The Phylogeographic Diversity of EBV and Admixed Ancestry in the Americas–Another Model of Disrupted Human-Pathogen Co-Evolution
Abstract
:1. Introduction
2. Phylogenetic Classification of EBV
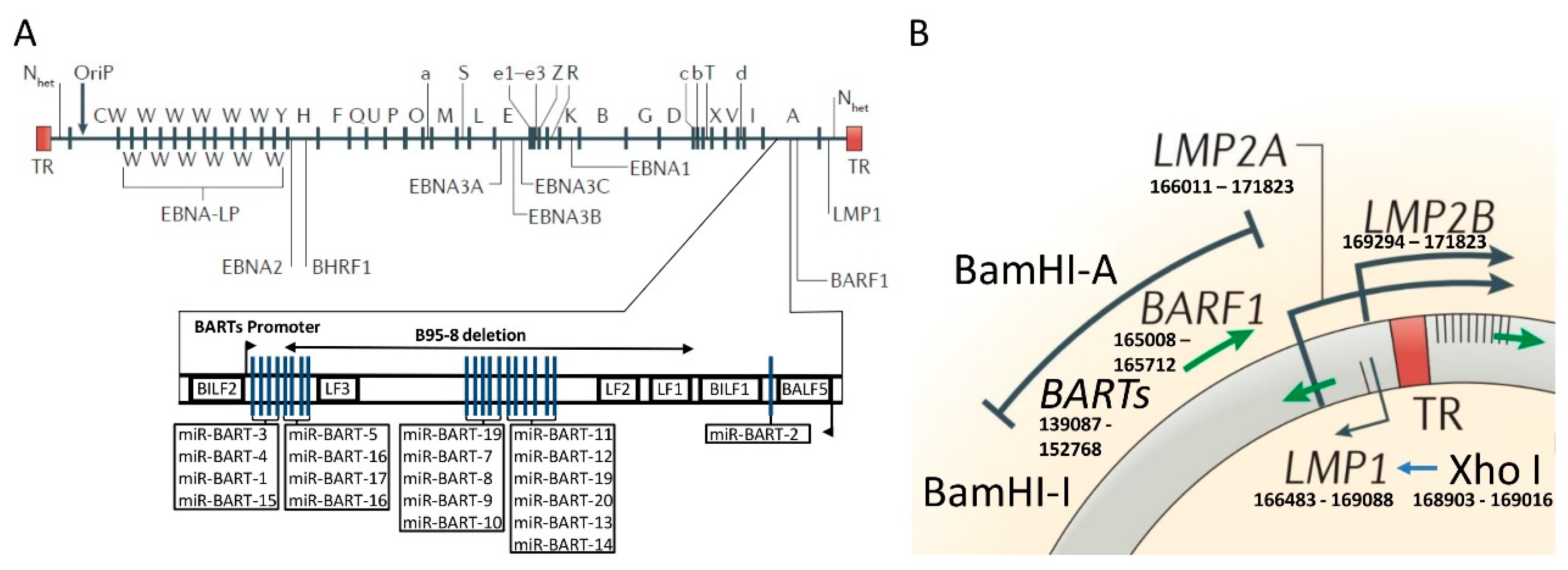
3. The Molecular Structure of the Cosegregated BamHI–I Fragment and XhoI Region of the EBV
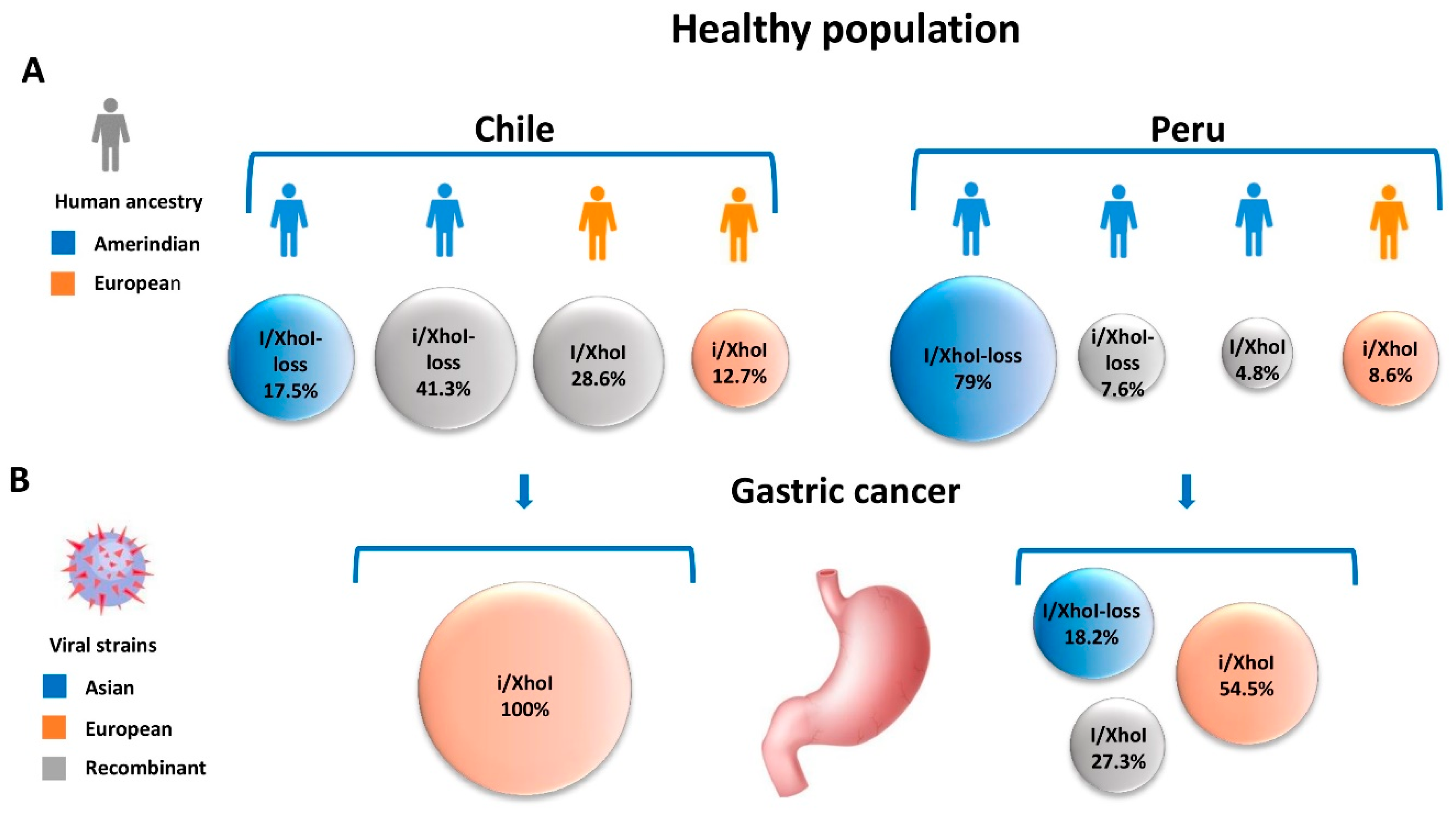
4. Human Ancestry in the Americas and EBV-Associated Gastric Carcinoma
5. Other Examples of “Disrupted Co-Evolution” in Cancer-Related Infectious Agents
6. Are Phylogeographic Variations of Epstein–Barr Virus Relevant to Other EBV-Associated Diseases?
7. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- The Cancer Genome Atlas Consortium. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishikawa, J.; Iizasa, H.; Yoshiyama, H.; Shimokuri, K.; Kobayashi, Y.; Sasaki, S.; Nakamura, M.; Yanai, H.; Sakai, K.; Suehiro, Y.; et al. Clinical Importance of Epstein(-)Barr Virus-Associated Gastric Cancer. Cancers 2018, 10, 167. [Google Scholar] [CrossRef] [PubMed]
- Sousa, H.; Pinto-Correia, A.L.; Medeiros, R.; Dinis-Ribeiro, M. Epstein-Barr virus is associated with gastric carcinoma: The question is what is the significance? World J. Gastroenterol. 2008, 14, 4347–4351. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, S.H.; Han, S.H.; An, J.S.; Lee, E.S.; Kim, Y.S. Clinicopathological and molecular characteristics of Epstein-Barr virus-associated gastric carcinoma: A meta-analysis. J. Gastroenterol. Hepatol. 2009, 24, 354–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, G.; Pfeiffer, R.; Camargo, M.C.; Rabkin, C.S. Meta-analysis shows that prevalence of Epstein-Barr virus-positive gastric cancer differs based on sex and anatomic location. Gastroenterology 2009, 137, 824–833. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Du, H.; Wang, Z.; Zhou, L.; Zhao, X.; Zeng, Y. Meta-analysis of the relationship between Epstein-Barr virus infection and clinicopathological features of patients with gastric carcinoma. Science China. Life Sci. 2010, 53, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Camargo, M.C.; Murphy, G.; Koriyama, C.; Pfeiffer, R.M.; Kim, W.H.; Herrera-Goepfert, R.; Corvalan, A.H.; Carrascal, E.; Abdirad, A.; Anwar, M.; et al. Determinants of Epstein-Barr virus-positive gastric cancer: An international pooled analysis. Br. J. Cancer 2011, 105, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Koriyama, C.; Akiba, S.; Iriya, K.; Yamaguti, T.; Hamada, G.S.; Itoh, T.; Eizuru, Y.; Aikou, T.; Watanabe, S.; Tsugane, S.; et al. Epstein-Barr virus-associated gastric carcinoma in Japanese Brazilians and non-Japanese Brazilians in Sao Paulo. Jpn. J. Cancer res. 2001, 92, 911–917. [Google Scholar] [CrossRef]
- Gulley, M.L.; Pulitzer, D.R.; Eagan, P.A.; Schneider, B.G. Epstein-Barr virus infection is an early event in gastric carcinogenesis and is independent of bcl-2 expression and p53 accumulation. Hum. Pathol. 1996, 27, 20–27. [Google Scholar] [CrossRef]
- Vo, Q.N.; Geradts, J.; Gulley, M.L.; Boudreau, D.A.; Bravo, J.C.; Schneider, B.G. Epstein-Barr virus in gastric adenocarcinomas: Association with ethnicity and CDKN2A promoter methylation. J. Clin. Pathol. 2002, 55, 669–675. [Google Scholar] [CrossRef]
- Alarcon, A.; Figueroa, U.; Espinoza, B.; Sandoval, A.; Carrasco-Aviño, G.; Aguayo, F.R.; Corvalan, A.H. Epstein-Barr Virus–Associated Gastric Carcinoma: The Americas’ Perspective; Lunet, N., Ed.; Intech: London, UK, 2017. [Google Scholar] [CrossRef]
- Baer, R.; Bankier, A.T.; Biggin, M.D.; Deininger, P.L.; Farrell, P.J.; Gibson, T.J.; Hatfull, G.; Hudson, G.S.; Satchwell, S.C.; Seguin, C.; et al. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 1984, 310, 207–211. [Google Scholar] [CrossRef] [PubMed]
