The Transcriptional Roles of ALK Fusion Proteins in Tumorigenesis
Abstract
1. Introduction
2. Nucleophosmin 1-Anaplastic Lymphoma Kinase (NPM1-ALK)
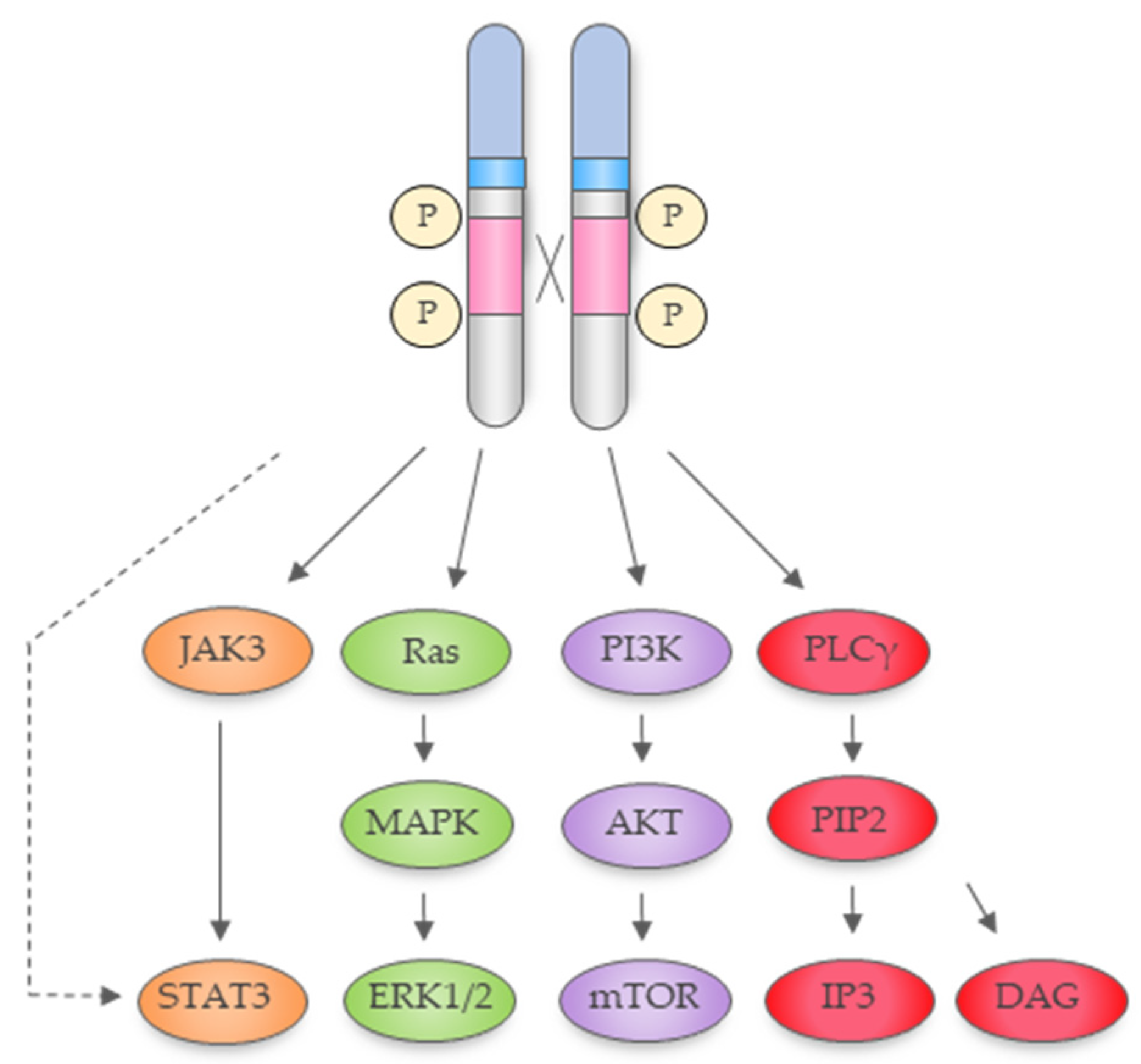
2.1. NPM1-ALK Is a Constitutively Active Tyrosine Kinase Activating a Plethora of Signal Transduction Pathways
2.1.1. Ras-Extracellular Signal-Regulated Kinase Pathway
2.1.2. The Janus Kinase 3−Signal Transducer and Activator of Transcription 3 Pathway
2.1.3. Phospholipase C Gamma (PLCγ) Pathway
2.1.4. Phosphatidylinositol 3-Kinase–Protein kinase B (Akt) Pathway
2.1.5. Epigenetic Pathways
2.2. NPM1-ALK Mediates DNA Methylation
2.3. NPM1-ALK Activates microRNAs (MiRNAs; miRs)
2.4. Long Non-Coding RNAs Are Expressed in ALCL
2.5. NPM1-ALK Driven Transcription Factors
2.6. NPM1-ALK Interacts with Nuclear Proteins
3. NPM1 as an ALK-Translocation Fusion Partner
3.1. Known Functions of Nucleophosmin 1
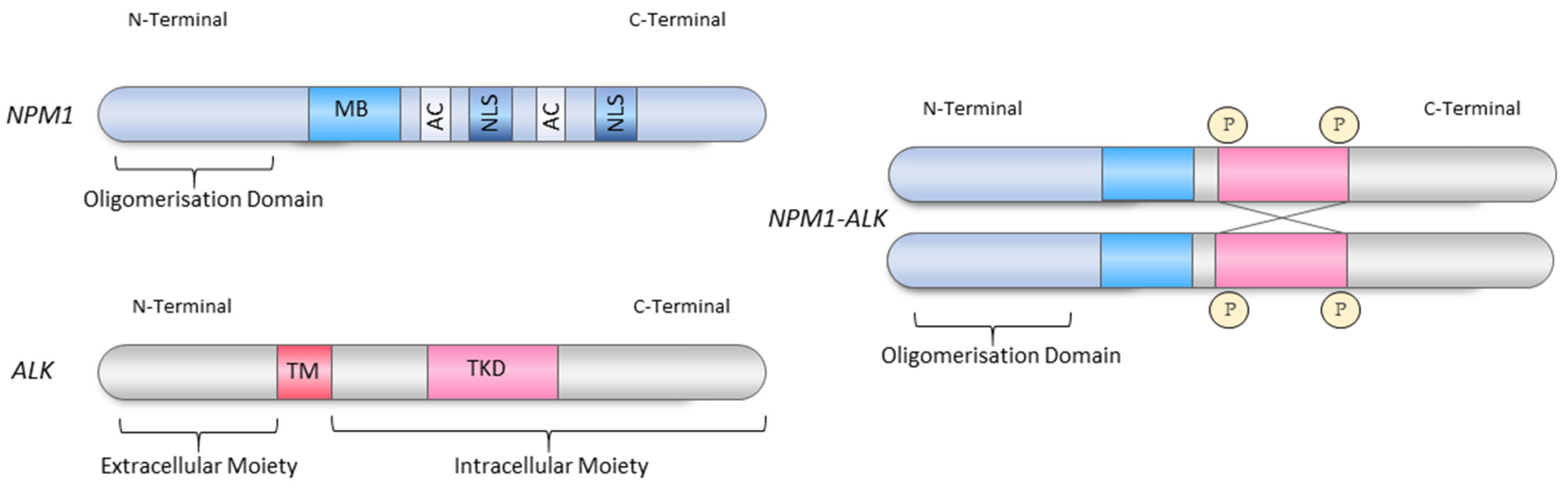
3.2. Structure of Nucleophosmin 1
3.3. The Roles of the Retained NPM1 Domains in the NPM1-ALK Fusion Protein
3.4. Is Heterozygous Loss of NPM1 a Key Factor Driving Lymphomagenesis?
4. Other ALK Fusion Proteins Are Causative of Cancer
4.1. Do other ALK Fusion Proteins Have a Role in the Nucleus?
4.2. EML4 Is an ALK-Translocation Fusion Partner
4.2.1. Structure and Function of the EML Protein Family
4.2.2. EML4-ALK Variants
4.2.3. Does Localisation of EML4-ALK Affect Its Function?
4.2.4. EML4-ALK Mediated Signalling
5. Expression of Full-Length ALK in Other Cancers
6. Conclusions
Funding
Conflicts of Interest
References
- Morris, S.W.; Kirstein, M.N.; Valentine, M.B.; Dittmer, K.G.; Shapiro, D.N.; Saltman, D.L.; Look, A.T. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science 1994, 263, 1281–1284. [Google Scholar] [CrossRef]
- Iwahara, T.; Fujimoto, J.; Wen, D.; Cupples, R.; Bucay, N.; Arakawa, T.; Mori, S.; Ratzkin, B.; Yamamoto, T. Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene 1997, 14, 439–449. [Google Scholar] [CrossRef]
- Bischof, D.; Pulford, K.; Mason, D.Y.; Morris, S.W. Role of the nucleophosmin (NPM) portion of the non-Hodgkin’s lymphoma-associated NPM-anaplastic lymphoma kinase fusion protein in oncogenesis. Mol. Cell. Biol. 1997, 17, 2312–2325. [Google Scholar] [CrossRef]
- Soda, M.; Choi, Y.L.; Enomoto, M.; Takada, S.; Yamashita, Y.; Ishikawa, S.; Fujiwara, S.; Watanabe, H.; Kurashina, K.; Hatanaka, H.; et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature 2007, 448, 561–566. [Google Scholar] [CrossRef]
- Colleoni, G.W.B.; Bridge, J.A.; Garicochea, B.; Liu, J.; Filippa, D.A.; Ladanyi, M. ATIC-ALK: A novel variant ALK gene fusion in anaplastic large cell lymphoma resulting from the recurrent cryptic chromosomal inversion, inv(2)(p23q35). Am. J. Pathol. 2000, 156, 781–789. [Google Scholar] [CrossRef]
- Cools, J.; Wlodarska, I.; Somers, R.; Mentens, N.; Pedeutour, F.; Maes, B.; De Wolf-Peeters, C.; Pauwels, P.; Hagemeijer, A.; Marynen, P. Identification of novel fusion partners of ALK, the anaplastic lymphoma kinase, in anaplastic large-cell lymphoma and inflammatory myofibroblastic tumor. Genes Chromosom. Cancer 2002, 34, 354–362. [Google Scholar] [CrossRef]
- Hernández, L.; Pinyol, M.; Hernández, S.; Beà, S.; Pulford, K.; Rosenwald, A.; Lamant, L.; Falini, B.; Ott, G.; Mason, D.Y.; et al. TRK-fused gene (TFG) is a new partner of ALK in anaplastic large cell lymphoma producing two structurally different TFG-ALK translocations. Blood 1999, 94, 3265–3268. [Google Scholar]
- Lawrence, B.; Perez-Atayde, A.; Hibbard, M.K.; Rubin, B.P.; Dal Cin, P.; Pinkus, J.L.; Pinkus, G.S.; Xiao, S.; Yi, E.S.; Fletcher, C.D.M.; et al. TPM3-ALK and TPM4-ALK Oncogenes in Inflammatory Myofibroblastic Tumors. Am. J. Pathol. 2000, 157, 377–384. [Google Scholar] [CrossRef]
- Feldman, A.L.; Vasmatzis, G.; Asmann, Y.W.; Davila, J.; Middha, S.; Eckloff, B.W.; Johnson, S.H.; Porcher, J.C.; Ansell, S.M.; Caride, A. Novel TRAF1-ALK fusion identified by deep RNA sequencing of anaplastic large cell lymphoma. Genes Chromosom. Cancer 2013, 52, 1097–1102. [Google Scholar] [CrossRef]
- Armstrong, F.; Duplantier, M.-M.; Trempat, P.; Hieblot, C.; Lamant, L.; Espinos, E.; Racaud-Sultan, C.; Allouche, M.; Campo, E.; Delsol, G.; et al. Differential effects of X-ALK fusion proteins on proliferation, transformation and invasion properties of NIH3T3 cells. Oncogene 2004, 23, 6071–6082. [Google Scholar] [CrossRef]
- Palacios, G.; Shaw, T.I.; Li, Y.; Singh, R.K.; Valentine, M.; Sandlund, J.T.; Lim, M.S.; Mullighan, C.G.; Leventaki, V. Novel ALK fusion in anaplastic large cell lymphoma involving EEF1G, a subunit of the eukaryotic elongation factor-1 complex. Leukemia 2017, 31, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Young, L.C.; Togashi, Y.; Soda, M.; Hatano, S.; Inamura, K.; Takada, S.; Ueno, T.; Yamashita, Y.; Satoh, Y.; et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin. Cancer Res. 2009, 15, 3143–3149. [Google Scholar] [CrossRef] [PubMed]
- Togashi, Y.; Soda, M.; Sakata, S.; Sugawara, E.; Hatano, S.; Asaka, R.; Nakajima, T.; Mano, H.; Takeuchi, K. KLC1-ALK: A Novel Fusion in Lung Cancer Identified Using a Formalin-Fixed Paraffin-Embedded Tissue Only. PLoS ONE 2012, 7, e31323. [Google Scholar] [CrossRef] [PubMed]
- Kelly, L.M.; Barila, G.; Liu, P.; Evdokimova, V.N.; Trivedi, S.; Panebianco, F.; Gandhi, M.; Carty, S.E.; Hodak, S.P.; Luo, J.; et al. Identification of the transforming STRN-ALK fusion as a potential therapeutic target in the aggressive forms of thyroid cancer. Proc. Natl. Acad. Sci. USA 2014, 111, 4233–4238. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Kim, P.; Jung, Y.; Keum, J.; Kim, S.-N.; Choi, Y.S.; Do, I.-G.; Lee, J.; Choi, S.-J.; Kim, S.; et al. Discovery of ALK-PTPN3 gene fusion from human non-small cell lung carcinoma cell line using next generation RNA sequencing. Genes Chromosom. Cancer 2012, 51, 590–597. [Google Scholar] [CrossRef]
