Rev. 2004, 202, 139–156. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.T.; Segal, B.M.; Nakanishi, K.; Okamura, H.; Shevach, E.M. The costimulatory effect of IL-18 on the induction of antigen-specific IFN-gamma production by resting T cells is IL-12 dependent and is mediated by up-regulation of the IL-12 receptor beta2 subunit. Eur. J. Immunol. 2000, 30, 1113–1119. [Google Scholar] [CrossRef]
- Smeltz, R.B.; Chen, J.; Ehrhardt, R.; Shevach, E.M. Role of IFN-gamma in Th1 differentiation: IFN-gamma regulates IL-18R alpha expression by preventing the negative effects of IL-4 and by inducing/maintaining IL-12 receptor beta 2 expression. J. Immunol. 2002, 168, 6165–6172. [Google Scholar] [CrossRef] [PubMed]
- Wandinger, K.P.; Sturzebecher, C.S.; Bielekova, B.; Detore, G.; Rosenwald, A.; Staudt, L.M.; McFarland, H.F.; Martin, R. Complex immunomodulatory effects of interferon-beta in multiple sclerosis include the upregulation of T helper 1-associated marker genes. Ann. Neurol. 2001, 50, 349–357. [Google Scholar] [CrossRef]
- Yoshida, H.; Hunter, C.A. The immunobiology of interleukin-27. Annu. Rev. Immunol. 2015, 33, 417–443. [Google Scholar] [CrossRef]
- Szabo, S.J.; Dighe, A.S.; Gubler, U.; Murphy, K.M. Regulation of the interleukin (IL)-12R beta 2 subunit expression in developing T helper 1 (Th1) and Th2 cells. J. Exp. Med. 1997, 185, 817–824. [Google Scholar] [CrossRef]
- Tugues, S.; Burkhard, S.H.; Ohs, I.; Vrohlings, M.; Nussbaum, K.; Vom Berg, J.; Kulig, P.; Becher, B. New insights into IL-12-mediated tumor suppression. Cell Death Differ. 2015, 22, 237–246. [Google Scholar] [CrossRef]
- Xu, M.; Mizoguchi, I.; Morishima, N.; Chiba, Y.; Mizuguchi, J.; Yoshimoto, T. Regulation of antitumor immune responses by the IL-12 family cytokines, IL-12, IL-23, and IL-27. Clin. Dev. Immunol. 2010, 2010, 832454. [Google Scholar] [CrossRef]
- Berraondo, P.; Etxeberria, I.; Ponz-Sarvise, M.; Melero, I. Revisiting interleukin-12 as a cancer immunotherapy agent. Clin. Cancer Res. 2018, 24, 2716–2718. [Google Scholar] [CrossRef]
- Croxford, A.L.; Mair, F.; Becher, B. IL-23: One cytokine in control of autoimmunity. Eur. J. Immunol. 2012, 42, 2263–2273. [Google Scholar] [CrossRef]
- Hawkes, J.E.; Yan, B.Y.; Chan, T.C.; Krueger, J.G. Discovery of the IL-23/IL-17 signaling pathway and the treatment of psoriasis. J. Immunol. 2018, 201, 1605–1613. [Google Scholar] [CrossRef] [PubMed]
- Schon, M.P.; Erpenbeck, L. The interleukin-23/interleukin-17 axis links adaptive and innate immunity in psoriasis. Front. Immunol. 2018, 9, 1323. [Google Scholar] [CrossRef] [PubMed]
- Belladonna, M.L.; Renauld, J.C.; Bianchi, R.; Vacca, C.; Fallarino, F.; Orabona, C.; Fioretti, M.C.; Grohmann, U.; Puccetti, P. IL-23 and IL-12 have overlapping, but distinct, effects on murine dendritic cells. J. Immunol. 2002, 168, 5448–5454. [Google Scholar] [CrossRef] [PubMed]
- Parham, C.; Chirica, M.; Timans, J.; Vaisberg, E.; Travis, M.; Cheung, J.; Pflanz, S.; Zhang, R.; Singh, K.P.; Vega, F.; et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J. Immunol. 2002, 168, 5699–5708. [Google Scholar] [CrossRef] [PubMed]