- de Jesus, O.; Smith, P.R.; Spender, L.C.; Elgueta Karstegl, C.; Niller, H.H.; Huang, D.; Farrell, P.J. Updated Epstein-Barr virus (EBV) DNA sequence and analysis of a promoter for the BART (CST, BARF0) RNAs of EBV. J. Gen. Virol. 2003, 84, 1443–1450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, H.; Li, T.; Hung, G.C.; Li, B.; Tsai, S.; Lo, S.C. Identification and characterization of EBV genomes in spontaneously immortalized human peripheral blood B lymphocytes by NGS technology. BMC Genom. 2013, 14, 804. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, W.; Pan, Y.; Ji, J.; Lu, Z.; Ke, Y. Genome-wide analysis of Epstein-Barr virus (EBV) isolated from EBV-associated gastric carcinoma (EBVaGC). Oncotarget 2015, 7, 4903. [Google Scholar] [CrossRef] [PubMed]
- Santpere, G.; Darre, F.; Blanco, S.; Alcami, A.; Villoslada, P.; Mar Alba, M.; Navarro, A. Genome-wide analysis of wild-type Epstein-Barr virus genomes derived from healthy individuals of the 1,000 Genomes Project. Genome Biol. Evol. 2014, 6, 846–860. [Google Scholar] [CrossRef]
- Neves, M.; Marinho-Dias, J.; Ribeiro, J.; Sousa, H. Epstein-Barr virus strains and variations: Geographic or disease-specific variants? J. Med. Virol. 2017, 89, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Sample, J.; Young, L.; Martin, B.; Chatman, T.; Kieff, E.; Rickinson, A.; Kieff, E. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J. Virol. 1990, 64, 4084–4092. [Google Scholar]
- Zimber, U.; Adldinger, H.K.; Lenoir, G.M.; Vuillaume, M.; Knebel-Doeberitz, M.V.; Laux, G.; Desgranges, C.; Wittmann, P.; Freese, U.K.; Schneider, U.; et al. Geographical prevalence of two types of Epstein-Barr virus. Virology 1986, 154, 56–66. [Google Scholar] [CrossRef]
- Young, L.S.; Yao, Q.Y.; Rooney, C.M.; Sculley, T.B.; Moss, D.J.; Rupani, H.; Laux, G.; Bornkamm, G.W.; Rickinson, A.B. New type B isolates of Epstein-Barr virus from Burkitt’s lymphoma and from normal individuals in endemic areas. J. Gen. Virol. 1987, 68 Pt 11, 2853–2862. [Google Scholar] [CrossRef]
- Young, L.S.; Murray, P.G. Epstein-Barr virus and oncogenesis: From latent genes to tumours. Oncogene 2003, 22, 5108–5121. [Google Scholar] [CrossRef]
- Hudson, G.S.; Gibson, T.J.; Barrell, B.G. The BamHI F region of the B95-8 Epstein-Barr virus genome. Virology 1985, 147, 99–109. [Google Scholar] [CrossRef]
- Corvalan, A.H.; Ding, S.; Koriyama, C.; Carrascal, E.; Carrasquilla, G.; Backhouse, C.; Urzua, L.; Argandona, J.; Palma, M.; Eizuru, Y.; et al. Association of a distinctive strain of Epstein-Barr virus with gastric cancer. Int. J. Cancer 2006, 118, 1736–1742. [Google Scholar] [CrossRef] [PubMed]
- Lung, M.L.; Lam, W.P.; Sham, J.; Choy, D.; Yong-Sheng, Z.; Guo, H.Y.; Ng, M.H. Detection and prevalence of the “f” variant of Epstein-Barr virus in southern China. Virology 1991, 185, 67–71. [Google Scholar] [CrossRef]
- Lung, M.L.; Chang, R.S.; Huang, M.L.; Guo, H.Y.; Choy, D.; Sham, J.; Tsao, S.Y.; Cheng, P.; Ng, M.H. Epstein-Barr virus genotypes associated with nasopharyngeal carcinoma in southern China. Virology 1990, 177, 44–53. [Google Scholar] [CrossRef]
- Chen, J.N.; Ding, Y.G.; Feng, Z.Y.; Li, H.G.; He, D.; Du, H.; Wu, B.; Shao, C.K. Association of distinctive Epstein-Barr virus variants with gastric carcinoma in Guangzhou, southern China. J. Med. Virol. 2010, 82, 658–667. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Wang, Y.; Liu, X.; Chao, Y.; Xing, X.; Zhao, C.; Liu, C.; Luo, B. Genotypic analysis of Epstein-Barr virus isolates associated with nasopharyngeal carcinoma in Northern China. Intervirology 2011, 54, 131–138. [Google Scholar] [CrossRef]
- Lung, M.L.; Chang, G.C. Detection of distinct Epstein-Barr virus genotypes in NPC biopsies from southern Chinese and Caucasians. Int. J. Cancer 1992, 52, 34–37. [Google Scholar] [CrossRef]
- Lung, M.L.; Chang, R.S.; Jones, J.H. Genetic polymorphism of natural Epstein-Barr virus isolates from infectious mononucleosis patients and healthy carriers. J. Virol. 1988, 62, 3862–3866. [Google Scholar]