- Iyevleva, A.G.; Raskin, G.A.; Tiurin, V.I.; Sokolenko, A.P.; Mitiushkina, N.V.; Aleksakhina, S.N.; Garifullina, A.R.; Strelkova, T.N.; Merkulov, V.O.; Ivantsov, A.O.; et al. Novel ALK fusion partners in lung cancer. Cancer Lett. 2015, 362, 116–121. [Google Scholar] [CrossRef]
- Jiang, J.; Wu, X.; Tong, X.; Wei, W.; Chen, A.; Wang, X.; Shao, Y.W.; Huang, J. GCC2-ALK as a targetable fusion in lung adenocarcinoma and its enduring clinical responses to ALK inhibitors. Lung Cancer 2018, 115, 5–11. [Google Scholar] [CrossRef]
- Duyster, J.; Bai, R.-Y.; Morris, S.W. Translocations involving anaplastic lymphoma kinase (ALK). Oncogene 2001, 20, 5623–5637. [Google Scholar] [CrossRef]
- Chiarle, R.; Voena, C.; Ambrogio, C.; Piva, R.; Inghirami, G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat. Rev. Cancer 2008, 8, 11–23. [Google Scholar] [CrossRef]
- Roukos, V.; Mathas, S. The origins of ALK translocations. Front. Biosci. 2015, 7, 260–268. [Google Scholar]
- Ceccon, M.; Merlo, M.E.B.; Mologni, L.; Poggio, T.; Varesio, L.M.; Menotti, M.; Bombelli, S.; Rigolio, R.; Manazza, A.D.; Di Giacomo, F.; et al. Excess of NPM-ALK oncogenic signaling promotes cellular apoptosis and drug dependency. Oncogene 2015, 35, 3854–3865. [Google Scholar] [CrossRef] [PubMed]
- Crockett, D.K.; Lin, Z.; Elenitoba-Johnson, K.S.; Lim, M.S. Identification of NPM-ALK interacting proteins by tandem mass spectrometry. Oncogene 2004, 23, 2617–2629. [Google Scholar] [CrossRef] [PubMed]
- Trigg, R.M.; Turner, S.D. ALK in Neuroblastoma: Biological and Therapeutic Implications. Cancers 2018, 10, 113. [Google Scholar] [CrossRef] [PubMed]
- Downward, J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 2003, 3, 11–22. [Google Scholar] [CrossRef]
- Fujimoto, J.; Shiota, M.; Iwahara, T.; Seki, N.; Satoh, H.; Mori, S.; Yamamoto, T. Characterization of the transforming activity of p80, a hyperphosphorylated protein in a Ki-1 lymphoma cell line with chromosomal translocation t(2;5). Proc. Natl. Acad. Sci. USA 1996, 93, 4181–4186. [Google Scholar] [CrossRef] [PubMed]
- Pulford, K.; Morris, S.W.; Mason, D.Y. Anaplastic lymphoma kinase proteins and malignancy. Curr. Opin. Hematol. 2001, 8, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Kasprzycka, M.; Liu, X.; Raghunath, P.N.; Wlodarski, P.; Wasik, M.A. Oncogenic tyrosine kinase NPM/ALK induces activation of the MEK/ERK signaling pathway independently of c-Raf. Oncogene 2007, 26, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Rane, S.G.; Reddy, E.P. Janus kinases: Components of multiple signaling pathways. Oncogene 2000, 19, 5662–5679. [Google Scholar] [CrossRef]
- Han, Y.; Amin, H.M.; Franko, B.; Frantz, C.; Shi, X.; Lai, R.; Weber-Nordt, R.; Dusanter-Fourt, I.; Dreyfus, F.; Groner, B.; et al. Loss of SHP1 enhances JAK3/STAT3 signaling and decreases proteosome degradation of JAK3 and NPM-ALK in ALK+ anaplastic large-cell lymphoma. Blood 2006, 108, 2796–2803. [Google Scholar] [CrossRef]
- Bowman, T.; Garcia, R.; Turkson, J.; Jove, R. STATs in oncogenesis. Oncogene 2000, 19, 2474–2488. [Google Scholar] [CrossRef]
- Zamo, A.; Chiarle, R.; Piva, R.; Howes, J.; Fan, Y.; Chilosi, M.; Levy, D.E.; Inghirami, G. Anaplastic lymphoma kinase (ALK) activates Stat3 and protects hematopoietic cells from cell death. Oncogene 2002, 21, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Chiarle, R.; Simmons, W.J.; Cai, H.; Dhall, G.; Zamo, A.; Raz, R.; Karras, J.G.; Levy, D.E.; Inghirami, G. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat. Med. 2005, 11, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Falasca, M.; Logan, S.K.; Lehto, V.P.; Baccante, G.; Lemmon, M.A.; Schlessinger, J. Activation of phospholipase C gamma by PI 3-kinase-induced PH domain-mediated membrane targeting. EMBO J. 1998, 17, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Honorat, J.-F.; Ragab, A.; Lamant, L.; Delsol, G.; Ragab-Thomas, J. SHP1 tyrosine phosphatase negatively regulates NPM-ALK tyrosine kinase signaling. Blood 2006, 107, 4130–4138. [Google Scholar] [CrossRef] [PubMed]
- Bai, R.Y.; Dieter, P.; Peschel, C.; Morris, S.W.; Duyster, J. Nucleophosmin-anaplastic lymphoma kinase of large-cell anaplastic lymphoma is a constitutively active tyrosine kinase that utilizes phospholipase C-gamma to mediate its mitogenicity. Mol. Cell. Biol. 1998, 18, 6951–6961. [Google Scholar] [CrossRef] [PubMed]
- Chang, F.; Lee, J.T.; Navolanic, P.M.; Steelman, L.S.; Shelton, J.G.; Blalock, W.L.; Franklin, R.A.; McCubrey, J.A. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: A target for cancer chemotherapy. Leukemia 2003, 17, 590–603. [Google Scholar] [CrossRef] [PubMed]
- Slupianek, A.; Nieborowska-Skorska, M.; Hoser, G.; Morrione, A.; Majewski, M.; Xue, L.; Morris, S.W.; Wasik, M.A.; Skorski, T. Role of Phosphatidylinositol 3-Kinase-Akt Pathway in Nucleophosmin/Anaplastic Lymphoma Kinase-mediated Lymphomagenesis. Cancer Res. 2001, 61, 2194–2199. [Google Scholar]
- Datta, S.R.; Dudek, H.; Tao, X.; Masters, S.; Fu, H.; Gotoh, Y.; Greenberg, M.E. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 1997, 91, 231–241. [Google Scholar] [CrossRef]
- Bai, R.Y.; Ouyang, T.; Miething, C.; Morris, S.W.; Peschel, C.; Duyster, J. Nucleophosmin-anaplastic lymphoma kinase associated with anaplastic large-cell lymphoma activates the phosphatidylinositol 3-kinase/Akt antiapoptotic signaling pathway. Blood 2000, 96, 4319–4327. [Google Scholar]
- Jin, B.; Li, Y.; Robertson, K.D. DNA methylation: Superior or subordinate in the epigenetic hierarchy? Genes Cancer 2011, 2, 607–617. [Google Scholar] [CrossRef]