- Mathur, A.N.; Chang, H.C.; Zisoulis, D.G.; Stritesky, G.L.; Yu, Q.; O’Malley, J.T.; Kapur, R.; Levy, D.E.; Kansas, G.S.; Kaplan, M.H. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J. Immunol. 2007, 178, 4901–4907. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.O.; Panopoulos, A.D.; Nurieva, R.; Chang, S.H.; Wang, D.; Watowich, S.S.; Dong, C. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 2007, 282, 9358–9363. [Google Scholar] [CrossRef]
- Kortylewski, M.; Xin, H.; Kujawski, M.; Lee, H.; Liu, Y.; Harris, T.; Drake, C.; Pardoll, D.; Yu, H. Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Cancer Cell. 2009, 15, 114–123. [Google Scholar] [CrossRef]
- Ngiow, S.F.; Teng, M.W.; Smyth, M.J. A balance of interleukin-12 and -23 in cancer. Trends Immunol. 2013, 34, 548–555. [Google Scholar] [CrossRef]
- Yan, J.; Smyth, M.J.; Teng, M.W.L. Interleukin (IL)-12 and IL-23 and their conflicting roles in cancer. Cold Spring Harb. Perspect. Biol. 2018, 10, a028530. [Google Scholar] [CrossRef]
- Ishizaki, M.; Akimoto, T.; Muromoto, R.; Yokoyama, M.; Ohshiro, Y.; Sekine, Y.; Maeda, H.; Shimoda, K.; Oritani, K.; Matsuda, T. Involvement of tyrosine kinase-2 in both the IL-12/Th1 and IL-23/Th17 axes in vivo. J. Immunol. 2011, 187, 181–189. [Google Scholar] [CrossRef]
- Nemoto, M.; Hattori, H.; Maeda, N.; Akita, N.; Muramatsu, H.; Moritani, S.; Kawasaki, T.; Maejima, M.; Ode, H.; Hachiya, A.; et al. Compound heterozygous TYK2 mutations underlie primary immunodeficiency with T-cell lymphopenia. Sci. Rep. 2018, 8, 6956. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, W.; O’Garra, A. IL-10 family cytokines IL-10 and IL-22: From basic science to clinical translation. Immunity 2019, 50, 871–891. [Google Scholar] [CrossRef] [PubMed]
- Couper, K.N.; Blount, D.G.; Riley, E.M. IL-10: The master regulator of immunity to infection. J. Immunol. 2008, 180, 5771–5777. [Google Scholar] [CrossRef] [PubMed]
- Hedrich, C.M.; Bream, J.H. Cell type-specific regulation of IL-10 expression in inflammation and disease. Immunol. Res. 2010, 47, 185–206. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, M.; O’Garra, A. The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 2010, 10, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Gabrysova, L.; Howes, A.; Saraiva, M.; O’Garra, A. The regulation of IL-10 expression. Curr. Top. Microbiol. Immunol. 2014, 380, 157–190. [Google Scholar] [CrossRef]
- Hutchins, A.P.; Diez, D.; Miranda-Saavedra, D. The IL-10/STAT3-mediated anti-inflammatory response: recent developments and future challenges. Brief. Funct Genom. 2013, 12, 489–498. [Google Scholar] [CrossRef]
- Murray, P.J. Understanding and exploiting the endogenous interleukin-10/STAT3-mediated anti-inflammatory response. Curr. Opin. Pharmacol. 2006, 6, 379–386. [Google Scholar] [CrossRef]
- Walter, M.R. The molecular basis of IL-10 function: from receptor structure to the onset of signaling. Curr. Top. Microbiol. Immunol. 2014, 380, 191–212. [Google Scholar] [CrossRef]
- Mosser, D.M.; Zhang, X. Interleukin-10: New perspectives on an old cytokine. Immunol. Rev. 2008, 226, 205–218. [Google Scholar] [CrossRef]
- Muhl, H. Pro-inflammatory signaling by IL-10 and IL-22: Bad habit stirred up by interferons? Front. Immunol. 2013, 4, 18. [Google Scholar] [CrossRef] [PubMed]