- Sidagis, J.; Ueno, K.; Tokunaga, M.; Ohyama, M.; Eizuru, Y. Molecular epidemiology of Epstein-Barr virus (EBV) in EBV-related malignancies. Int. J. Cancer 1997, 72, 72–76. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Hamid, M.; Chen, J.J.; Constantine, N.; Massoud, M.; Raab-Traub, N. EBV strain variation: Geographical distribution and relation to disease state. Virology 1992, 190, 168–175. [Google Scholar] [CrossRef]
- Hu, L.F.; Zabarovsky, E.R.; Chen, F.; Cao, S.L.; Ernberg, I.; Klein, G.; Winberg, G. Isolation and sequencing of the Epstein-Barr virus BNLF-1 gene (LMP1) from a Chinese nasopharyngeal carcinoma. J. Gen. Virol. 1991, 72 Pt 10, 2399–2409. [Google Scholar] [CrossRef] [Green Version]
- Khanim, F.; Yao, Q.Y.; Niedobitek, G.; Sihota, S.; Rickinson, A.B.; Young, L.S. Analysis of Epstein-Barr virus gene polymorphisms in normal donors and in virus-associated tumors from different geographic locations. Blood 1996, 88, 3491–3501. [Google Scholar] [PubMed]
- Adhikari, K.; Chacon-Duque, J.C.; Mendoza-Revilla, J.; Fuentes-Guajardo, M.; Ruiz-Linares, A. The Genetic Diversity of the Americas. Annu. Rev. Genom. Hum. Genet. 2017, 18, 277–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ordonez, P.; Koriyama, C.; Ding, S.; Yoshiwara, E.; Corvalan, A.H.; Takano, J.; Chirinos, J.L.; Watanabe, J.; Miyagui, J.; Hidalgo, H.; et al. Identification of the distinctive type i/XhoI+ strain of Epstein-Barr virus in gastric carcinoma in Peru. Anticancer Res. 2011, 31, 3607–3613. [Google Scholar] [PubMed]
- Sans, M. Admixture studies in Latin America: From the 20th to the 21st century. Hum. Boil. 2000, 72, 155–177. [Google Scholar]
- Eyheramendy, S.; Martinez, F.I.; Manevy, F.; Vial, C.; Repetto, G.M. Genetic structure characterization of Chileans reflects historical immigration patterns. Nat. Commun. 2015, 6, 6472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halliburton, I.W. Intertypic recombinants of herpes simplex viruses. J. Gen. Virol. 1980, 48, 1–23. [Google Scholar] [CrossRef]
- Young, L.S.; Yap, L.F.; Murray, P.G. Epstein-Barr virus: More than 50 years old and still providing surprises. Nature reviews. Cancer 2016, 16, 789–802. [Google Scholar] [CrossRef]
- Barth, S.; Pfuhl, T.; Mamiani, A.; Ehses, C.; Roemer, K.; Kremmer, E.; Jaker, C.; Hock, J.; Meister, G.; Grasser, F.A. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008, 36, 666–675. [Google Scholar] [CrossRef]
- Yao, Y.; Xu, M.; Liang, L.; Zhang, H.; Xu, R.; Feng, Q.; Feng, L.; Luo, B.; Zeng, Y.X. Genome-wide analysis of Epstein-Barr virus identifies variants and genes associated with gastric carcinoma and population structure. Tumour Boil. 2017, 39, 1010428317714195. [Google Scholar] [CrossRef]
- Price, A.M.; Luftig, M.A. To be or not IIb: A multi-step process for Epstein-Barr virus latency establishment and consequences for B cell tumorigenesis. PLoS Pathog. 2015, 11, e1004656. [Google Scholar] [CrossRef] [PubMed]
- Gilligan, K.; Sato, H.; Rajadurai, P.; Busson, P.; Young, L.; Rickinson, A.; Tursz, T.; Raab-Traub, N. Novel transcription from the Epstein-Barr virus terminal EcoRI fragment, DIJhet, in a nasopharyngeal carcinoma. J. Virol. 1990, 64, 4948–4956. [Google Scholar] [PubMed]
- Sadler, R.H.; Raab-Traub, N. Structural analyses of the Epstein-Barr virus BamHI A transcripts. J. Virol. 1995, 69, 1132–1141. [Google Scholar] [PubMed]
- Hayes, D.P.; Brink, A.A.; Vervoort, M.B.; Middeldorp, J.M.; Meijer, C.J.; van den Brule, A.J. Expression of Epstein-Barr virus (EBV) transcripts encoding homologues to important human proteins in diverse EBV associated diseases. Mol. Pathol. 1999, 52, 97–103. [Google Scholar] [CrossRef] [PubMed]
- zur Hausen, A.; Brink, A.A.; Craanen, M.E.; Middeldorp, J.M.; Meijer, C.J.; van den Brule, A.J. Unique transcription pattern of Epstein-Barr virus (EBV) in EBV-carrying gastric adenocarcinomas: Expression of the transforming BARF1 gene. Cancer Res. 2000, 60, 2745–2748. [Google Scholar] [PubMed]