- Hassler, M.R.; Pulverer, W.; Lakshminarasimhan, R.; Redl, E.; Hacker, J.; Garland, G.D.; Merkel, O.; Schiefer, A.-I.; Simonitsch-Klupp, I.; Kenner, L.; et al. Insights into the Pathogenesis of Anaplastic Large-Cell Lymphoma through Genome-wide DNA Methylation Profiling. Cell Rep. 2016, 17, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Merkel, O.; Hamacher, F.; Laimer, D.; Sifft, E.; Trajanoski, Z.; Scheideler, M.; Egger, G.; Hassler, M.R.; Thallinger, C.; Schmatz, A.; et al. Identification of differential and functionally active miRNAs in both anaplastic lymphoma kinase (ALK)+ and ALK− anaplastic large-cell lymphoma. Proc. Natl. Acad. Sci. USA 2010, 107, 16228–16233. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, T.; Bai, R.-Y.; Bassermann, F.; von Klitzing, C.; Klumpen, S.; Miething, C.; Morris, S.W.; Peschel, C.; Duyster, J. Identification and characterization of a nuclear interacting partner of anaplastic lymphoma kinase (NIPA). J. Biol. Chem. 2003, 278, 30028–30036. [Google Scholar] [CrossRef] [PubMed]
- Galietta, A.; Gunby, R.H.; Redaelli, S.; Stano, P.; Carniti, C.; Bachi, A.; Tucker, P.W.; Tartari, C.J.; Huang, C.-J.; Colombo, E.; et al. NPM/ALK binds and phosphorylates the RNA/DNA-binding protein PSF in anaplastic large-cell lymphoma. Blood 2007, 110, 2600–2609. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bestor, T.; Laudano, A.; Mattaliano, R.; Ingram, V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol. 1988, 203, 971–983. [Google Scholar] [CrossRef]
- Okano, M.; Xie, S.; Li, E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998, 19, 219–220. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, H.Y.; Liu, X.; Wasik, M.A. STAT5A is epigenetically silenced by the tyrosine kinase NPM1-ALK and acts as a tumor suppressor by reciprocally inhibiting NPM1-ALK expression. Nat. Med. 2007, 13, 1341–1348. [Google Scholar] [CrossRef] [PubMed]
- Nan, X.; Campoy, F.J.; Bird, A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 1997, 88, 471–481. [Google Scholar] [CrossRef]
- Lewis, J.D.; Meehan, R.R.; Henzel, W.J.; Maurer-Fogy, I.; Jeppesen, P.; Klein, F.; Bird, A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 1992, 69, 905–914. [Google Scholar] [CrossRef]
- Reddington, J.P.; Sproul, D.; Meehan, R.R. DNA methylation reprogramming in cancer: Does it act by re-configuring the binding landscape of Polycomb repressive complexes? Bioessays 2014, 36, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Cedar, H.; Bergman, Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat. Rev. Genet. 2009, 10, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, H.Y.; Liu, X.; Bhutani, G.; Kantekure, K.; Wasik, M. IL-2R common gamma-chain is epigenetically silenced by nucleophosphin-anaplastic lymphoma kinase (NPM-ALK) and acts as a tumor suppressor by targeting NPM-ALK. Proc. Natl. Acad. Sci. USA 2011, 108, 11977–11982. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, H.Y.; Woetmann, A.; Raghunath, P.N.; Odum, N.; Wasik, M.A. STAT3 induces transcription of the DNA methyltransferase 1 gene (DNMT1) in malignant T lymphocytes. Blood 2006, 108, 1058–1064. [Google Scholar] [CrossRef]
- Malcolm, T.I.M.; Villarese, P.; Fairbairn, C.J.; Lamant, L.; Trinquand, A.; Hook, C.E.; Burke, G.A.A.; Brugières, L.; Hughes, K.; Payet, D.; et al. Anaplastic large cell lymphoma arises in thymocytes and requires transient TCR expression for thymic egress. Nat. Commun. 2016, 7, 10087. [Google Scholar] [CrossRef]
- Nagasawa, T.; Zhang, Q.; Raghunath, P.N.; Wong, H.Y.; El-Salem, M.; Szallasi, A.; Marzec, M.; Gimotty, P.; Rook, A.H.; Vonderheid, E.C.; et al. Multi-gene epigenetic silencing of tumor suppressor genes in T-cell lymphoma cells; delayed expression of the p16 protein upon reversal of the silencing. Leuk. Res. 2006, 30, 303–312. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, H.Y.; Bhutani, G.; Liu, X.; Paessler, M.; Tobias, J.W.; Baldwin, D.; Swaminathan, K.; Milone, M.C.; Wasik, M.A. Lack of TNF expression protects anaplastic lymphoma kinase-positive T-cell lymphoma (ALK+ TCL) cells from apoptosis. Proc. Natl. Acad. Sci. USA 2009, 106, 15843–15848. [Google Scholar] [CrossRef]
- Akimzhanov, A.; Krenacs, L.; Schlegel, T.; Klein-Hessling, S.; Bagdi, E.; Stelkovics, E.; Kondo, E.; Chuvpilo, S.; Wilke, P.; Avots, A.; et al. Epigenetic Changes and Suppression of the Nuclear Factor of Activated T Cell 1 (NFATC1) Promoter in Human Lymphomas with Defects in Immunoreceptor Signaling. Am. J. Pathol. 2008, 172, 215–224. [Google Scholar] [CrossRef]
- Ambrogio, C.; Martinengo, C.; Voena, C.; Tondat, F.; Riera, L.; di Celle, P.F.; Inghirami, G.; Chiarle, R. NPM-ALK oncogenic tyrosine kinase controls T-cell identity by transcriptional regulation and epigenetic silencing in lymphoma cells. Cancer Res. 2009, 69, 8611–8619. [Google Scholar] [CrossRef]
- Piazza, R.; Magistroni, V.; Mogavero, A.; Andreoni, F.; Ambrogio, C.; Chiarle, R.; Mologni, L.; Bachmann, P.S.; Lock, R.B.; Collini, P.; et al. Epigenetic silencing of the proapoptotic gene BIM in anaplastic large cell lymphoma through an MeCP2/SIN3a deacetylating complex. Neoplasia 2013, 15, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human RISC Couples MicroRNA Biogenesis and Posttranscriptional Gene Silencing. Cell 2005, 123, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Baer, C.; Claus, R.; Plass, C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 2013, 73, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Jansson, M.D.; Lund, A.H. MicroRNA and cancer. Mol. Oncol. 2012, 6, 590–610. [Google Scholar] [CrossRef] [PubMed]