- Dennis, K.L.; Blatner, N.R.; Gounari, F.; Khazaie, K. Current status of interleukin-10 and regulatory T-cells in cancer. Curr. Opin. Oncol 2013, 25, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Mannino, M.H.; Zhu, Z.; Xiao, H.; Bai, Q.; Wakefield, M.R.; Fang, Y. The paradoxical role of IL-10 in immunity and cancer. Cancer Lett 2015, 367, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Kortylewski, M.; Pardoll, D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat. Rev. Immunol. 2007, 7, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Wilbers, R.H.P.; van Raaij, D.R.; Westerhof, L.B.; Bakker, J.; Smant, G.; Schots, A. Re-evaluation of IL-10 signaling reveals novel insights on the contribution of the intracellular domain of the IL-10R2 chain. PLoS ONE 2017, 12, e0186317. [Google Scholar] [CrossRef] [PubMed]
- Parks, O.B.; Pociask, D.A.; Hodzic, Z.; Kolls, J.K.; Good, M. Interleukin-22 signaling in the regulation of intestinal health and disease. Front. Cell Dev. Biol. 2015, 3, 85. [Google Scholar] [CrossRef] [PubMed]
- Zenewicz, L.A. IL-22: There is a gap in our knowledge. Immunohorizons 2018, 2, 198–207. [Google Scholar] [CrossRef]
- Wolk, K.; Witte, E.; Witte, K.; Warszawska, K.; Sabat, R. Biol.ogy of interleukin-22. Semin. Immunopathol. 2010, 32, 17–31. [Google Scholar] [CrossRef]
- Zenewicz, L.A.; Flavell, R.A. Recent advances in IL-22 biology. Int. Immunol. 2011, 23, 159–163. [Google Scholar] [CrossRef]
- Dudakov, J.A.; Hanash, A.M.; van den Brink, M.R. Interleukin-22: Immunobiology and pathology. Annu. Rev. Immunol. 2015, 33, 747–785. [Google Scholar] [CrossRef]
- Eidenschenk, C.; Rutz, S.; Liesenfeld, O.; Ouyang, W. Role of IL-22 in microbial host defense. Curr. Top. Microbiol. Immunol. 2014, 380, 213–236. [Google Scholar] [CrossRef] [PubMed]
- Sabat, R.; Ouyang, W.; Wolk, K. Therapeutic opportunities of the IL-22-IL-22R1 system. Nat. Rev. Drug Discov 2014, 13, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, P.; Gronke, K.; Diefenbach, A. A catch-22: Interleukin-22 and cancer. Eur. J. Immunol. 2018, 48, 15–31. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Savan, R. The role of the IL-22/IL-22R1 axis in cancer. Cytokine Growth Factor Rev. 2014, 25, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Markota, A.; Endres, S.; Kobold, S. Targeting interleukin-22 for cancer therapy. Hum. Vaccin Immunother. 2018, 14, 2012–2015. [Google Scholar] [CrossRef] [PubMed]
- Niess, J.H.; Hruz, P.; Kaymak, T. The interleukin-20 cytokines in intestinal diseases. Front. Immunol. 2018, 9, 1373. [Google Scholar] [CrossRef]
- Donnelly, R.P.; Sheikh, F.; Kotenko, S.V.; Dickensheets, H. The expanded family of class II cytokines that share the IL-10 receptor-2 (IL-10R2) chain. J. Leukoc. Biol. 2004, 76, 314–321. [Google Scholar] [CrossRef]
- Sheikh, F.; Baurin, V.V.; Lewis-Antes, A.; Shah, N.K.; Smirnov, S.V.; Anantha, S.; Dickensheets, H.; Dumoutier, L.; Renauld, J.C.; Zdanov, A.; et al. Cutting edge: IL-26 signals through a novel receptor complex composed of IL-20 receptor 1 and IL-10 receptor 2. J. Immunol. 2004, 172, 2006–2010. [Google Scholar] [CrossRef]
- Donnelly, R.P.; Sheikh, F.; Dickensheets, H.; Savan, R.; Young, H.A.; Walter, M.R. Interleukin-26: An IL-10-related cytokine produced by Th17 cells. Cytokine Growth Factor Rev. 2010, 21, 393–401. [Google Scholar] [CrossRef]
- Larochette, V.; Miot, C.; Poli, C.; Beaumont, E.; Roingeard, P.; Fickenscher, H.; Jeannin, P.; Delneste, Y. IL-26, a cytokine with roles in extracellular DNA-induced inflammation and microbial defense. Front. Immunol. 2019, 10, 204. [Google Scholar] [CrossRef]