- Chang, M.S.; Kim, D.H.; Roh, J.K.; Middeldorp, J.M.; Kim, Y.S.; Kim, S.; Han, S.; Kim, C.W.; Lee, B.L.; Kim, W.H.; et al. Epstein-Barr virus-encoded BARF1 promotes proliferation of gastric carcinoma cells through regulation of NF-kappaB. J. Virol. 2013, 87, 10515–10523. [Google Scholar] [CrossRef] [PubMed]
- Brink, A.A.; Vervoort, M.B.; Middeldorp, J.M.; Meijer, C.J.; van den Brule, A.J. Nucleic acid sequence-based amplification, a new method for analysis of spliced and unspliced Epstein-Barr virus latent transcripts, and its comparison with reverse transcriptase PCR. J. Clin. Microbial. 1998, 36, 3164–3169. [Google Scholar]
- Wei, M.X.; Ooka, T. A transforming function of the BARF1 gene encoded by Epstein-Barr virus. EMBO J. 1989, 8, 2897–2903. [Google Scholar] [CrossRef]
- Takada, K. Role of EBER and BARF1 in nasopharyngeal carcinoma (NPC) tumorigenesis. Semin. Cancer Biol. 2012, 22, 162–165. [Google Scholar] [CrossRef]
- Wei, M.X.; Moulin, J.C.; Decaussin, G.; Berger, F.; Ooka, T. Expression and tumorigenicity of the Epstein-Barr virus BARF1 gene in human Louckes B-lymphocyte cell line. Cancer Res. 1994, 54, 1843–1848. [Google Scholar]
- Sheng, W.; Decaussin, G.; Ligout, A.; Takada, K.; Ooka, T. Malignant transformation of Epstein-Barr virus-negative Akata cells by introduction of the BARF1 gene carried by Epstein-Barr virus. J. Virol. 2003, 77, 3859–3865. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Tsao, S.W.; Ooka, T.; Nicholls, J.M.; Cheung, H.W.; Fu, S.; Wong, Y.C.; Wang, X. Anti-apoptotic role of BARF1 in gastric cancer cells. Cancer Lett. 2006, 238, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Sall, A.; Caserta, S.; Jolicoeur, P.; Franqueville, L.; de Turenne-Tessier, M.; Ooka, T. Mitogenic activity of Epstein-Barr virus-encoded BARF1 protein. Oncogene 2004, 23, 4938–4944. [Google Scholar] [CrossRef] [PubMed]
- Wiech, T.; Nikolopoulos, E.; Lassman, S.; Heidt, T.; Schopflin, A.; Sarbia, M.; Werner, M.; Shimizu, Y.; Sakka, E.; Ooka, T.; et al. Cyclin D1 expression is induced by viral BARF1 and is overexpressed in EBV-associated gastric cancer. Virchows Arch. 2008, 452, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Strockbine, L.D.; Cohen, J.I.; Farrah, T.; Lyman, S.D.; Wagener, F.; DuBose, R.F.; Armitage, R.J.; Spriggs, M.K. The Epstein-Barr virus BARF1 gene encodes a novel, soluble colony-stimulating factor-1 receptor. J. Virol. 1998, 72, 4015–4021. [Google Scholar] [PubMed]
- Sapi, E.; Flick, M.B.; Gilmore-Hebert, M.; Rodov, S.; Kacinski, B.M. Transcriptional regulation of the c-fms (CSF-1R) proto-oncogene in human breast carcinoma cells by glucocorticoids. Oncogene 1995, 10, 529–542. [Google Scholar] [PubMed]
- Lin, E.Y.; Nguyen, A.V.; Russell, R.G.; Pollard, J.W. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J. Exp. Med. 2001, 193, 727–740. [Google Scholar] [CrossRef]
- Hoebe, E.K.; Le Large, T.Y.; Tarbouriech, N.; Oosterhoff, D.; De Gruijl, T.D.; Middeldorp, J.M.; Greijer, A.E. Epstein-Barr virus-encoded BARF1 protein is a decoy receptor for macrophage colony stimulating factor and interferes with macrophage differentiation and activation. Viral Immunol. 2012, 25, 461–470. [Google Scholar] [CrossRef]
- Hoebe, E.K.; Le Large, T.Y.; Greijer, A.E.; Middeldorp, J.M. BamHI-A rightward frame 1, an Epstein-Barr virus-encoded oncogene and immune modulator. Rev. Med. Virol. 2013, 23, 367–383. [Google Scholar] [CrossRef] [Green Version]
- Mohidin, T.B.; Ng, C.C. BARF1 gene silencing triggers caspase-dependent mitochondrial apoptosis in Epstein-Barr virus-positive malignant cells. J. Biosci. 2015, 40, 41–51. [Google Scholar] [CrossRef]
- Cai, X.; Schafer, A.; Lu, S.; Bilello, J.P.; Desrosiers, R.C.; Edwards, R.; Raab-Traub, N.; Cullen, B.R. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006, 2, e23. [Google Scholar] [CrossRef] [PubMed]