- Merkel, O.; Hamacher, F.; Griessl, R.; Grabner, L.; Schiefer, A.-I.; Prutsch, N.; Baer, C.; Egger, G.; Schlederer, M.; Krenn, P.W.; et al. Oncogenic role of miR-155 in anaplastic large cell lymphoma lacking the t(2;5) translocation. J. Pathol. 2015, 236, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Iqbal, J.; Teruya-Feldstein, J.; Shen, Y.; Dabrowska, M.J.; Dybkaer, K.; Lim, M.S.; Piva, R.; Barreca, A.; Pellegrino, E.; et al. microRNA expression profiling identifies molecular signatures associated with anaplastic large cell lymphoma. Blood 2013, 122, 2083–2092. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, H.; Suzuki, H.I.; Nishimori, H.; Noguchi, M.; Yao, T.; Komatsu, N.; Mano, H.; Sugimoto, K.; Miyazono, K. miR-135b mediates NPM-ALK-driven oncogenicity and renders IL-17-producing immunophenotype to anaplastic large cell lymphoma. Blood 2011, 118, 6881–6892. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-H.; Xiao, X.; Zhang, Y.-N.; Wang, Y.-M.; Feng, L.-M.; Wu, Y.-M.; Zhang, Y.-X. MicroRNA miR-886-5p inhibits apoptosis by down-regulating Bax expression in human cervical carcinoma cells. Gynecol. Oncol. 2011, 120, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.S.; Jeon, H.-S.; Sun, Z.; Aubry, M.C.; Tang, H.; Park, C.-H.; Rakhshan, F.; Schultz, D.A.; Kolbert, C.P.; Lupu, R.; et al. Increased miR-708 Expression in NSCLC and Its Association with Poor Survival in Lung Adenocarcinoma from Never Smokers. Clin. Cancer Res. 2012, 18, 3658–3667. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.-J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef]
- Faraoni, I.; Antonetti, F.R.; Cardone, J.; Bonmassar, E. miR-155 gene: A typical multifunctional microRNA. Biochim. Biophys. Acta Mol. Basis Dis. 2009, 1792, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Serra, P.; Esteller, M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene 2012, 31, 1609–1622. [Google Scholar] [CrossRef] [PubMed]
- Thomas, N.S.B. The STAT3-DNMT1 connection. JAK-STAT 2012, 1, 257–260. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bissels, U.; Bosio, A.; Wagner, W. MicroRNAs are shaping the hematopoietic landscape. Haematologica 2012, 97, 160–167. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Jiang, X.; Chen, J. The role of miR-150 in normal and malignant hematopoiesis. Oncogene 2014, 33, 3887–3893. [Google Scholar] [CrossRef] [PubMed]
- Hoareau-Aveilla, C.; Valentin, T.; Daugrois, C.; Quelen, C.; Mitou, G.; Quentin, S.; Jia, J.; Spicuglia, S.; Ferrier, P.; Ceccon, M.; et al. Reversal of microRNA-150 silencing disadvantages crizotinib-resistant NPM-ALK(+) cell growth. J. Clin. Investig. 2015, 125, 3505–3518. [Google Scholar] [CrossRef] [PubMed]
- Vance, K.W.; Ponting, C.P. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014, 30, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Kung, J.T.Y.; Colognori, D.; Lee, J.T. Long noncoding RNAs: Past, present, and future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [PubMed]
- Fragliasso, V.; Verma, A.; Bareja, R.; Heavican, T.; Iqbal, J.; Chan, W.C.; Merli, F.; Ciarrocchi, A.; Elemento, O.; Inghirami, G. Novel Long Non Coding RNA Blackmamba Is Associated to ALK-anaplastic Large Cell Lymphoma. Blood 2016, 128, 461. [Google Scholar]
- Schleussner, N.; Merkel, O.; Costanza, M.; Liang, H.-C.; Hummel, F.; Romagnani, C.; Durek, P.; Anagnostopoulos, I.; Hummel, M.; Jöhrens, K.; et al. The AP-1-BATF and -BATF3 module is essential for growth, survival and TH17/ILC3 skewing of anaplastic large cell lymphoma. Leukemia 2018, 32, 1994–2007. [Google Scholar] [CrossRef]
- Schiefer, A.-I.; Vesely, P.; Hassler, M.R.; Egger, G.; Kenner, L. The role of AP-1 and epigenetics in ALCL. Front. Biosci. 2015, 7, 226–235. [Google Scholar]
- Chung, I.-H.; Lu, P.-H.; Lin, Y.-H.; Tsai, M.-M.; Lin, Y.-W.; Yeh, C.-T.; Lin, K.-H. The long non-coding RNA LINC01013 enhances invasion of human anaplastic large-cell lymphoma. Sci. Rep. 2017, 7, 295. [Google Scholar] [CrossRef] [PubMed]
- Prensner, J.R.; Chen, W.; Iyer, M.K.; Cao, Q.; Ma, T.; Han, S.; Sahu, A.; Malik, R.; Wilder-Romans, K.; Navone, N.; et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014, 74, 1651–1660. [Google Scholar] [CrossRef] [PubMed]
- Aguilo, F.; Di Cecilia, S.; Walsh, M.J. Long Non-coding RNA ANRIL and Polycomb in Human Cancers and Cardiovascular Disease. Curr. Top. Microbiol. Immunol. 2016, 394, 29–39. [Google Scholar] [PubMed]
- Fu, W.; Lu, Y.; Hu, B.; Liang, W.; Zhu, X.; Yang, H.; Li, G.; Zhang, J.; Fu, W.; Lu, Y.; et al. Long noncoding RNA Hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget 2016, 7, 4712–4723. [Google Scholar] [CrossRef] [PubMed]
- Glasmacher, E.; Agrawal, S.; Chang, A.B.; Murphy, T.L.; Zeng, W.; Vander Lugt, B.; Khan, A.A.; Ciofani, M.; Spooner, C.J.; Rutz, S.; et al. A Genomic Regulatory Element That Directs Assembly and Function of Immune-Specific AP-1-IRF Complexes. Science 2012, 338, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.D.; Yeung, D.; Hadfield, K.; Cook, S.J.; Alexander, D.R. The NPM-ALK tyrosine kinase mimics TCR signalling pathways, inducing NFAT and AP-1 by RAS-dependent mechanisms. Cell. Signal. 2007, 19, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Leventaki, V.; Drakos, E.; Medeiros, L.J.; Lim, M.S.; Elenitoba-Johnson, K.S.; Claret, F.X.; Rassidakis, G.Z. NPM-ALK oncogenic kinase promotes cell-cycle progression through activation of JNK/cJun signaling in anaplastic large-cell lymphoma. Blood 2007, 110, 1621–1630. [Google Scholar] [CrossRef]
- Qin, F.-X.; Shao, H.-Y.; Chen, X.-C.; Tan, S.; Zhang, H.-J.; Miao, Z.-Y.; Wang, L.; Chen, H.; Zhang, L. Knockdown of NPM1 by RNA interference inhibits cells proliferation and induces apoptosis in leukemic cell line. Int. J. Med. Sci. 2011, 8, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Maggi, L.B.; Brady, S.N.; Apicelli, A.J.; Dai, M.-S.; Lu, H.; Weber, J.D. Nucleophosmin is essential for ribosomal protein L5 nuclear export. Mol. Cell. Biol. 2006, 26, 3798–3809. [Google Scholar] [CrossRef] [PubMed]
