Integrative Omic Profiling Reveals Unique Hypoxia Induced Signatures in Gastric Cancer Associated Myofibroblasts
Abstract
:1. Introduction
2. Results
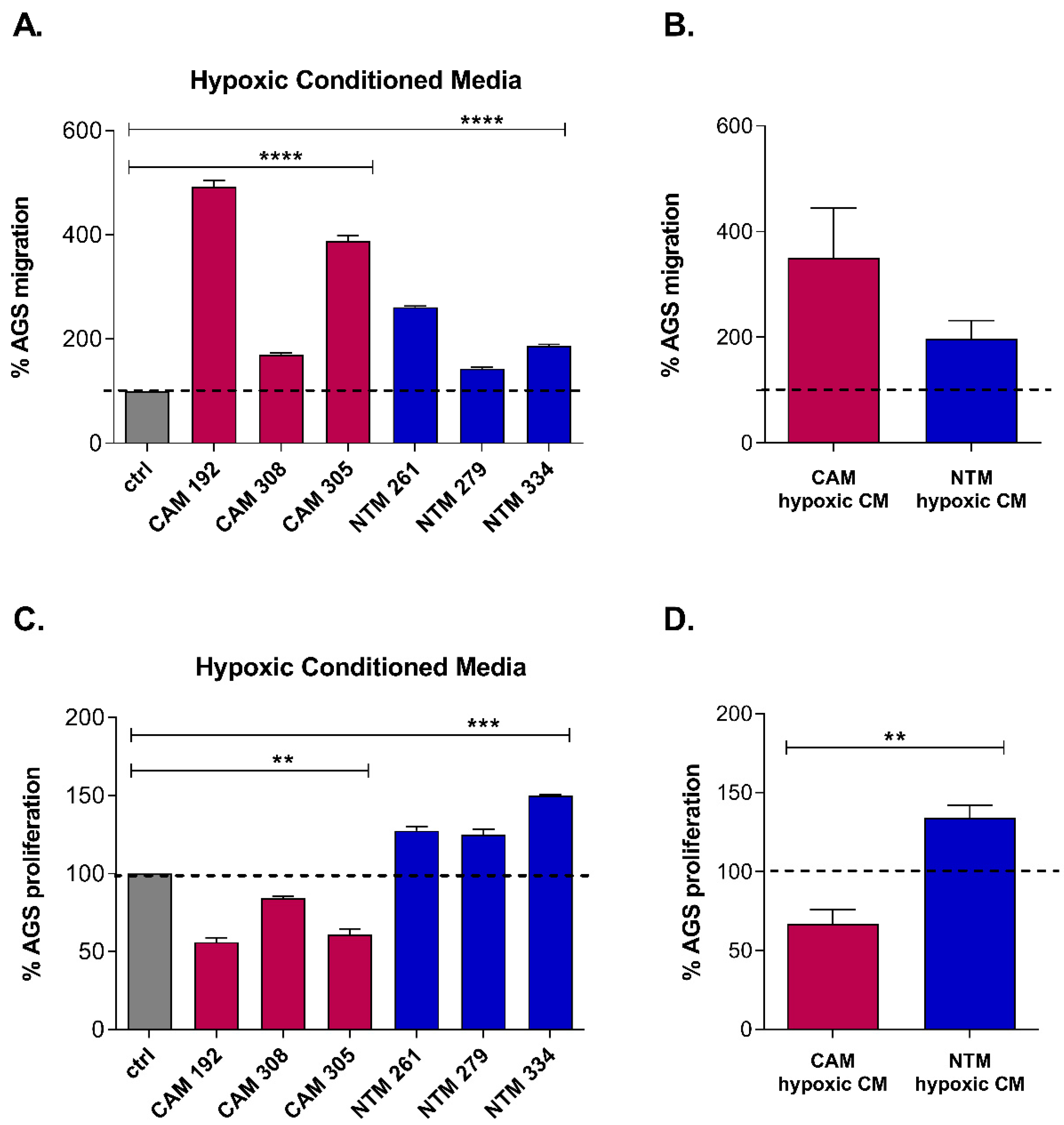
2.1. Hypoxia Enhances CAM-Induced Cancer Cell Migration and Promotes CAM-Like Properties in NTMs
2.2. Hypoxia Confers Differential Changes in Gene Expression in Gastric Myofibroblasts Purified from Different Tissue Microenvironments
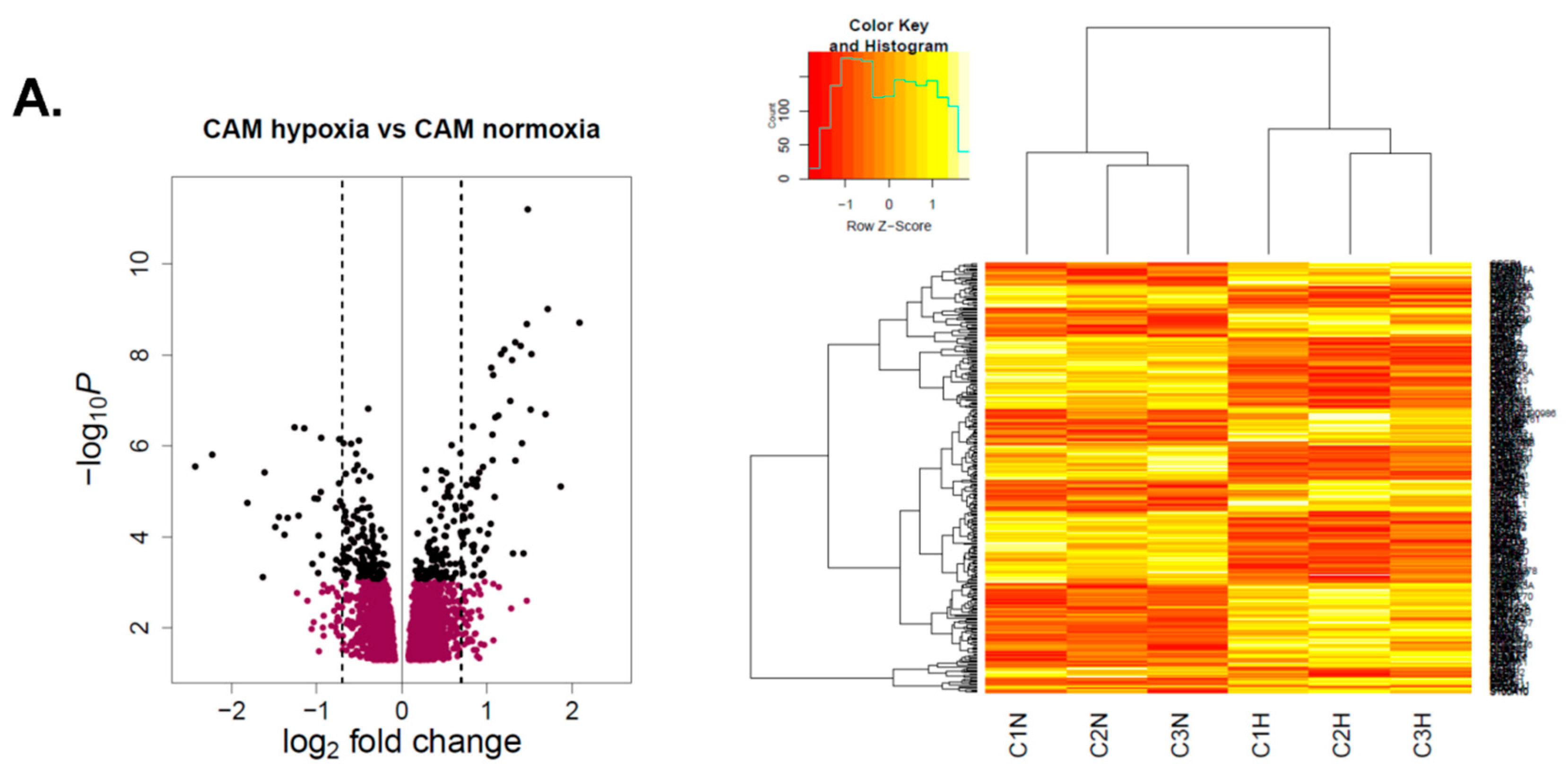
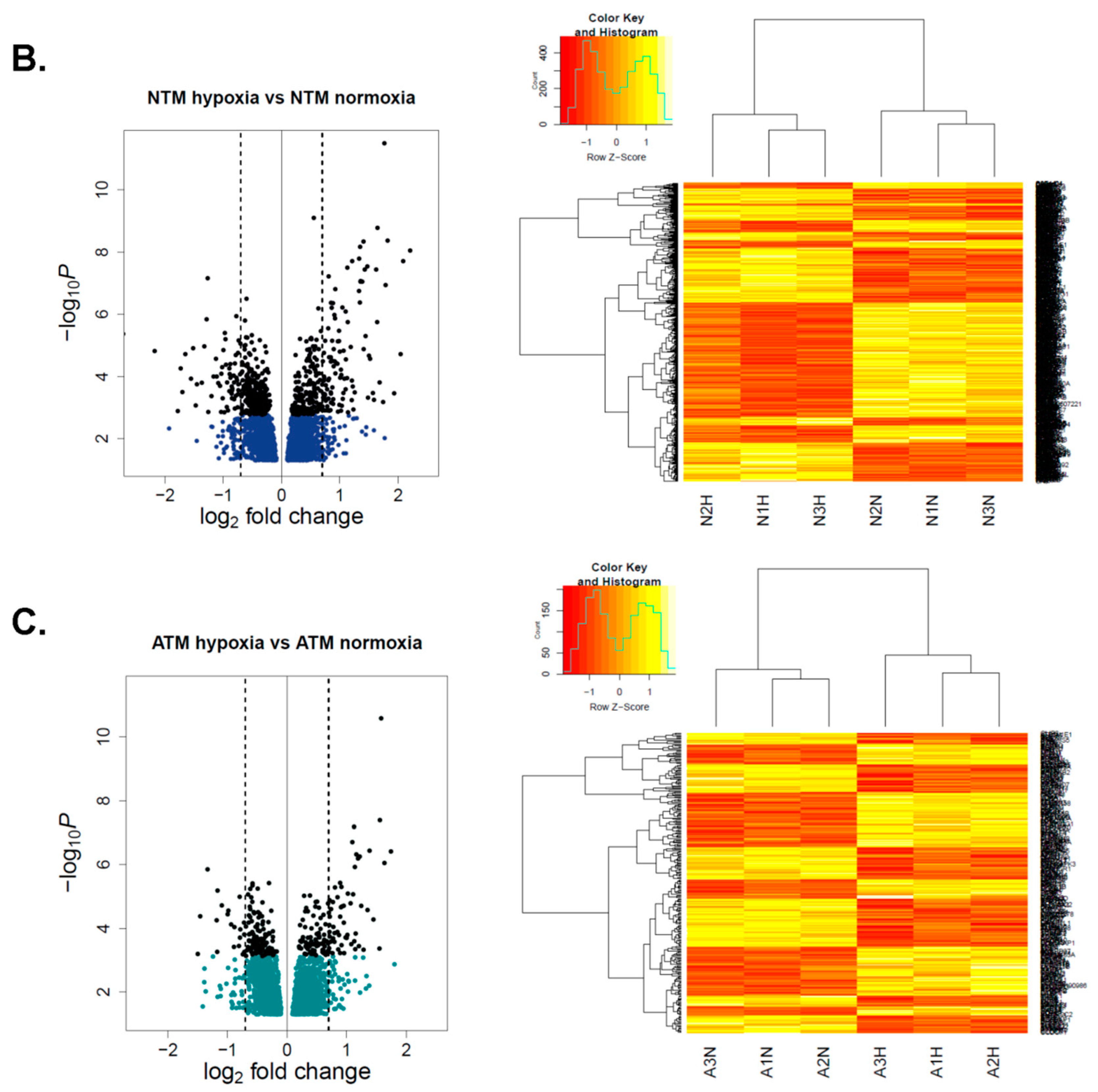
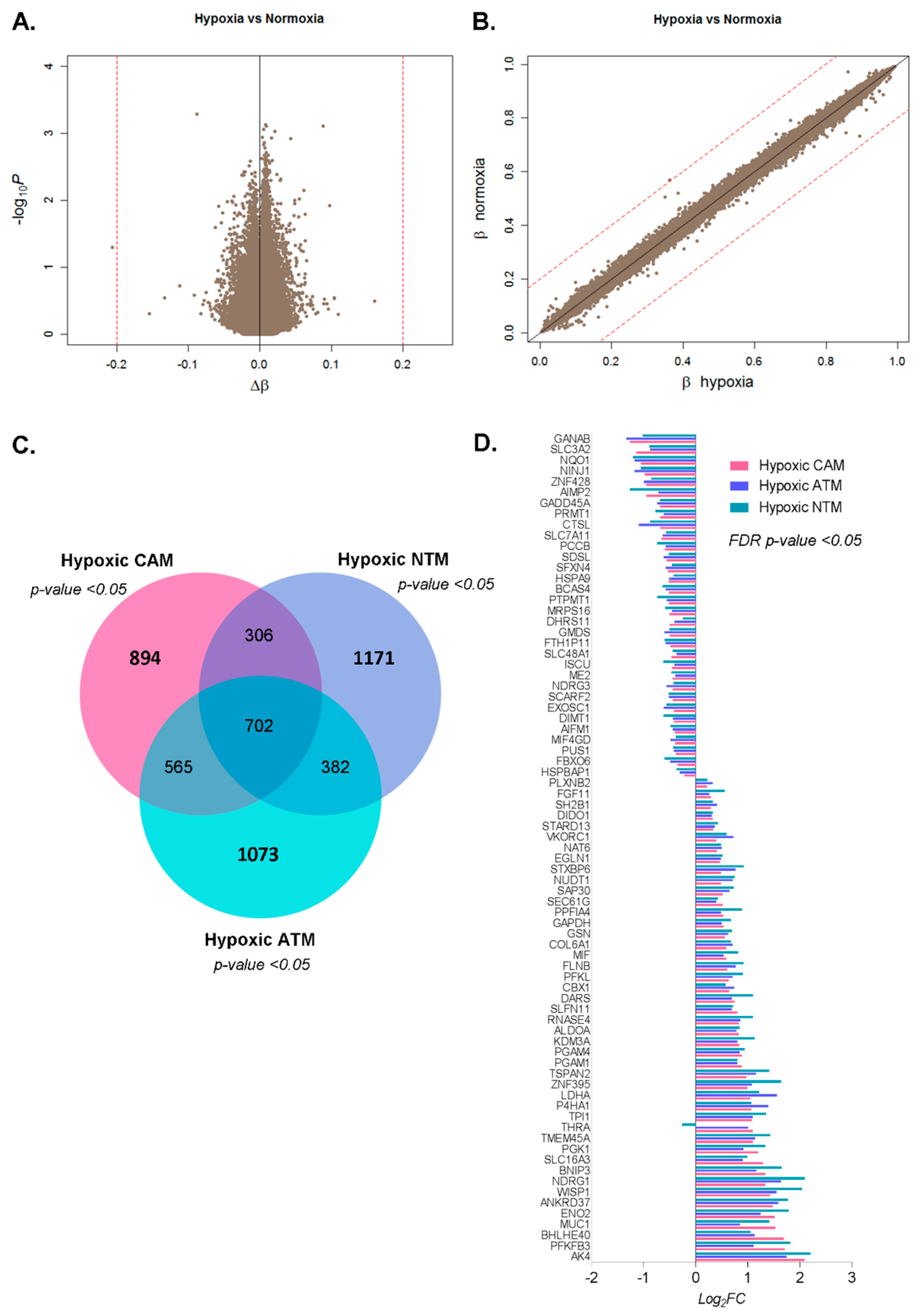
2.2.1. Differential Hypoxia-Induced Myofibroblast Phenotypes Are Not Due to Changes in DNA Methylation
2.2.2. Hypoxia-Induced Gene Expression Signatures
2.2.3. CAMs, ATMs, and NTMs Express Unique Gene Signatures Under Hypoxia
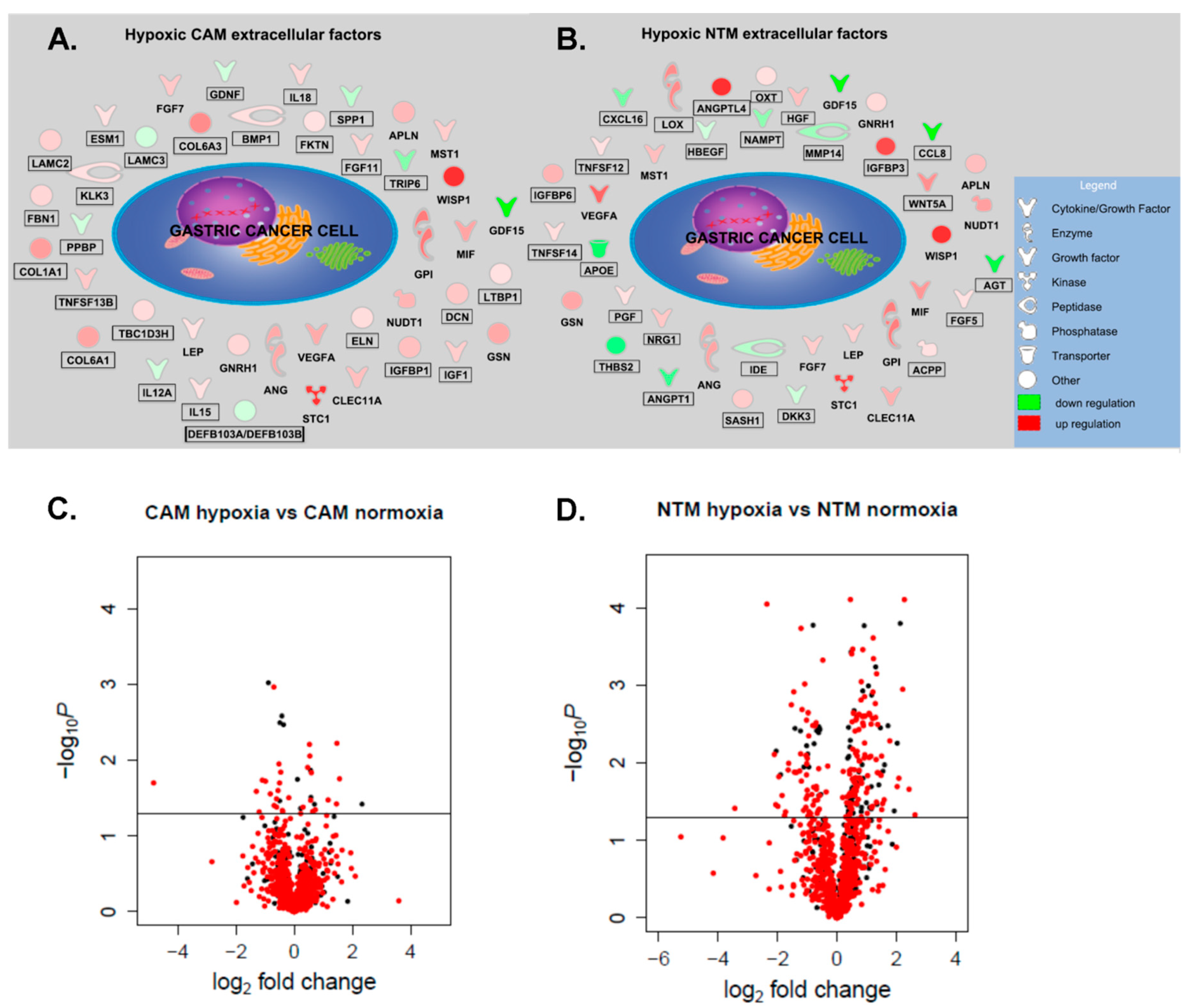
2.3. Analysis of Hypoxia-Induced CAM and NTM Secretomes
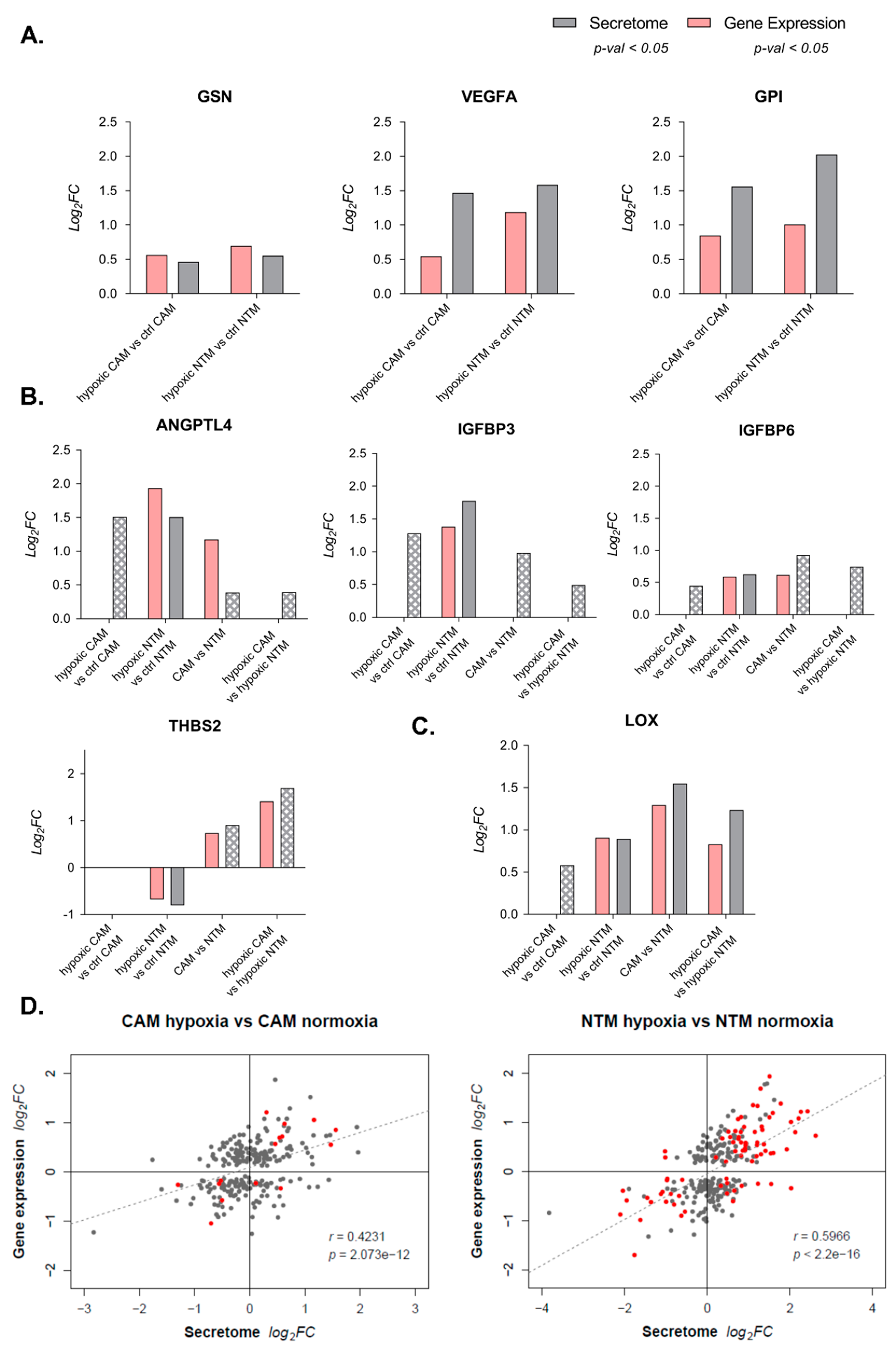
2.4. Gene Expression Profiles Enable Prediction of Secreted Factors
2.5. Secretion Is Not Strongly Regulated by Transcriptome Changes
3. Discussion
4. Materials and Methods
4.1. Primary Gastric Myofibroblasts
4.2. Gastric Cancer Cell Culture
4.3. Hypoxic Conditioned Media Preparation
4.4. Cancer Cell Migration Assay
4.5. Cancer Cell Proliferation Assay
4.6. Integrated Multi-Omics Myofibroblast Profiling
4.6.1. Illumina Human HT-12 v4 Array
4.6.2. Illumina Infinium HumanMethylation450k Array