- Grundhoff, A.; Sullivan, C.S.; Ganem, D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 2006, 12, 733–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.Y.; Pfuhl, T.; Motsch, N.; Barth, S.; Nicholls, J.; Grasser, F.; Meister, G. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J. Virol. 2009, 83, 3333–3341. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Cosmopoulos, K.; Pegtel, M.; Hopmans, E.; Murray, P.; Middeldorp, J.; Shapiro, M.; Thorley-Lawson, D.A. A novel persistence associated EBV miRNA expression profile is disrupted in neoplasia. PLoS Pathog. 2011, 7, e1002193. [Google Scholar] [CrossRef] [PubMed]
- Seto, E.; Moosmann, A.; Gromminger, S.; Walz, N.; Grundhoff, A.; Hammerschmidt, W. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 2010, 6, e1001063. [Google Scholar] [CrossRef] [PubMed]
- Albanese, M.; Tagawa, T. MicroRNAs of Epstein-Barr Virus Control Innate and Adaptive Antiviral Immunity. J. Virol. 2017, 91, e01667-16. [Google Scholar] [CrossRef] [PubMed]
- Polakovicova, I.; Jerez, S.; Wichmann, I.A.; Sandoval-Borquez, A.; Carrasco-Veliz, N.; Corvalan, A.H. Role of microRNAs and Exosomes in Helicobacter pylori and Epstein-Barr Virus Associated Gastric Cancers. Front. Microbiol. 2018, 9, 636. [Google Scholar] [CrossRef]
- Homburger, J.R.; Moreno-Estrada, A.; Gignoux, C.R.; Nelson, D.; Sanchez, E.; Ortiz-Tello, P.; Pons-Estel, B.A.; Acevedo-Vasquez, E.; Miranda, P.; Langefeld, C.D.; et al. Genomic Insights into the Ancestry and Demographic History of South America. PLoS Genet. 2015, 11, e1005602. [Google Scholar] [CrossRef]
- Moreno-Estrada, A.; Gignoux, C.R.; Fernandez-Lopez, J.C.; Zakharia, F.; Sikora, M.; Contreras, A.V.; Acuna-Alonzo, V.; Sandoval, K.; Eng, C.; Romero-Hidalgo, S.; et al. Human genetics. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science 2014, 344, 1280–1285. [Google Scholar] [CrossRef]
- Shriner, D. Overview of Admixture Mapping. Curr. Protoc. Hum. Genet. 2017, 94, 1–23. [Google Scholar] [CrossRef]
- Moreno-Mayar, J.V.; Vinner, L.; de Barros Damgaard, P.; de la Fuente, C.; Chan, J.; Spence, J.P.; Allentoft, M.E.; Vimala, T.; Racimo, F.; Pinotti, T.; et al. Early human dispersals within the Americas. Science 2018. [Google Scholar] [CrossRef] [PubMed]
- Bryc, K.; Velez, C.; Karafet, T.; Moreno-Estrada, A.; Reynolds, A.; Auton, A.; Hammer, M.; Bustamante, C.D.; Ostrer, H. Colloquium paper: Genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proc. Natl. Acad. Sci. USA 2010, 107 (Suppl. 2), 8954–8961. [Google Scholar] [CrossRef] [PubMed]
- Ashktorab, H.; Kupfer, S.S.; Brim, H.; Carethers, J.M. Racial Disparity in Gastrointestinal Cancer Risk. Gastroenterology 2017, 153, 910–923. [Google Scholar] [CrossRef] [PubMed]
- Dong, E.; Duan, L.; Wu, B.U. Racial and Ethnic Minorities at Increased Risk for Gastric Cancer in a Regional US Population Study. Clin. Gastroenterol. Hepatol. 2017, 15, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Moore, S.P.; Hassler, S.; Ellison-Loschmann, L.; Forman, D.; Bray, F. The burden of stomach cancer in indigenous populations: A systematic review and global assessment. Gut 2014, 63, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Friborg, J.T.; Melbye, M. Cancer patterns in Inuit populations. Lancet Oncol. 2008, 9, 892–900. [Google Scholar] [CrossRef]
- Circumpolar Inuit Cancer Review Working Group; Kelly, J.; Lanier, A.; Santos, M.; Healey, S.; Louchini, R.; Friborg, J.; Young, K.; Ng, C. Cancer among the circumpolar Inuit, 1989-2003. II. Patterns and trends. Int. J. Circumpolar Health 2008, 67, 408–420. [Google Scholar]
- Amiri, M.; Janssen, F.; Kunst, A.E. The decline in stomach cancer mortality: Exploration of future trends in seven European countries. Eur. J. Epidemiol. 2011, 26, 23–28. [Google Scholar] [CrossRef]
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics, 2013. CA: A Cancer J. Clin. 2013, 63, 11–30. [Google Scholar] [CrossRef]