- Savkur, R.; Olson, M.O.J. Preferential cleavage in pre-ribosomal RNA byprotein B23 endoribonuclease. Nucleic Acids Res. 1998, 26, 4508–4515. [Google Scholar] [CrossRef] [PubMed]
- Murano, K.; Okuwaki, M.; Hisaoka, M.; Nagata, K. Transcription Regulation of the rRNA Gene by a Multifunctional Nucleolar Protein, B23/Nucleophosmin, through Its Histone Chaperone Activity. Mol. Cell. Biol. 2008, 28, 3114–3126. [Google Scholar] [CrossRef] [PubMed]
- Okuwaki, M.; Matsumoto, K.; Tsujimoto, M.; Nagata, K. Function of nucleophosmin/B23, a nucleolar acidic protein, as a histone chaperone. FEBS Lett. 2001, 506, 272–276. [Google Scholar] [CrossRef]
- Box, J.K.; Paquet, N.; Adams, M.N.; Boucher, D.; Bolderson, E.; O’Byrne, K.J.; Richard, D.J. Nucleophosmin: From structure and function to disease development. BMC Mol. Biol. 2016, 17, 19. [Google Scholar] [CrossRef] [PubMed]
- Okuwaki, M. The Structure and Functions of NPM1/Nucleophsmin/B23, a Multifunctional Nucleolar Acidic Protein. J. Biochem. 2007, 143, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.; Matsui, T.; Karp, J.M.; Onikubo, T.; Cahill, S.; Brenowitz, M.; Cowburn, D.; Girvin, M.; Shechter, D. Dynamic intramolecular regulation of the histone chaperone nucleoplasmin controls histone binding and release. Nat. Commun. 2017, 8, 2215. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Budhu, A.; Forgues, M.; Wang, X.W. Temporal and spatial control of nucleophosmin by the Ran–Crm1 complex in centrosome duplication. Nat. Cell Biol. 2005, 7, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Szebeni, A.; Herrera, J.E.; Olson, M.O.J. Interaction of nucleolar protein B23 with peptides related to nuclear localization signals and its modulation by phosphorylation. Biochemistry 1995, 34, 8037–8042. [Google Scholar] [CrossRef]
- Szebeni, A.; Mehrotra, B.; Baumann, A.; Adam, S.A.; Wingfield, P.T.; Olson, M.O.J. Nucleolar Protein B23 Stimulates Nuclear Import of the HIV-1 Rev Protein and NLS-Conjugated Albumin. Biochemistry 1997, 36, 3941–3949. [Google Scholar] [CrossRef]
- Fankhauser, C.; Izaurralde, E.; Adachi, Y.; Wingfield, P.; Laemmli, U.K. Specific complex of human immunodeficiency virus type 1 rev and nucleolar B23 proteins: Dissociation by the Rev response element. Mol. Cell. Biol. 1991, 11, 2567–2575. [Google Scholar] [CrossRef]
- Li, Y.P. Protein B23 is an important human factor for the nucleolar localization of the human immunodeficiency virus protein Tat. J. Virol. 1997, 71, 4098–4102. [Google Scholar] [PubMed]
- Valdez, B.C.; Perlaky, L.; Henning, D.; Saijo, Y.; Chan, P.K.; Busch, H. Identification of the nuclear and nucleolar localization signals of the protein p120. Interaction with translocation protein B23. J. Biol. Chem. 1994, 269, 23776–23783. [Google Scholar] [PubMed]
- Swaminathan, V.; Kishore, A.H.; Febitha, K.K.; Kundu, T.K. Human Histone Chaperone Nucleophosmin Enhances Acetylation-Dependent Chromatin Transcription. Mol. Cell. Biol. 2005, 25, 7534–7545. [Google Scholar] [CrossRef] [PubMed]
- Gadad, S.S.; Senapati, P.; Syed, S.H.; Rajan, R.E.; Shandilya, J.; Swaminathan, V.; Chatterjee, S.; Colombo, E.; Dimitrov, S.; Pelicci, P.G.; et al. The Multifunctional Protein Nucleophosmin (NPM1) Is a Human Linker Histone H1 Chaperone. Biochemistry 2011, 50, 2780–2789. [Google Scholar] [CrossRef] [PubMed]
- Akey, C.W.; Luger, K. Histone chaperones and nucleosome assembly. Curr. Opin. Struct. Biol. 2003, 13, 6–14. [Google Scholar] [CrossRef]
- McBryant, S.J.; Park, Y.-J.; Abernathy, S.M.; Laybourn, P.J.; Nyborg, J.K.; Luger, K. Preferential binding of the histone (H3-H4)2 tetramer by NAP1 is mediated by the amino-terminal histone tails. J. Biol. Chem. 2003, 278, 44574–44583. [Google Scholar] [CrossRef]
- Lindström, M.S. NPM1/B23: A Multifunctional Chaperone in Ribosome Biogenesis and Chromatin Remodeling. Biochem. Res. Int. 2011, 2011, 195209. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Boone, D.; Hann, S.R. Nucleophosmin interacts directly with c-Myc and controls c-Myc-induced hyperproliferation and transformation. Proc. Natl. Acad. Sci. USA 2008, 105, 18794–18799. [Google Scholar] [CrossRef] [PubMed]
- Hingorani, K.; Szebeni, A.; Olson, M.O.J. Mapping the Functional Domains of Nucleolar Protein B23. J. Biol. Chem. 2000, 275, 24451–24457. [Google Scholar] [CrossRef]
- Choi, J.W.; Lee, S.B.; Kim, C.K.; Lee, K.-H.; Cho, S.-W.; Ahn, J.-Y. Lysine 263 residue of NPM/B23 is essential for regulating ATP binding and B23 stability. FEBS Lett. 2008, 582, 1073–1080. [Google Scholar] [CrossRef]
- McDuff, F.K.E.; Hook, C.E.; Tooze, R.M.; Huntly, B.J.; Pandolfi, P.P.; Turner, S.D. Determining the contribution of NPM1 heterozygosity to NPM-ALK-induced lymphomagenesis. Lab. Investig. 2011, 91, 1298–1303. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, H.H.; Kim, H.S.; Kang, J.Y.; Lee, B.I.; Ha, J.Y.; Yoon, H.J.; Lim, S.O.; Jung, G.; Suh, S.W. Crystal structure of human nucleophosmin-core reveals plasticity of the pentamer-pentamer interface. Proteins Struct. Funct. Bioinform. 2007, 69, 672–678. [Google Scholar] [CrossRef] [PubMed]
- Dhar, S.K.; St. Clair, D.K. Nucleophosmin Blocks Mitochondrial Localization of p53 and Apoptosis. J. Biol. Chem. 2009, 284, 16409–16418. [Google Scholar] [CrossRef] [PubMed]
- Okuda, M.; Horn, H.F.; Tarapore, P.; Tokuyama, Y.; Smulian, A.G.; Chan, P.K.; Knudsen, E.S.; Hofmann, I.A.; Snyder, J.D.; Bove, K.E.; et al. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell 2000, 103, 127–140. [Google Scholar] [CrossRef]