4.6.3. Secretome Profiling
4.7. Gene Ontology Enrichment Analysis
4.8. Gene Set Enrichment Analysis
4.9. Ingenuity Pathway Analysis
4.10. R/Bioconductor
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Bhowmick, N.A.; Neilson, E.G.; Moses, H.L. Stromal fibroblasts in cancer initiation and progression. Nature 2004, 432, 332–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, M.; Louise Jones, J. Jekyll and Hyde: The role of the microenvironment on the progression of cancer. J. Pathol. 2011, 223, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Balabanova, S.; Holmberg, C.; Steele, I.; Ebrahimi, B.; Rainbow, L.; Burdyga, T.; McCaig, C.; Tiszlavicz, L.; Lertkowit, N.; Giger, O.T.; et al. The neuroendocrine phenotype of gastric myofibroblasts and its loss with cancer progression. Carcinogenesis 2014, 35, 1798–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmberg, C.; Quante, M.; Steele, I.; Kumar, J.D.; Balabanova, S.; Duval, C.; Czepan, M.; Rakonczay, Z., Jr.; Tiszlavicz, L.; Nemeth, I.; et al. Release of TGFbetaig-h3 by gastric myofibroblasts slows tumor growth and is decreased with cancer progression. Carcinogenesis 2012, 33, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Najgebauer, H.; Liloglou, T.; Jithesh, P.V.; Giger, O.T.; Varro, A.; Sanderson, C.M. Integrated omics profiling reveals novel patterns of epigenetic programming in cancer-associated myofibroblasts. Carcinogenesis 2019. [Google Scholar] [CrossRef] [PubMed]