- Mathias, R.A.; Taub, M.A.; Gignoux, C.R.; Fu, W.; Musharoff, S.; O’Connor, T.D.; Vergara, C.; Torgerson, D.G.; Pino-Yanes, M.; Shringarpure, S.S.; et al. A continuum of admixture in the Western Hemisphere revealed by the African Diaspora genome. Nat. Commun. 2016, 7, 12522. [Google Scholar] [CrossRef] [Green Version]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.P.; Forman, D.; Pineros, M.; Fernandez, S.M.; de Oliveira Santos, M.; Bray, F. Cancer in indigenous people in Latin America and the Caribbean: A review. Cancer Med. 2014, 3, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Pineros, M.; Ferlay, J.; Murillo, R. Cancer incidence estimates at the national and district levels in Colombia. Salud Publica Mex. 2006, 48, 455–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Silva, E.M.; Fernandes, M.R.; de Carvalho, D.C.; Leitao, L.P.C.; Cavalcante, G.C.; Pereira, E.E.B.; Modesto, A.A.C.; Guerreiro, J.F.; de Assumpcao, P.P.; Dos Santos, S.E.B.; et al. Correction to: Effect of genetic ancestry to the risk of susceptibility to gastric cancer in a mixed population of the Brazilian Amazon. BMC Res. Notes 2017, 10, 761. [Google Scholar] [CrossRef] [PubMed]
- Kodaman, N.; Sobota, R.S.; Mera, R.; Schneider, B.G.; Williams, S.M. Disrupted human–pathogen co-evolution: A model for disease. Front. Genet. 2014, 5, 290. [Google Scholar] [CrossRef] [PubMed]
- Linz, B.; Balloux, F.; Moodley, Y.; Manica, A.; Liu, H.; Roumagnac, P.; Falush, D.; Stamer, C.; Prugnolle, F.; van der Merwe, S.W.; et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature 2007, 445, 915–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorell, K.; Yahara, K.; Berthenet, E.; Lawson, D.J.; Mikhail, J.; Kato, I.; Mendez, A.; Rizzato, C.; Bravo, M.M.; Suzuki, R.; et al. Rapid evolution of distinct Helicobacter pylori subpopulations in the Americas. PLoS Genet. 2017, 13, e1006546. [Google Scholar] [CrossRef]
- Munoz-Ramirez, Z.Y.; Mendez-Tenorio, A.; Kato, I.; Bravo, M.M.; Rizzato, C.; Thorell, K.; Torres, R.; Aviles-Jimenez, F.; Camorlinga, M.; Canzian, F.; et al. Whole Genome Sequence and Phylogenetic Analysis Show Helicobacter pylori Strains from Latin America Have Followed a Unique Evolution Pathway. Front. Cell. Infect. Microbiol. 2017, 7, 50. [Google Scholar] [CrossRef]
- de Sablet, T.; Piazuelo, M.B.; Shaffer, C.L.; Schneider, B.G.; Asim, M.; Chaturvedi, R.; Bravo, L.E.; Sicinschi, L.A.; Delgado, A.G.; Mera, R.M.; et al. Phylogeographic origin of Helicobacter pylori is a determinant of gastric cancer risk. Gut 2011, 60, 1189–1195. [Google Scholar] [CrossRef] [Green Version]
- Van Doorslaer, K.; Chen, Z.; Bernard, H.U.; Chan, P.K.S.; DeSalle, R.; Dillner, J.; Forslund, O.; Haga, T.; McBride, A.A.; Villa, L.L.; et al. ICTV Virus Taxonomy Profile: Papillomaviridae. J. Gen. Virol. 2018, 99, 989–990. [Google Scholar] [CrossRef]
- Ho, L.; Chan, S.Y.; Burk, R.D.; Das, B.C.; Fujinaga, K.; Icenogle, J.P.; Kahn, T.; Kiviat, N.; Lancaster, W.; Mavromara-Nazos, P.; et al. The genetic drift of human papillomavirus type 16 is a means of reconstructing prehistoric viral spread and the movement of ancient human populations. J. Virol. 1993, 67, 6413–6423. [Google Scholar] [PubMed]
- Ong, C.K.; Chan, S.Y.; Campo, M.S.; Fujinaga, K.; Mavromara-Nazos, P.; Labropoulou, V.; Pfister, H.; Tay, S.K.; ter Meulen, J.; Villa, L.L.; et al. Evolution of human papillomavirus type 18: An ancient phylogenetic root in Africa and intratype diversity reflect coevolution with human ethnic groups. J. Virol. 1993, 67, 6424–6431. [Google Scholar] [PubMed]