- Braum, O.; Pirzer, H.; Fickenscher, H. Interleukin-26, a highly cationic T-cell cytokine targeting epithelial cells. Antiinflamm. Antiallergy Agents Med. Chem. 2012, 11, 221–229. [Google Scholar] [CrossRef] [PubMed]
- You, W.; Tang, Q.; Zhang, C.; Wu, J.; Gu, C.; Wu, Z.; Li, X. IL-26 promotes the proliferation and survival of human gastric cancer cells by regulating the balance of STAT1 and STAT3 activation. PLoS ONE 2013, 8, e63588. [Google Scholar] [CrossRef] [PubMed]
- Truong, A.D.; Hong, Y.; Hoang, C.T.; Lee, J.; Hong, Y.H. Chicken IL-26 regulates immune responses through the JAK/STAT and NF-kappaB signaling pathways. Dev. Comp. Immunol. 2017, 73, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Iversen, M.B.; Paludan, S.R. Mechanisms of type III interferon expression. J. Interferon Cytokine Res. 2010, 30, 573–578. [Google Scholar] [CrossRef] [PubMed]
- Odendall, C.; Kagan, J.C. The unique regulation and functions of type III interferons in antiviral immunity. Curr. Opin. Virol. 2015, 12, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Wack, A.; Terczynska-Dyla, E.; Hartmann, R. Guarding the frontiers: the biology of type III interferons. Nat. Immunol. 2015, 16, 802–809. [Google Scholar] [CrossRef]
- Zanoni, I.; Granucci, F.; Broggi, A. Interferon (IFN)-lambda Takes the Helm: Immunomodulatory roles of type III IFNs. Front. Immunol. 2017, 8, 1661. [Google Scholar] [CrossRef]
- Stanifer, M.L.; Pervolaraki, K.; Boulant, S. Differential regulation of type I and type III interferon signaling. Int. J. Mol. Sci. 2019, 20, 1445. [Google Scholar] [CrossRef]
- Lazear, H.M.; Nice, T.J.; Diamond, M.S. Interferon-lambda: Immune functions at barrier surfaces and beyond. Immunity 2015, 43, 15–28. [Google Scholar] [CrossRef]
- Lasfar, A.; Zloza, A.; Silk, A.W.; Lee, L.Y.; Cohen-Solal, K.A. Interferon lambda: Toward a dual role in cancer. J. Interferon Cytokine Res. 2019, 39, 22–29. [Google Scholar] [CrossRef]
- Stiff, A.; Carson, W., III. Investigations of interferon-lambda for the treatment of cancer. J. Innate Immun 2015, 7, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Finotti, G.; Tamassia, N.; Cassatella, M.A. Interferon-lambdas and plasmacytoid dendritic cells: A close relationship. Front. Immunol. 2017, 8, 1015. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Takaoka, A.; Taniguchi, T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 2006, 25, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Michalska, A.; Blaszczyk, K.; Wesoly, J.; Bluyssen, H.A.R. A positive feedback amplifier circuit that regulates interferon (IFN)-stimulated gene expression and controls type I and type II IFN responses. Front. Immunol. 2018, 9, 1135. [Google Scholar] [CrossRef]
- Mostafavi, S.; Yoshida, H.; Moodley, D.; LeBoite, H.; Rothamel, K.; Raj, T.; Ye, C.J.; Chevrier, N.; Zhang, S.Y.; Feng, T.; et al. Parsing the interferon transcriptional network and its disease associations. Cell 2016, 164, 564–578. [Google Scholar] [CrossRef]
- Vogl, C.; Flatt, T.; Fuhrmann, B.; Hofmann, E.; Wallner, B.; Stiefvater, R.; Kovarik, P.; Strobl, B.; Muller, M. Transcriptome analysis reveals a major impact of JAK protein tyrosine kinase 2 (Tyk2) on the expression of interferon-responsive and metabolic genes. BMC Genom. 2010, 11, 199. [Google Scholar] [CrossRef]