- He, X.J.; Tao, H.Q.; Hu, Z.M.; Ma, Y.Y.; Xu, J.; Wang, H.J.; Xia, Y.J.; Li, L.; Fei, B.Y.; Li, Y.Q.; et al. Expression of galectin-1 in carcinoma-associated fibroblasts promotes gastric cancer cell invasion through upregulation of integrin β1. Cancer Sci. 2014. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Gao, J.; Wang, S.; Ye, N.; Chong, Y.; Huang, Y.; Wang, J.; Li, B.; Yin, W.; Wang, D. Cancer-associated fibroblasts promote angiogenesis in gastric cancer through galectin-1 expression. Tumour Biol. 2015, 37, 1889–1899. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Fukui, H.; Hara, K.; Zhang, X.; Kitayama, Y.; Eda, H.; Tomita, T.; Oshima, T.; Kikuchi, S.; Watari, J.; et al. FGF9 from cancer-associated fibroblasts is a possible mediator of invasion and anti-apoptosis of gastric cancer cells. BMC Cancer 2015, 15, 333. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.S.; Yang, X.H.; Chen, X.; Wang, X.D.; Hua, J.; Zhou, D.L.; Zhou, B.; Song, Z.S. MicroRNA-106b in cancer-associated fibroblasts from gastric cancer promotes cell migration and invasion by targeting PTEN. FEBS Lett. 2014, 588, 2162–2169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhi, K.; Shen, X.; Zhang, H.; Bi, J. Cancer-associated fibroblasts are positively correlated with metastatic potential of human gastric cancers. J. Exp. Clin. Cancer Res. 2010, 29, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasegawa, T.; Yashiro, M.; Nishii, T.; Matsuoka, J.; Fuyuhiro, Y.; Morisaki, T.; Fukuoka, T.; Shimizu, K.; Shimizu, T.; Miwa, A.; et al. Cancer-associated fibroblasts might sustain the stemness of scirrhous gastric cancer cells via transforming growth factor-β signaling. Int. J. Cancer 2014, 134, 1785–1795. [Google Scholar] [CrossRef] [PubMed]
- Satoyoshi, R.; Kuriyama, S.; Aiba, N.; Yashiro, M.; Tanaka, M. Asporin activates coordinated invasion of scirrhous gastric cancer and cancer-associated fibroblasts. Oncogene 2014, 34, 650. [Google Scholar] [CrossRef] [PubMed]
- Gilkes, D.M.; Semenza, G.L.; Wirtz, D. Hypoxia and the extracellular matrix: Drivers of tumour metastasis. Nat. Rev. Cancer 2014, 14, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Dayan, F.; Mazure, N.M.; Brahimi-Horn, M.C.; Pouysségur, J. A dialogue between the hypoxia-inducible factor and the tumor microenvironment. Cancer Microenviron. 2008, 1, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Giaccia, A.J.; Schipani, E. Role of carcinoma-associated fibroblasts and hypoxia in tumor progression. In Diverse Effects of Hypoxia on Tumor Progression; Simon, M.C., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 31–45. [Google Scholar]