- Tornesello, M.L.; Duraturo, M.L.; Salatiello, I.; Buonaguro, L.; Losito, S.; Botti, G.; Stellato, G.; Greggi, S.; Piccoli, R.; Pilotti, S.; et al. Analysis of human papillomavirus type-16 variants in Italian women with cervical intraepithelial neoplasia and cervical cancer. J. Med. Virol. 2004, 74, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.F.; Koutsky, L.A.; Galloway, D.A.; Kuypers, J.; Hughes, J.P.; Wheeler, C.M.; Holmes, K.K.; Kiviat, N.B. Genomic variation of human papillomavirus type 16 and risk for high grade cervical intraepithelial neoplasia. J. Natl. Cancer Inst. 1997, 89, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Correia, S.; Palser, A.; Elgueta Karstegl, C.; Middeldorp, J.M.; Ramayanti, O.; Cohen, J.I.; Hildesheim, A.; Fellner, M.D.; Wiels, J.; White, R.E.; et al. Natural Variation of Epstein-Barr Virus Genes, Proteins, and Primary MicroRNA. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.M.; Yu, K.J.; Mbulaiteye, S.M.; Hildesheim, A.; Bhatia, K. The extent of genetic diversity of Epstein-Barr virus and its geographic and disease patterns: A need for reappraisal. Virus Res. 2009, 143, 209–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neves, M.; Marinho-Dias, J.; Ribeiro, J.; Esteves, M.; Maltez, E.; Baldaque, I.; Breda, E.; Monteiro, E.; Medeiros, R.; Sousa, H. Characterization of Epstein-Barr virus strains and LMP1-deletion variants in Portugal. J. Med. Virol. 2015, 87, 1382–1388. [Google Scholar] [CrossRef] [PubMed]
- Cheung, S.T.; Leung, S.F.; Lo, K.W.; Chiu, K.W.; Tam, J.S.; Fok, T.F.; Johnson, P.J.; Lee, J.C.; Huang, D.P. Specific latent membrane protein 1 gene sequences in type 1 and type 2 Epstein-Barr virus from nasopharyngeal carcinoma in Hong Kong. Int. J. Cancer 1998, 76, 399–406. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.J.; Lay, J.D.; Chen, C.L.; Chen, J.Y.; Liu, M.Y.; Su, I.J. Genomic analysis of Epstein-Barr virus in nasal and peripheral T-cell lymphoma: A comparison with nasopharyngeal carcinoma in an endemic area. J. Med. Virol. 1996, 50, 314–321. [Google Scholar] [CrossRef]
- Lin, S.X.; Zong, Y.S.; Zhang, M.; Han, A.J.; Zhong, B.L.; Liang, Y.J. Study of sequence variations of Epstein-Barr virus LMP1 gene in nasopharyngeal carcinoma. Zhonghua Bing Li Xue Za Zhi = Chin. J. Pathol. 2005, 34, 791–795. [Google Scholar]
- Tan, E.L.; Peh, S.C.; Sam, C.K. Analyses of Epstein-Barr virus latent membrane protein-1 in Malaysian nasopharyngeal carcinoma: High prevalence of 30-bp deletion, Xho1 polymorphism and evidence of dual infections. J. Med. Virol. 2003, 69, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.S.; Su, I.J.; Chung, P.J.; Shu, C.H.; Ng, C.K.; Wu, S.J.; Liu, S.T. Detection of an Epstein-Barr-virus variant in T-cell-lymphoma tissues identical to the distinct strain observed in nasopharyngeal carcinoma in the Taiwanese population. Int. J. Cancer 1995, 62, 673–677. [Google Scholar] [CrossRef] [PubMed]
- Ayadi, W.; Feki, L.; Khabir, A.; Boudawara, T.; Ghorbel, A.; Charfeddine, I.; Daoud, J.; Frikha, M.; Hammami, A.; Karray-Hakim, H. Polymorphism analysis of Epstein-Barr virus isolates of nasopharyngeal carcinoma biopsies from Tunisian patients. Virus Genes 2007, 34, 137–145. [Google Scholar] [CrossRef] [PubMed]
- da Costa, V.G.; Marques-Silva, A.C.; Moreli, M.L. The Epstein-Barr virus latent membrane protein-1 (LMP1) 30-bp deletion and XhoI-polymorphism in nasopharyngeal carcinoma: A meta-analysis of observational studies. Syst. Rev. 2015, 4, 46. [Google Scholar] [CrossRef] [PubMed]
- Cheung, S.T.; Lo, K.W.; Leung, S.F.; Chan, W.Y.; Choi, P.H.; Johnson, P.J.; Lee, J.C.; Huang, D.P. Prevalence of LMP1 deletion variant of Epstein-Barr virus in nasopharyngeal carcinoma and gastric tumors in Hong Kong. Int. J. Cancer 1996, 66, 711–712. [Google Scholar] [CrossRef] [Green Version]
- Chang, K.P.; Hao, S.P.; Lin, S.Y.; Ueng, S.H.; Pai, P.C.; Tseng, C.K.; Hsueh, C.; Hsieh, M.S.; Yu, J.S.; Tsang, N.M. The 30-bp deletion of Epstein-Barr virus latent membrane protein-1 gene has no effect in nasopharyngeal carcinoma. Laryngoscope 2006, 116, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.H.; Campo, E.; N.L., H.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J.; Vardiman, J.W. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; IARC: Lyon, France, 2008. [Google Scholar]