- Pervushin, K.; Riek, R.; Wider, G.; Wüthrich, K.; Pytel, N.J.; Satumba, J.; Nourse, A.; Park, C.-G.; Babu, M.M.; White, S.W.; et al. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA 1997, 94, 12366–12371. [Google Scholar] [CrossRef]
- Hwang, S.R.; Murga-Zamalloa, C.; Brown, N.; Basappa, J.; McDonnell, S.R.; Mendoza-Reinoso, V.; Basrur, V.; Wilcox, R.; Elenitoba-Johnson, K.; Lim, M.S. Pyrimidine tract-binding protein 1 mediates pyruvate kinase M2-dependent phosphorylation of signal transducer and activator of transcription 3 and oncogenesis in anaplastic large cell lymphoma. Lab. Investig. 2017, 97, 962. [Google Scholar] [CrossRef][Green Version]
- Cheng, K.; Sportoletti, P.; Ito, K.; Clohessy, J.G.; Teruya-Feldstein, J.; Kutok, J.L.; Pandolfi, P.P. The cytoplasmic NPM mutant induces myeloproliferation in a transgenic mouse model. Blood 2010, 115, 3341–3345. [Google Scholar] [CrossRef]
- Hallberg, B.; Palmer, R.H. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat. Rev. Cancer 2013, 13, 685–700. [Google Scholar] [CrossRef]
- Koivunen, J.P.; Mermel, C.; Zejnullahu, K.; Murphy, C.; Lifshits, E.; Holmes, A.J.; Choi, H.G.; Kim, J.; Chiang, D.; Thomas, R.; et al. EML4-ALK Fusion Gene and Efficacy of an ALK Kinase Inhibitor in Lung Cancer. Clin. Cancer Res. 2008, 14, 4275–4283. [Google Scholar] [CrossRef]
- Wiesner, T.; Lee, W.; Obenauf, A.C.; Ran, L.; Murali, R.; Zhang, Q.F.; Wong, E.W.P.; Hu, W.; Scott, S.N.; Shah, R.H.; et al. Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature 2015, 526, 453–457. [Google Scholar] [CrossRef]
- Takakura, Y.; Yamaguchi, N.; Honda, T.; Morii, M.; Yuki, R.; Nakayama, Y.; Yamaguchi, N. The Truncated Isoform of the Receptor Tyrosine Kinase ALK Generated by Alternative Transcription Initiation (ALKATI) Induces Chromatin Structural Changes in the Nucleus in a Kinase Activity-Dependent Manner. Biol. Pharm. Bull. 2017, 40, 1968–1975. [Google Scholar] [CrossRef] [PubMed]
- Beylot-Barry, M.; Groppi, A.; Vergier, B.; Pulford, K.; Merlio, J.P.; French Study Group of Cutaneous Lymphoma. The Characterization of t(2;5) Reciprocal Transcripts and Genomic Breakpoints in CD30+ Cutaneous Lymphoproliferations. Blood 1998, 91, 4668–4676. [Google Scholar] [PubMed]
- Penzel, R.; Schirmacher, P.; Warth, A. A Novel EML4-ALK Variant: Exon 6 of EML4 Fused to Exon 19 of ALK. J. Thorac. Oncol. 2012, 7, 1198–1199. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.; Li, L.; Guan, Y.; Soriano, R.; Rivers, C.S.; Mohan, S.; Pandita, A.; Tang, J.; Modrusan, Z. Exon Array Profiling Detects EML4-ALK Fusion in Breast, Colorectal, and Non-Small Cell Lung Cancers. Mol. Cancer Res. 2009, 7, 1466–1476. [Google Scholar] [CrossRef] [PubMed]
- Nogales, E. Structural Insights into Microtubule Function. Annu. Rev. Biochem. 2000, 69, 277–302. [Google Scholar] [CrossRef] [PubMed]
- Hirokawa, N. Microtubule organization and dynamics dependent on microtubule-associated proteins. Curr. Opin. Cell Biol. 1994, 6, 74–81. [Google Scholar] [CrossRef]
- Ahmad, F.J.; Yu, W.; McNally, F.J.; Baas, P.W. An essential role for katanin in severing microtubules in the neuron. J. Cell Biol. 1999, 145, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Cassimeris, L. The oncoprotein 18/stathmin family of microtubule destabilizers. Curr. Opin. Cell Biol. 2002, 14, 18–24. [Google Scholar] [CrossRef]
- Suprenant, K.A.; Dean, K.; McKee, J.; Hake, S. EMAP, an echinoderm microtubule-associated protein found in microtubule-ribosome complexes. J. Cell Sci. 1993, 104, 445–450. [Google Scholar]
- Suprenant, K.A.; Tuxhorn, J.A.; Daggett, M.A.; Ahrens, D.P.; Hostetler, A.; Palange, J.M.; VanWinkle, C.E.; Livingston, B.T. Conservation of the WD-repeat, microtubule-binding protein, EMAP, in sea urchins, humans, and the nematode C. elegans. Dev. Genes Evol. 2000, 210, 2–10. [Google Scholar] [CrossRef]
- Eichenmuller, B.; Everley, P.; Palange, J.; Lepley, D.; Suprenant, K.A. The human EMAP-like protein-70 (ELP70) is a microtubule destabilizer that localizes to the mitotic apparatus. J. Biol. Chem. 2002, 277, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Richards, M.W.; Law, E.W.P.; Rennalls, L.P.; Busacca, S.; O’Regan, L.; Fry, A.M.; Fennell, D.A.; Bayliss, R. Crystal structure of EML1 reveals the basis for Hsp90 dependence of oncogenic EML4-ALK by disruption of an atypical propeller domain. Proc. Natl. Acad. Sci. USA 2014, 111, 5195–5200. [Google Scholar] [CrossRef] [PubMed]
- Richards, M.W.; O’Regan, L.; Roth, D.; Montgomery, J.M.; Straube, A.; Fry, A.M.; Bayliss, R. Microtubule association of EML proteins and the EML4-ALK variant 3 oncoprotein require an N-terminal trimerization domain. Biochem. J. 2015, 467, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Houtman, S.H.; Rutteman, M.; De Zeeuw, C.I.; French, P.J. Echinoderm microtubule-associated protein like protein 4, a member of the echinoderm microtubule-associated protein family, stabilizes microtubules. Neuroscience 2007, 144, 1373–1382. [Google Scholar] [CrossRef] [PubMed]
- Pollmann, M.; Parwaresch, R.; Adam-Klages, S.; Kruse, M.-L.; Buck, F.; Heidebrecht, H.-J. Human EML4, a novel member of the EMAP family, is essential for microtubule formation. Exp. Cell Res. 2006, 312, 3241–3251. [Google Scholar] [CrossRef] [PubMed]
- Radtke, J.; Rezaie, S.G.; Kugler, C.; Zabel, P.; Schultz, H.; Vollmer, E.; Goldmann, T.; Lang, D.S. Expression analysis of EML4 in normal lung tissue and non-small cell lung cancer (NSCLC) in the absence and presence of chemotherapeutics. Rom. J. Morphol. Embryol. 2010, 51, 647–653. [Google Scholar] [PubMed]
- Soda, M.; Takada, S.; Takeuchi, K.; Choi, Y.L.; Enomoto, M.; Ueno, T.; Haruta, H.; Hamada, T.; Yamashita, Y.; Ishikawa, Y.; et al. A mouse model for EML4-ALK-positive lung cancer. Proc. Natl. Acad. Sci. USA 2008, 105, 19893–19897. [Google Scholar] [CrossRef] [PubMed]