- Leonard, W.J.; Lin, J.X.; O’Shea, J.J. The gammac family of cytokines: Basic biology to therapeutic ramifications. Immunity 2019, 50, 832–850. [Google Scholar] [CrossRef]
- Robinson, T.O.; Schluns, K.S. The potential and promise of IL-15 in immuno-oncogenic therapies. Immunol. Lett. 2017, 190, 159–168. [Google Scholar] [CrossRef]
- Santana Carrero, R.M.; Beceren-Braun, F.; Rivas, S.C.; Hegde, S.M.; Gangadharan, A.; Plote, D.; Pham, G.; Anthony, S.M.; Schluns, K.S. IL-15 is a component of the inflammatory milieu in the tumor microenvironment promoting antitumor responses. Proc. Natl. Acad. Sci. USA 2019, 116, 599–608. [Google Scholar] [CrossRef]
- Waldmann, T.A. The shared and contrasting roles of IL2 and IL15 in the life and death of normal and neoplastic lymphocytes: Implications for cancer therapy. Cancer Immunol. Res. 2015, 3, 219–227. [Google Scholar] [CrossRef]
- Yoshimura, A.; Naka, T.; Kubo, M. SOCS proteins, cytokine signalling and immune regulation. Nat. Rev. Immunol. 2007, 7, 454–465. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Diaz, A.; Shin, D.S.; Moreno, B.H.; Saco, J.; Escuin-Ordinas, H.; Rodriguez, G.A.; Zaretsky, J.M.; Sun, L.; Hugo, W.; Wang, X.; et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell. Rep. 2017, 19, 1189–1201. [Google Scholar] [CrossRef] [PubMed]
- Alspach, E.; Lussier, D.M.; Schreiber, R.D. Interferon gamma and its important roles in promoting and inhibiting spontaneous and therapeutic cancer immunity. Cold Spring Harb. Perspect. Biol. 2019, 11, a028480. [Google Scholar] [CrossRef] [PubMed]
- Castro, F.; Cardoso, A.P.; Goncalves, R.M.; Serre, K.; Oliveira, M.J. Interferon-gamma at the crossroads of tumor immune surveillance or evasion. Front. Immunol. 2018, 9, 847. [Google Scholar] [CrossRef]
- Ivashkiv, L.B. IFNgamma: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat. Rev. Immunol. 2018, 18, 545–558. [Google Scholar] [CrossRef]
- Budczies, J.; Bockmayr, M.; Klauschen, F.; Endris, V.; Frohling, S.; Schirmacher, P.; Denkert, C.; Stenzinger, A. Mutation patterns in genes encoding interferon signaling and antigen presentation: A pan-cancer survey with implications for the use of immune checkpoint inhibitors. Genes Chromosomes Cancer 2017, 56, 651–659. [Google Scholar] [CrossRef]
- Gao, J.; Shi, L.Z.; Zhao, H.; Chen, J.; Xiong, L.; He, Q.; Chen, T.; Roszik, J.; Bernatchez, C.; Woodman, S.E.; et al. Loss of IFN-gamma pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 2016, 167, 397–404. [Google Scholar] [CrossRef]
- Shin, D.S.; Zaretsky, J.M.; Escuin-Ordinas, H.; Garcia-Diaz, A.; Hu-Lieskovan, S.; Kalbasi, A.; Grasso, C.S.; Hugo, W.; Sandoval, S.; Torrejon, D.Y.; et al. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov. 2017, 7, 188–201. [Google Scholar] [CrossRef]
- Amatya, N.; Garg, A.V.; Gaffen, S.L. IL-17 signaling: The yin and the yang. Trends Immunol. 2017, 38, 310–322. [Google Scholar] [CrossRef]
- McGeachy, M.J.; Cua, D.J.; Gaffen, S.L. The IL-17 family of cytokines in health and disease. Immunity 2019, 50, 892–906. [Google Scholar] [CrossRef]
- Chang, S.H. T helper 17 (Th17) cells and interleukin-17 (IL-17) in cancer. Arch. Pharm Res. 2019, 42, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Knochelmann, H.M.; Dwyer, C.J.; Bailey, S.R.; Amaya, S.M.; Elston, D.M.; Mazza-McCrann, J.M.; Paulos, C.M. When worlds collide: Th17 and Treg cells in cancer and autoimmunity. Cell. Mol. Immunol. 2018, 15, 458–469. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Bechara, R.; Zhao, J.; McGeachy, M.J.; Gaffen, S.L. IL-17 receptor-based signaling and implications for disease. Nat. Immunol. 2019, 20, 1594–1602. [Google Scholar] [CrossRef] [PubMed]