- Comito, G.; Giannoni, E.; Di Gennaro, P.; Segura, C.P.; Gerlini, G.; Chiarugi, P. Stromal fibroblasts synergize with hypoxic oxidative stress to enhance melanoma aggressiveness. Cancer Lett. 2012, 324, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Hypoxia-inducible factor 1: Master regulator of O-2 homeostasis. Curr. Opin. Genet. Dev. 1998, 8, 588–594. [Google Scholar] [CrossRef]
- Wang, G.L.; Jiang, B.H.; Rue, E.A.; Semenza, G.L. Hypoxia-inducible factor-1 is a basic-helix-loop-helix-pas heterodimer regulated by cellular o-2 tension. Proc. Natl. Acad. Sci. USA 1995, 92, 5510–5514. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Thomas, P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013, 41, D377–D386. [Google Scholar] [CrossRef] [PubMed]
- Meinken, J.; Walker, G.; Cooper, C.R.; Min, X.J. MetazSecKB: The human and animal secretome and subcellular proteome knowledgebase. Database 2015, 2015, bav077. [Google Scholar] [CrossRef] [PubMed]
- Naba, A.; Clauser, K.R.; Hoersch, S.; Liu, H.; Carr, S.A.; Hynes, R.O. The matrisome: In silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol. Cell. Proteom. 2012, 11. [Google Scholar] [CrossRef]
- Mathivanan, S.; Fahner, C.J.; Reid, G.E.; Simpson, R.J. ExoCarta 2012: Database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012, 40, D1241–D1244. [Google Scholar] [CrossRef] [PubMed]
- Nieman, K.M.; Kenny, H.A.; Penicka, C.V.; Ladanyi, A.; Buell-Gutbrod, R.; Zillhardt, M.R.; Romero, I.L.; Carey, M.S.; Mills, G.B.; Hotamisligil, G.S.; et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 2011, 