- Sanders, H.R.; Li, H.-R.; Bruey, J.-M.; Scheerle, J.A.; Meloni-Ehrig, A.M.; Kelly, J.C.; Novick, C.; Albitar, M. Exon scanning by reverse transcriptase–polymerase chain reaction for detection of known and novel EML4–ALK fusion variants in non–small cell lung cancer. Cancer Genet. 2011, 204, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.L.; Takeuchi, K.; Soda, M.; Inamura, K.; Togashi, Y.; Hatano, S.; Enomoto, M.; Hamada, T.; Haruta, H.; Watanabe, H.; et al. Identification of Novel Isoforms of the EML4-ALK Transforming Gene in Non-Small Cell Lung Cancer. Cancer Res. 2008, 68, 4971–4976. [Google Scholar] [CrossRef]
- Soda, M.; Isobe, K.; Inoue, A.; Maemondo, M.; Oizumi, S.; Fujita, Y.; Gemma, A.; Yamashita, Y.; Ueno, T.; Takeuchi, K.; et al. A prospective PCR-based screening for the EML4-ALK oncogene in non-small cell lung cancer. Clin. Cancer Res. 2012, 18, 5682–5689. [Google Scholar] [CrossRef]
- Takeuchi, K.; Choi, Y.L.; Soda, M.; Inamura, K.; Togashi, Y.; Hatano, S.; Enomoto, M.; Takada, S.; Yamashita, Y.; Satoh, Y.; et al. Multiplex Reverse Transcription-PCR Screening for EML4-ALK Fusion Transcripts. Clin. Cancer Res. 2008, 14, 6618–6624. [Google Scholar] [CrossRef] [PubMed]
- Esfahani, K.; Cohen, V. HSP90 as a novel molecular target in non-small-cell lung cancer. Lung Cancer 2016, 7, 11–17. [Google Scholar] [PubMed]
- Lin, J.J.; Zhu, V.W.; Yoda, S.; Yeap, B.Y.; Schrock, A.B.; Dagogo-Jack, I.; Jessop, N.A.; Jiang, G.Y.; Le, L.P.; Gowen, K.; et al. Impact of EML4-ALK Variant on Resistance Mechanisms and Clinical Outcomes in ALK-Positive Lung Cancer. J. Clin. Oncol. 2018, 36, 1199. [Google Scholar] [CrossRef] [PubMed]
- Woo, C.G.; Seo, S.; Kim, S.W.; Jang, S.J.; Park, K.S.; Song, J.Y.; Lee, B.; Richards, M.W.; Bayliss, R.; Lee, D.H.; et al. Differential protein stability and clinical responses of EML4-ALK fusion variants to various ALK inhibitors in advanced ALK rearranged non-small cell lung cancer. Ann. Oncol. 2016, 28, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Heuckmann, J.M.; Balke-Want, H.; Malchers, F.; Peifer, M.; Sos, M.L.; Koker, M.; Meder, L.; Lovly, C.M.; Heukamp, L.C.; Pao, W.; et al. Differential Protein Stability and ALK Inhibitor Sensitivity of EML4-ALK Fusion Variants. Clin. Cancer Res. 2012, 18, 4682–4690. [Google Scholar] [CrossRef] [PubMed]
- Gasparini, P.; Cascione, L.; Landi, L.; Carasi, S.; Lovat, F.; Tibaldi, C.; Alì, G.; D’Incecco, A.; Minuti, G.; Chella, A.; et al. microRNA classifiers are powerful diagnostic/prognostic tools in ALK-, EGFR-, and KRAS -driven lung cancers. Proc. Natl. Acad. Sci. USA 2015, 112, 14924–14929. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Deng, X.; Yoshioka, Y.; Vougiouklakis, T.; Park, J.-H.; Suzuki, T.; Dohmae, N.; Ueda, K.; Hamamoto, R.; Nakamura, Y. Effects of SMYD2-mediated EML4-ALK methylation on the signaling pathway and growth in non-small-cell lung cancer cells. Cancer Sci. 2017, 108, 1203–1209. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Scarborough, H.; Kim, J.; Rozhok, A.I.; Chen, Y.A.; Zhang, X.; Song, L.; Bai, Y.; Fang, B.; Liu, R.Z.; et al. Coupling an EML4-ALK-centric interactome with RNA interference identifies sensitizers to ALK inhibitors. Sci. Signal. 2016, 9, rs12. [Google Scholar] [CrossRef]
- Hong, S.; Chen, N.; Fang, W.; Zhan, J.; Liu, Q.; Kang, S.; He, X.; Liu, L.; Zhou, T.; Huang, J.; et al. Upregulation of PD-L1 by EML4-ALK fusion protein mediates the immune escape in ALK positive NSCLC: Implication for optional anti-PD-1/PD-L1 immune therapy for ALK-TKIs sensitive and resistant NSCLC patients. Oncoimmunology 2016, 5, e1094598. [Google Scholar] [CrossRef] [PubMed]
- Brodeur, G.M. Neuroblastoma: Biological insights into a clinical enigma. Nat. Rev. Cancer 2003, 3, 203–216. [Google Scholar] [CrossRef]
- Lee, C.C.; Jia, Y.; Li, N.; Sun, X.; Ng, K.; Ambing, E.; Gao, M.-Y.; Hua, S.; Chen, C.; Kim, S.; et al. Crystal structure of the ALK (anaplastic lymphoma kinase) catalytic domain. Biochem. J. 2010, 430, 425–437. [Google Scholar] [CrossRef] [PubMed]
- Bresler, S.C.; Weiser, D.A.; Huwe, P.J.; Park, J.H.; Krytska, K.; Ryles, H.; Laudenslager, M.; Rappaport, E.F.; Wood, A.C.; McGrady, P.W.; et al. ALK Mutations Confer Differential Oncogenic Activation and Sensitivity to ALK Inhibition Therapy in Neuroblastoma. Cancer Cell 2014, 26, 682–694. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Wolfstetter, G.; Siaw, J.; Chand, D.; Hugosson, F.; Palmer, R.H.; Hallberg, B.; Guan, J.; Wolfstetter, G.; Siaw, J.; et al. Anaplastic lymphoma kinase L1198F and G1201E mutations identified in anaplastic thyroid cancer patients are not ligand-independent. Oncotarget 2017, 8, 11566–11578. [Google Scholar] [CrossRef] [PubMed]
- Takita, J. The role of anaplastic lymphoma kinase in pediatric cancers. Cancer Sci. 2017, 108, 1913–1920. [Google Scholar] [CrossRef] [PubMed]
- Osajima-Hakomori, Y.; Miyake, I.; Ohira, M.; Nakagawara, A.; Nakagawa, A.; Sakai, R. Biological Role of Anaplastic Lymphoma Kinase in Neuroblastoma. Am. J. Pathol. 2005, 167, 213–222. [Google Scholar] [CrossRef]
- Katayama, R.; Shaw, A.T.; Khan, T.M.; Mino-Kenudson, M.; Solomon, B.J.; Halmos, B.; Jessop, N.A.; Wain, J.C.; Yeo, A.T.; Benes, C.; et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci. Transl. Med. 2012, 4, 120ra17. [Google Scholar] [CrossRef] [PubMed]
- Doebele, R.C.; Pilling, A.B.; Aisner, D.L.; Kutateladze, T.G.; Le, A.T.; Weickhardt, A.J.; Kondo, K.L.; Linderman, D.J.; Heasley, L.E.; Franklin, W.A.; et al. Mechanisms of Resistance to Crizotinib in Patients with ALK Gene Rearranged Non-Small Cell Lung Cancer. Clin. Cancer Res. 2012, 18, 1472–1482. [Google Scholar] [CrossRef]