- Dejima, T.; Shibata, K.; Yamada, H.; Hara, H.; Iwakura, Y.; Naito, S.; Yoshikai, Y. Protective role of naturally occurring interleukin-17A-producing gammadelta T cells in the lung at the early stage of systemic candidiasis in mice. Infect. Immun. 2011, 79, 4503–4510. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.S.; Wong, N.; Rao, G.; Avery, D.T.; Torpy, J.; Hambridge, T.; Bustamante, J.; Okada, S.; Stoddard, J.L.; Deenick, E.K.; et al. Monogenic mutations differentially affect the quantity and quality of T follicular helper cells in patients with human primary immunodeficiencies. J. Allergy Clin. Immunol. 2015, 136, 993–1006. [Google Scholar] [CrossRef] [PubMed]
- Ubel, C.; Graser, A.; Koch, S.; Rieker, R.J.; Lehr, H.A.; Muller, M.; Finotto, S. Role of Tyk-2 in Th9 and Th17 cells in allergic asthma. Sci. Rep. 2014, 4, 5865. [Google Scholar] [CrossRef]
- Wöss, K.; Simonovic, N.; Strobl, B.; Macho-Maschler, S.; Müller, M. TYK2: An upstream kinase of STATs in cancer. Cancers 2019, 11, 1728. [Google Scholar] [CrossRef]
- Ide, H.; Nakagawa, T.; Terado, Y.; Kamiyama, Y.; Muto, S.; Horie, S. Tyk2 expression and its signaling enhances the invasiveness of prostate cancer cells. Biochem. Biophys. Res. Commun. 2008, 369, 292–296. [Google Scholar] [CrossRef]
- Schuster, C.; Muller, M.; Freissmuth, M.; Sexl, V.; Stoiber, D. Commentary on H. Ide et al., Tyk2 expression and its signaling enhances the invasiveness of prostate cancer cells. Biochem. Biophys. Res. Commun. 2008, 366, 869–870. [Google Scholar] [CrossRef]
- Ubel, C.; Mousset, S.; Trufa, D.; Sirbu, H.; Finotto, S. Establishing the role of tyrosine kinase 2 in cancer. Oncoimmunology 2013, 2, e22840. [Google Scholar] [CrossRef]
- Banfield, C.; Scaramozza, M.; Zhang, W.; Kieras, E.; Page, K.M.; Fensome, A.; Vincent, M.; Dowty, M.E.; Goteti, K.; Winkle, P.J.; et al. The Safety, tolerability, pharmacokinetics, and pharmacodynamics of a TYK2/JAK1 inhibitor (PF-06700841) in healthy subjects and patients with plaque psoriasis. J. Clin. Pharmacol. 2018, 58, 434–447. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Chen, X.; Zhang, H.; Xie, T.; Ye, X.Y. Selective Tyk2 inhibitors as potential therapeutic agents: a patent review (2015–2018). Expert Opin. Ther. Pat. 2019, 29, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Papp, K.; Gordon, K.; Thaci, D.; Morita, A.; Gooderham, M.; Foley, P.; Girgis, I.G.; Kundu, S.; Banerjee, S. Phase 2 trial of selective tyrosine kinase 2 inhibition in psoriasis. N. Engl. J. Med. 2018, 29, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, A.; Lahtz, C.; Nagao, T.; Song, J.Y.; Chan, W.C.; Lee, H.; Yue, C.; Look, T.; Muelfarth, R.; Li, W.; et al. CTLA4 promotes Tyk2-STAT3 dependent B cell oncogenecity. Cancer Res. 2017, 77, 5118–5128. [Google Scholar] [CrossRef] [PubMed]
- Reader, J.; Williams, N.; Bojdo, J.; Worthington, J.; Mitchell, T. Immunotherapeutic effects of the TYK2 inhibitor SAR-20351 in syngeneic tumor models [abstract]. Mol. Cancer Ther. 2019, 18, C086. [Google Scholar] [CrossRef]