- Ansell, S.M. Hodgkin Lymphoma: Diagnosis and Treatment. Mayo Clin. Proc. 2015, 90, 1574–1583. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, Y.; Choi, J.W.; Kim, Y.S. Prevalence and prognostic significance of Epstein-Barr virus infection in classical Hodgkin’s lymphoma: A meta-analysis. Arch. Med. Res. 2014, 45, 417–431. [Google Scholar] [CrossRef]
- Zhou, X.G.; Sandvej, K.; Li, P.J.; Ji, X.L.; Yan, Q.H.; Zhang, X.P.; Da, J.P.; Hamilton-Dutoit, S.J. Epstein--Barr virus gene polymorphisms in Chinese Hodgkin’s disease cases and healthy donors: Identification of three distinct virus variants. J. Gen. Virol. 2001, 82, 1157–1167. [Google Scholar] [CrossRef]
- Garcia-Cosio, M.; Santon, A.; Martin, P.; Reguero, M.E.; Cristobal, E.; Bellas, C. Analysis of Epstein-Barr virus strains and variants in classical Hodgkin’s lymphoma by laser microdissection. Histol. Histopathol. 2008, 23, 209–217. [Google Scholar] [CrossRef]
- Kim, I.; Park, E.R.; Park, S.H.; Lin, Z.; Kim, Y.S. Characteristics of Epstein-Barr virus isolated from the malignant lymphomas in Korea. J. Med. Virol. 2002, 67, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Sandvej, K.; Peh, S.C.; Andresen, B.S.; Pallesen, G. Identification of potential hot spots in the carboxy-terminal part of the Epstein-Barr virus (EBV) BNLF-1 gene in both malignant and benign EBV-associated diseases: High frequency of a 30-bp deletion in Malaysian and Danish peripheral T-cell lymphomas. Blood 1994, 84, 4053–4060. [Google Scholar] [PubMed]
- Boyle, M.J.; Vasak, E.; Tschuchnigg, M.; Turner, J.J.; Sculley, T.; Penny, R.; Cooper, D.A.; Tindall, B.; Sewell, W.A. Subtypes of Epstein-Barr virus (EBV) in Hodgkin’s disease: Association between B-type EBV and immunocompromise. Blood 1993, 81, 468–474. [Google Scholar] [PubMed]
- Guiretti, D.M.; Chabay, P.A.; Valva, P.; Stefanoff, C.G.; Barros, M.H.; De Matteo, E.; Renault, I.Z.; Preciado, M.V.; Hassan, R. Structural variability of the carboxy-terminus of Epstein-Barr virus encoded latent membrane protein 1 gene in Hodgkin’s lymphomas. J. Med. Virol. 2007, 79. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Chen, W.G.; Chen, Y.Y.; Murakami, I.; Chen, H.L.; Ohara, N.; Nose, S.; Hamaya, K.; Matsui, S.; Bacchi, M.M.; et al. Deletion of Epstein-Barr virus latent membrane protein 1 gene in Japanese and Brazilian gastric carcinomas, metastatic lesions, and reactive lymphocytes. Am. J. Pathol. 1998, 152, 191–198. [Google Scholar] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corvalán, A.H.; Ruedlinger, J.; de Mayo, T.; Polakovicova, I.; Gonzalez-Hormazabal, P.; Aguayo, F. The Phylogeographic Diversity of EBV and Admixed Ancestry in the Americas–Another Model of Disrupted Human-Pathogen Co-Evolution. Cancers 2019, 11, 217. https://doi.org/10.3390/cancers11020217
Corvalán AH, Ruedlinger J, de Mayo T, Polakovicova I, Gonzalez-Hormazabal P, Aguayo F. The Phylogeographic Diversity of EBV and Admixed Ancestry in the Americas–Another Model of Disrupted Human-Pathogen Co-Evolution. Cancers. 2019; 11(2):217. https://doi.org/10.3390/cancers11020217
Chicago/Turabian StyleCorvalán, Alejandro H., Jenny Ruedlinger, Tomas de Mayo, Iva Polakovicova, Patricio Gonzalez-Hormazabal, and Francisco Aguayo. 2019. "The Phylogeographic Diversity of EBV and Admixed Ancestry in the Americas–Another Model of Disrupted Human-Pathogen Co-Evolution" Cancers 11, no. 2: 217. https://doi.org/10.3390/cancers11020217
APA StyleCorvalán, A. H., Ruedlinger, J., de Mayo, T., Polakovicova, I., Gonzalez-Hormazabal, P., & Aguayo, F. (2019). The Phylogeographic Diversity of EBV and Admixed Ancestry in the Americas–Another Model of Disrupted Human-Pathogen Co-Evolution. Cancers, 11(2), 217. https://doi.org/10.3390/cancers11020217