- Prokoph, N.; Larose, H.; Lim, M.; Burke, G.; Turner, S.; Prokoph, N.; Larose, H.; Lim, M.S.; Burke, G.A.A.; Turner, S.D. Treatment Options for Paediatric Anaplastic Large Cell Lymphoma (ALCL): Current Standard and beyond. Cancers 2018, 10, 99. [Google Scholar] [CrossRef]
- Arbour, K.C.; Riely, G.J. Diagnosis and Treatment of Anaplastic Lymphoma Kinase-Positive Non-Small Cell Lung Cancer. Hematol. Oncol. Clin. N. Am. 2017, 31, 101–111. [Google Scholar] [CrossRef]
- Lantuejoul, S.; Rouquette, I.; Blons, H.; Le Stang, N.; Ilie, M.; Begueret, H.; Grégoire, V.; Hofman, P.; Gros, A.; Garcia, S.; et al. French multicentric validation of ALK rearrangement diagnostic in 547 lung adenocarcinomas. Eur. Respir. J. 2015, 46, 207–218. [Google Scholar] [CrossRef]
- Hout, D.; Schweitzer, B.; Lawrence, K.; Morris, S.; Tucker, T.; Mazzola, R.; Skelton, R.; McMahon, F.; Handshoe, J.; Lesperance, M.; et al. Performance of a RT-PCR Assay in Comparison to FISH and Immunohistochemistry for the Detection of ALK in Non-Small Cell Lung Cancer. Cancers 2017, 9, 99. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Wang, L.; Arcila, M.E.; Balasubramanian, S.; Greenbowe, J.R.; Ross, J.S.; Stephens, P.; Lipson, D.; Miller, V.A.; Kris, M.G.; et al. Broad, Hybrid Capture-Based Next-Generation Sequencing Identifies Actionable Genomic Alterations in Lung Adenocarcinomas Otherwise Negative for Such Alterations by Other Genomic Testing Approaches. Clin. Cancer Res. 2015, 21, 3631–3639. [Google Scholar] [CrossRef] [PubMed]
- Arcila, M.E.; Yu, H.A.; Drilon, A.E.; Riely, G.J.; Zehir, A.; Sadowska, J.; Hyman, D.M.; Kris, M.G.; Berger, M.F.; Ladanyi, M. Comprehensive assessment of targetable alterations in lung adenocarcinoma samples with limited material using MSK-IMPACT, a clinical, hybrid capture-based, next-generation sequencing (NGS) assay. J. Clin. Oncol. 2015, 33, e22160. [Google Scholar] [CrossRef]
- Peled, N.; Palmer, G.; Hirsch, F.R.; Wynes, M.W.; Ilouze, M.; Varella-Garcia, M.; Soussan-Gutman, L.; Otto, G.A.; Stephens, P.J.; Ross, J.S.; et al. Next-generation sequencing identifies and immunohistochemistry confirms a novel crizotinib-sensitive ALK rearrangement in a patient with metastatic non-small-cell lung cancer. J. Thorac. Oncol. 2012, 7, e14–e16. [Google Scholar] [CrossRef] [PubMed]
- Mino-Kenudson, M.; Chirieac, L.R.; Law, K.; Hornick, J.L.; Lindeman, N.; Mark, E.J.; Cohen, D.W.; Johnson, B.E.; Janne, P.A.; Iafrate, A.J.; et al. A Novel, Highly Sensitive Antibody Allows for the Routine Detection of ALK-Rearranged Lung Adenocarcinomas by Standard Immunohistochemistry. Clin. Cancer Res. 2010, 16, 1561–1571. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, A.; Di Lorito, A.; Pace, M.V.; Iezzi, M.; Felicioni, L.; D’Antuono, T.; Filice, G.; Guetti, L.; Mucilli, F.; Buttitta, F. ALK Protein Analysis by IHC Staining after Recent Regulatory Changes: A Comparison of Two Widely Used Approaches, Revision of the Literature, and a New Testing Algorithm. J. Thorac. Oncol. 2016, 11, 487–495. [Google Scholar] [CrossRef] [PubMed]

| ALK Fusion Proteins | Gene Name | Translocation | Localisation | Cancer Type | References |
|---|---|---|---|---|---|
| NPM1-ALK | Nucleophosmin 1 | t(2;5)(p23;q35) | Cytoplasm, Nucleus, Nucleolus | ALCL | [3] |
| EML4-ALK | Echinoderm Microtubule Associated Protein-Like 4) | inv(2)(p21p23)) | cytoplasm | NSCLC | [4] |
| ATIC-ALK | 5-Aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase | inv(2)(p23q35) | cytoplasm | ALCL | [5] |
| CLTC-ALK | Clathrin heavy chain | t(2;17)(p23;q23) | cytoplasm | ALCL | [6] |
| TPM3-ALK | Tropomyosin 3 | t(1;2)(q25;p23) | cytoplasm | ALCL | [7] |
| TPM4-ALK | Tropomyosin 4 | t(2;19)(p23;p13) | cytoplasm | ALCL | [8] |
| TFG-ALK | TRK-fused gene | t(2;3)(p23;q21) | cytoplasm | ALCL | [7] |
| TRAF1-ALK | TNF receptor associated factor 1 | t(2;9)(p23;q33) | cytoplasm | ALCL | [9] |
| RNF213-ALK | Ring finger protein 213 | t(2;17)(p23;q25) | cytoplasm | ALCL | [6] |
| MYH9-ALK | Myosin heavy chain 9 | t(2;22)(p23;q11) | cytoplasm | ALCL | [10] |
| MSN-ALK | Moesin | t(X;22)(q11;p23) | cytoplasm | ALCL | [7] |
| EEF1G-ALK | Eukaryotic translation elongation factor 1 gamma | t(2;11)(p23; q12.3) | cytoplasm | ALCL | [11] |
| KIF5B-ALK | Kinesin family member 5B | t(2;10)(p23;p11) | cytoplasm | NSCLC | [12] |
| KLC1-ALK | Kinesin light chain 1 | t(2;14)(p23;q32) | cytoplasm | NSCLC | [13] |
| STRN-ALK | Striatin | del(2)(p22p23) | cytoplasm | NSCLC | [14] |
| PTPN3-ALK | protein tyrosine phosphatase, non-receptor type 3 | t(2;9)(p23;q31) | cytoplasm | NSCLC | [15] |
| DCTN1-ALK | Dynactin subunit 1 | t(2;2)(p13;p23) | cytoplasm | NSCLC | [16] |
| GCC2-ALK | GRIP and coiled-coil domain-containing protein 2 | t(2;2)(p23;q12) | cytoplasm | NSCLC | [17] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ducray, S.P.; Natarajan, K.; Garland, G.D.; Turner, S.D.; Egger, G. The Transcriptional Roles of ALK Fusion Proteins in Tumorigenesis. Cancers 2019, 11, 1074. https://doi.org/10.3390/cancers11081074
Ducray SP, Natarajan K, Garland GD, Turner SD, Egger G. The Transcriptional Roles of ALK Fusion Proteins in Tumorigenesis. Cancers. 2019; 11(8):1074. https://doi.org/10.3390/cancers11081074
Chicago/Turabian StyleDucray, Stephen P., Karthikraj Natarajan, Gavin D. Garland, Suzanne D. Turner, and Gerda Egger. 2019. "The Transcriptional Roles of ALK Fusion Proteins in Tumorigenesis" Cancers 11, no. 8: 1074. https://doi.org/10.3390/cancers11081074
APA StyleDucray, S. P., Natarajan, K., Garland, G. D., Turner, S. D., & Egger, G. (2019). The Transcriptional Roles of ALK Fusion Proteins in Tumorigenesis. Cancers, 11(8), 1074. https://doi.org/10.3390/cancers11081074