- Howden, A.J.M.; Hukelmann, J.L.; Brenes, A.; Spinelli, L.; Sinclair, L.V.; Lamond, A.I.; Cantrell, D.A. Quantitative analysis of T cell proteomes and environmental sensors during T cell differentiation. Nat. Immunol. 2019, 20, 1542–1554. [Google Scholar] [CrossRef]
- Wagner, J.; Rapsomaniki, M.A.; Chevrier, S.; Anzeneder, T.; Langwieder, C.; Dykgers, A.; Rees, M.; Ramaswamy, A.; Muenst, S.; Soysal, S.D.; et al. A single-cell atlas of the tumor and immune ecosystem of human breast cancer. Cell 2019, 177, 1330–1345. [Google Scholar] [CrossRef]
- Wu, T.; Wu, X.; Wang, H.Y.; Chen, L. Immune contexture defined by single cell technology for prognosis prediction and immunotherapy guidance in cancer. Cancer Commun. 2019, 39, 21. [Google Scholar] [CrossRef]
- Si, Y.; Merz, S.F.; Jansen, P.; Wang, B.; Bruderek, K.; Altenhoff, P.; Mattheis, S.; Lang, S.; Gunzer, M.; Klode, J.; et al. Multidimensional imaging provides evidence for down-regulation of T cell effector function by MDSC in human cancer tissue. Sci. Immunol. 2019, 4, eaaw9159. [Google Scholar] [CrossRef]
- Johnson, H.M.; Noon-Song, E.; Ahmed, C.M. Noncanonical IFN signaling, steroids, and STATs: A probable role of V-ATPase. Mediators Inflamm. 2019, 2019, 4143604. [Google Scholar] [CrossRef]
- Zouein, F.A.; Duhe, R.J.; Booz, G.W. JAKs go nuclear: emerging role of nuclear JAK1 and JAK2 in gene expression and cell growth. Growth Factors 2011, 29, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Potla, R.; Koeck, T.; Wegrzyn, J.; Cherukuri, S.; Shimoda, K.; Baker, D.P.; Wolfman, J.; Planchon, S.M.; Esposito, C.; Hoit, B.; et al. Tyk2 tyrosine kinase expression is required for the maintenance of mitochondrial respiration in primary pro-B lymphocytes. Mol. Cell. Biol. 2006, 26, 8562–8571. [Google Scholar] [CrossRef] [PubMed]
- Avalle, L.; Poli, V. Nucleus, mitochondrion, or reticulum? STAT3 a la carte. Int. J. Mol. Sci. 2018, 19, 2820. [Google Scholar] [CrossRef] [PubMed]
- Meier, J.A.; Larner, A.C. Toward a new STATe: The role of STATs in mitochondrial function. Semin. Immunol. 2014, 26, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.H.; Zhao, Y.; Lam, Y.; Piekos, S.; Han, Y.C.; Reilly, C.; Joshi, P.; Hong, S.H.; Sung, C.O.; Giardina, C.; et al. BioTarget: A computational framework identifying cancer type specific transcriptional targets of immune response pathways. Sci. Rep. 2019, 9, 9029. [Google Scholar] [CrossRef]
- Liu, Y. A global immune gene expression signature for human cancers. Oncotarget 2019, 10, 1993–2005. [Google Scholar] [CrossRef]
- Stoney, R.; Robertson, D.L.; Nenadic, G.; Schwartz, J.M. Mapping biological process relationships and disease perturbations within a pathway network. NPJ Syst. Biol. Appl. 2018, 4, 22. [Google Scholar] [CrossRef]
| Cytokine Family | Receptor (R) Chain (⇔ TYK2) | Cytokine & Specific R Chain (⇔ JAK1 or 2) | STATs Activated 1 | Producing Cells | Responding Cells |
|---|---|---|---|---|---|
| Type I IFNs | IFNAR1 | IFNα/β2 IFNAR2 ⇔ JAK1 | STAT1, STAT23 STAT3-6 | all cells, pDCs (professional type I IFN producers) | all cells |
| IL-12/23 family | IL-12Rβ1 | IL-12 IL-12Rβ2 ⇔ JAK2 | STAT4 STAT1/3/5 | monocytes, macrophages, DCs | activated NK and T cells |
| IL-23 IL-23R ⇔ JAK2 | STAT3, STAT4 STAT1/5 | monocytes, macrophages, DCs | activated T cells, ILCs, NK cells, monocytes, macrophages, DCs | ||