17, 1498–1503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rysman, E.; Brusselmans, K.; Scheys, K.; Timmermans, L.; Derua, R.; Munck, S.; Van Veldhoven, P.P.; Waltregny, D.; Daniëls, V.W.; Machiels, J.; et al. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 2010, 70, 8117–8126. [Google Scholar] [CrossRef] [PubMed]
- Kagan, H.M.; Li, W. Lysyl oxidase: Properties, specificity, and biological roles inside and outside of the cell. J. Cell. Biochem. 2003, 88, 660–672. [Google Scholar] [CrossRef] [PubMed]
- Erler, J.T.; Bennewith, K.L.; Cox, T.R.; Lang, G.; Bird, D.; Koong, A.; Le, Q.T.; Giaccia, A.J. Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell 2009, 15, 35–44. [Google Scholar] [CrossRef] [PubMed]
- McCaig, C.; Duval, C.; Hemers, E.; Steele, I.; Pritchard, M.; Przemeck, S.; Dimaline, R.; Ahmed, S.; Bodger, K.; Kerrigan, D.D.; et al. The role of matrix metalloproteinase-7 in redefining the gastric microenvironment in response to Helicobacter pylori. Gastroenterology 2006, 130, 1754–1763. [Google Scholar] [CrossRef] [PubMed]
- Salic, A.; Mitchison, T.J. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 2415–2420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, P.; Kibbe, W.A.; Lin, S.M. lumi: A pipeline for processing Illumina microarray. Bioinformatics 2008, 24, 1547–1548. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.M.; Du, P.; Huber, W.; Kibbe, W.A. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008, 36, e11. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K. Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Gentleman, R., Huber, W., Irizarry, R., Dudoit, S., Eds.; Springer: New York, NY, USA, 2005; pp. 397–420. [Google Scholar]
- Smyth, G.K.; Michaud, J.; Scott, H.S. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 2005, 21, 2067–2075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B-Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Assenov, Y.; Müller, F.; Lutsik, P.; Walter, J.; Lengauer, T.; Bock, C. Comprehensive analysis of DNA methylation data with RnBeads. Nat. Methods 2014, 11, 1138–1140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the maxquant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Taverner, T.; Karpievitch, Y.V.; Polpitiya, A.D.; Brown, J.N.; Dabney, A.R.; Anderson, G.A.; Smith, R.D. DanteR: An extensible R-based tool for quantitative analysis of -omics data. Bioinformatics 2012, 28, 2404–2406. [Google Scholar] [CrossRef] [PubMed]
- Jarnuczak, A.F.; Vizcaíno, J.A. Using the PRIDE Database and ProteomeXchange for Submitting and Accessing Public Proteomics Datasets. Curr. Protoc. Bioinformatics 2017. [Google Scholar] [CrossRef]
- Khatri, P.; Drăghici, S. Ontological analysis of gene expression data: Current tools, limitations, and open problems. Bioinformatics (Oxford, England) 2005, 21, 3587–3595. [Google Scholar] [CrossRef] [PubMed]