| IL-10 family | IL-10R2 | IL-10 IL-10R1 ⇔ JAK1 | STAT3 STAT1/5 | Monocytes 4, macrophages 4, DCs NK cells 5, T cells, B cells | all subsets of leukocytes |
| IL-22 IL-22R1 ⇔ JAK1 | STAT3 STAT1/5 | T cells, ILCs, NK/T cells, macrophages, neutrophils, DC, fibroblasts | epithelial cells, keratinocytes, hepatocytes | ||
| IL-266 IL-20R1 ⇔ JAK1 | STAT3 STAT1 | Th17 cells, NK cells, macrophages, fibroblasts | non-hematopoietic and hematopoietic cell subsets 7 | ||
| type III IFNs/IFNλ IL-28R ⇔ JAK1 | STAT1, STAT23 STAT3-6 | all cells 8 | epithelial cells, DCs, neutrophils a.o. 9 |
| Mutation | Disease | IL-10 | IL-12 | IL-22 | IL-23 | IFN-I | IFN-III | ||
|---|---|---|---|---|---|---|---|---|---|
| mouse | LOF of TYK2 | Tyk2−/− | impaired tumor surveillance; susceptible to microbial infections; resistance/protection during inflammatory challenges | normal [64,66]; impaired [85] | impaired 1 [76,86,87] 2; [64,66,85,88,89,90,91,92,93,94] | impaired [95,96] | impaired [91,94,96,97] | impaired [98] 2; [64,66,74,99,100,101] | n.d. |
| Tyk2mut | susceptible to parasite infection; protection during inflammatory challenge | n.d. | impaired [68,102] | n.d. | impaired [68,102] | impaired [68,99] | n.d. | ||
| Tyk2Pmut | virus-induced diabetes | n.d. | n.d. | n.d. | n.d. | impaired [69] | n.d. | ||
| KI-TYK2 | Tyk2K923E; Tyk2K923R | impaired tumor surveillance; susceptible to virus infection; restoration of obesity | n.d. | impaired [76] 2 | n.d. | n.d. | impaired [74,75] | n.d. | |
| Tyk2P1124A | protection during inflammatory challenge | n.d. | impaired [79,82] | n.d. | impaired [79,82] | impaired [79,82] | n.d. | ||
| human | LOF of TYK2 | patient 1 | bacterial, viral and fungal infections, HIES | impaired [72,103] | impaired [72,103] | n.d. | impaired [72] | impaired [72] | n.d. |
| patients 2–10 | bacterial and viral infections, no HIES 3 | impaired [42,43,103] | impaired [42,43,73,103] | n.d. | impaired [43] | impaired [42,43] | impaired [43]; normal [42] | ||
| promoter variants | virus-induced type I diabetes | n.d. | n.d. | n.d. | n.d. | impaired [70,71] | n.d. | ||
| KI-TYK2 | TYK2P1104A | mycobacterial infection; autoimmune protected/susceptible | normal [77] 4 [79]/[78] | normal [77,78,81] 5; impaired [79] | n.d. | impaired [77,79,82] 6; normal [78] 5 | normal [77]; impaired [78,79,82] | n.d. | |
| TYK2I684S | healthy; autoimmune protected | normal [77] | normal [77]; impaired [81] | n.d. | normal [77] | normal [77] | n.d. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karjalainen, A.; Shoebridge, S.; Krunic, M.; Simonović, N.; Tebb, G.; Macho-Maschler, S.; Strobl, B.; Müller, M. TYK2 in Tumor Immunosurveillance. Cancers 2020, 12, 150. https://doi.org/10.3390/cancers12010150
Karjalainen A, Shoebridge S, Krunic M, Simonović N, Tebb G, Macho-Maschler S, Strobl B, Müller M. TYK2 in Tumor Immunosurveillance. Cancers. 2020; 12(1):150. https://doi.org/10.3390/cancers12010150
Chicago/Turabian StyleKarjalainen, Anzhelika, Stephen Shoebridge, Milica Krunic, Natalija Simonović, Graham Tebb, Sabine Macho-Maschler, Birgit Strobl, and Mathias Müller. 2020. "TYK2 in Tumor Immunosurveillance" Cancers 12, no. 1: 150. https://doi.org/10.3390/cancers12010150
APA StyleKarjalainen, A., Shoebridge, S., Krunic, M., Simonović, N., Tebb, G., Macho-Maschler, S., Strobl, B., & Müller, M. (2020). TYK2 in Tumor Immunosurveillance. Cancers, 12(1), 150. https://doi.org/10.3390/cancers12010150