- Eden, E.; Navon, R.; Steinfeld, I.; Lipson, D.; Yakhini, Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed]
- Huber, W.; Carey, V.J.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, B.S.; Bravo, H.C.; Davis, S.; Gatto, L.; Girke, T.; et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 2015, 12, 115–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Najgebauer, H.; Jarnuczak, A.F.; Varro, A.; Sanderson, C.M. Integrative Omic Profiling Reveals Unique Hypoxia Induced Signatures in Gastric Cancer Associated Myofibroblasts. Cancers 2019, 11, 263. https://doi.org/10.3390/cancers11020263
Najgebauer H, Jarnuczak AF, Varro A, Sanderson CM. Integrative Omic Profiling Reveals Unique Hypoxia Induced Signatures in Gastric Cancer Associated Myofibroblasts. Cancers. 2019; 11(2):263. https://doi.org/10.3390/cancers11020263
Chicago/Turabian StyleNajgebauer, Hanna, Andrew F. Jarnuczak, Andrea Varro, and Christopher M. Sanderson. 2019. "Integrative Omic Profiling Reveals Unique Hypoxia Induced Signatures in Gastric Cancer Associated Myofibroblasts" Cancers 11, no. 2: 263. https://doi.org/10.3390/cancers11020263
APA StyleNajgebauer, H., Jarnuczak, A. F., Varro, A., & Sanderson, C. M. (2019). Integrative Omic Profiling Reveals Unique Hypoxia Induced Signatures in Gastric Cancer Associated Myofibroblasts. Cancers, 11(2), 263. https://doi.org/10.3390/cancers11020263




