Hypoxic Environment and Paired Hierarchical 3D and 2D Models of Pediatric H3.3-Mutated Gliomas Recreate the Patient Tumor Complexity
Abstract
1. Background
2. Results
2.1. Clinical Features of the H3.3-K28M-Mutated High-Grade Gliomas
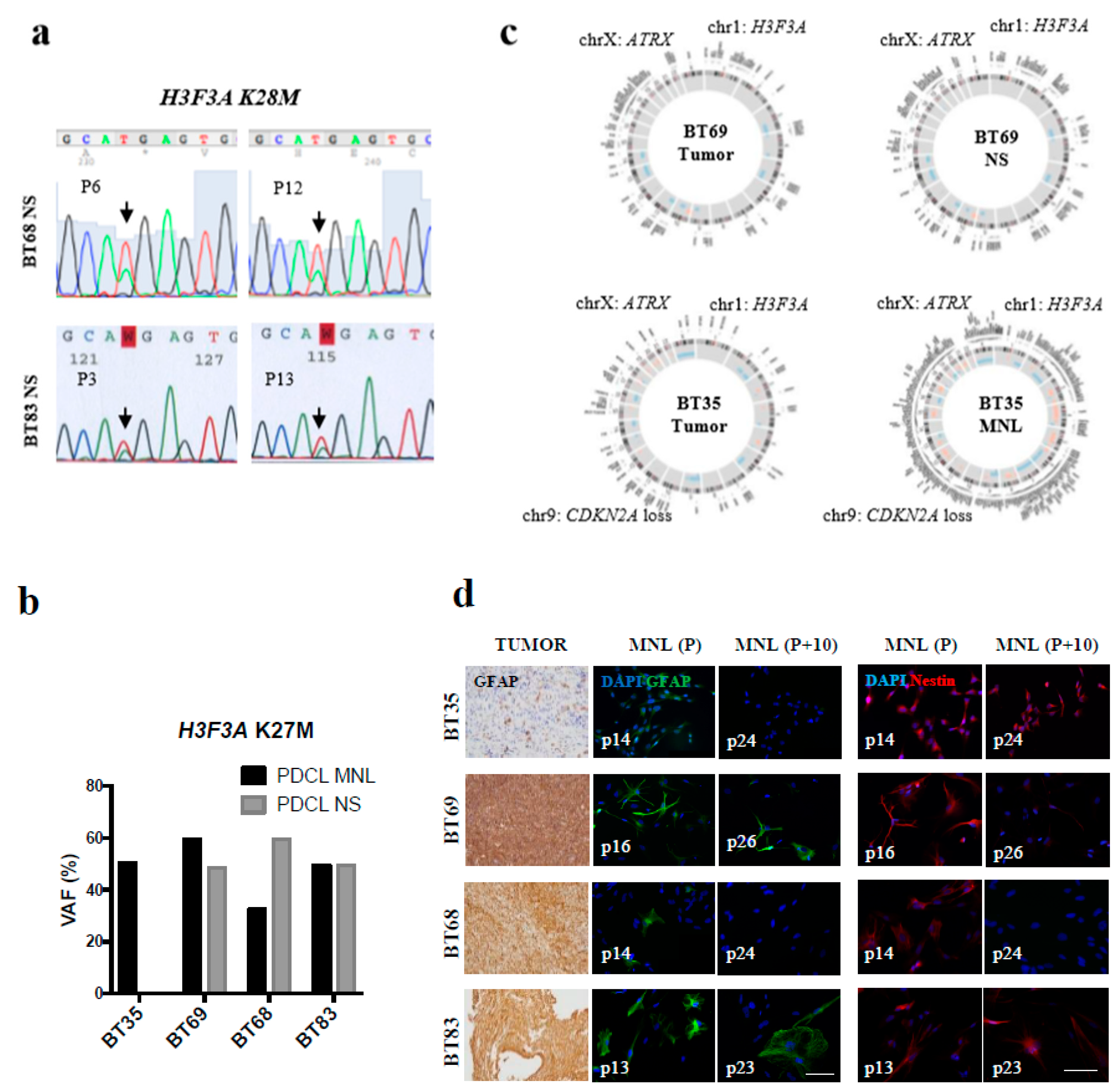
2.2. Early-Passage Culture of PDCLs Preserved the Genotype and Phenotype of Primary High-Grade Gliomas
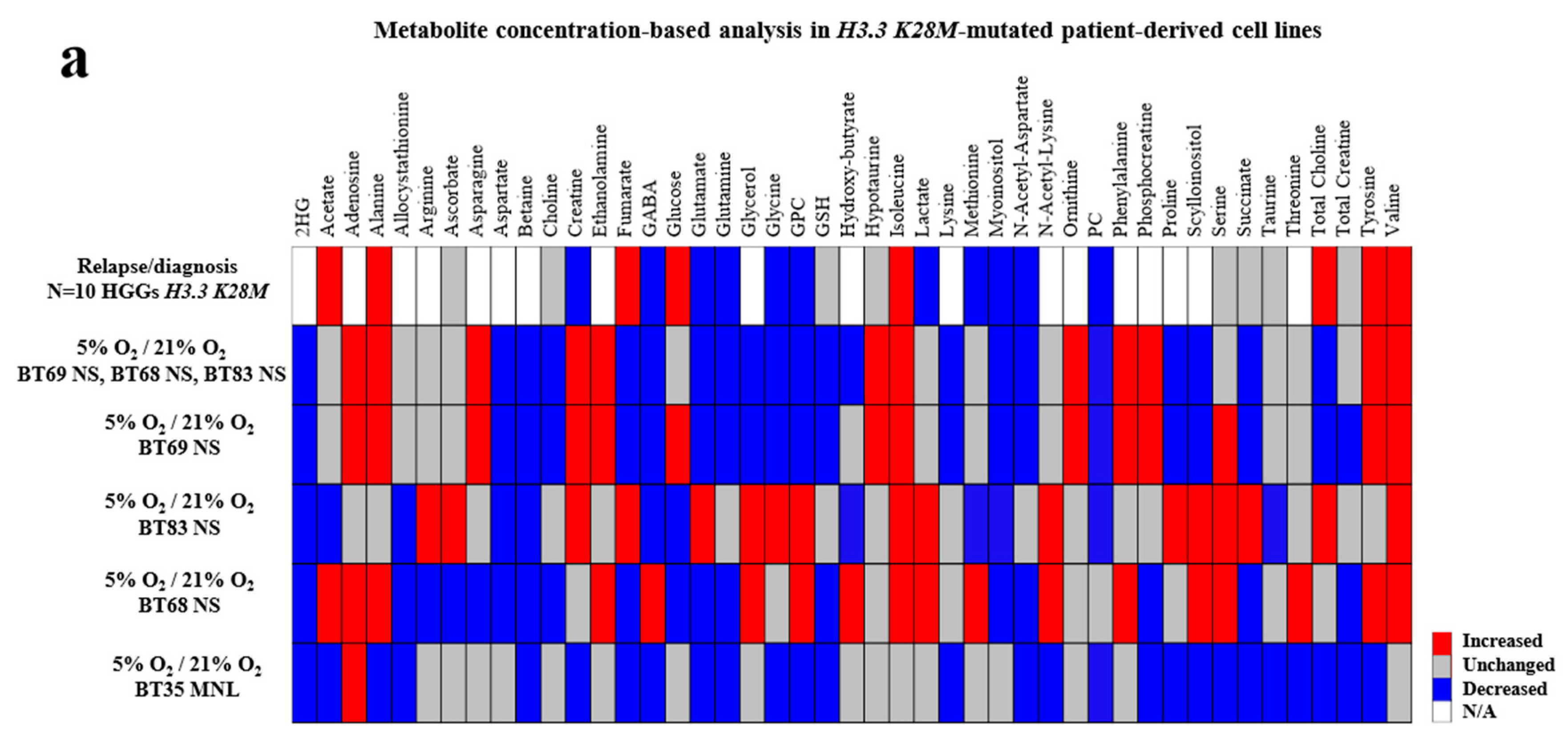
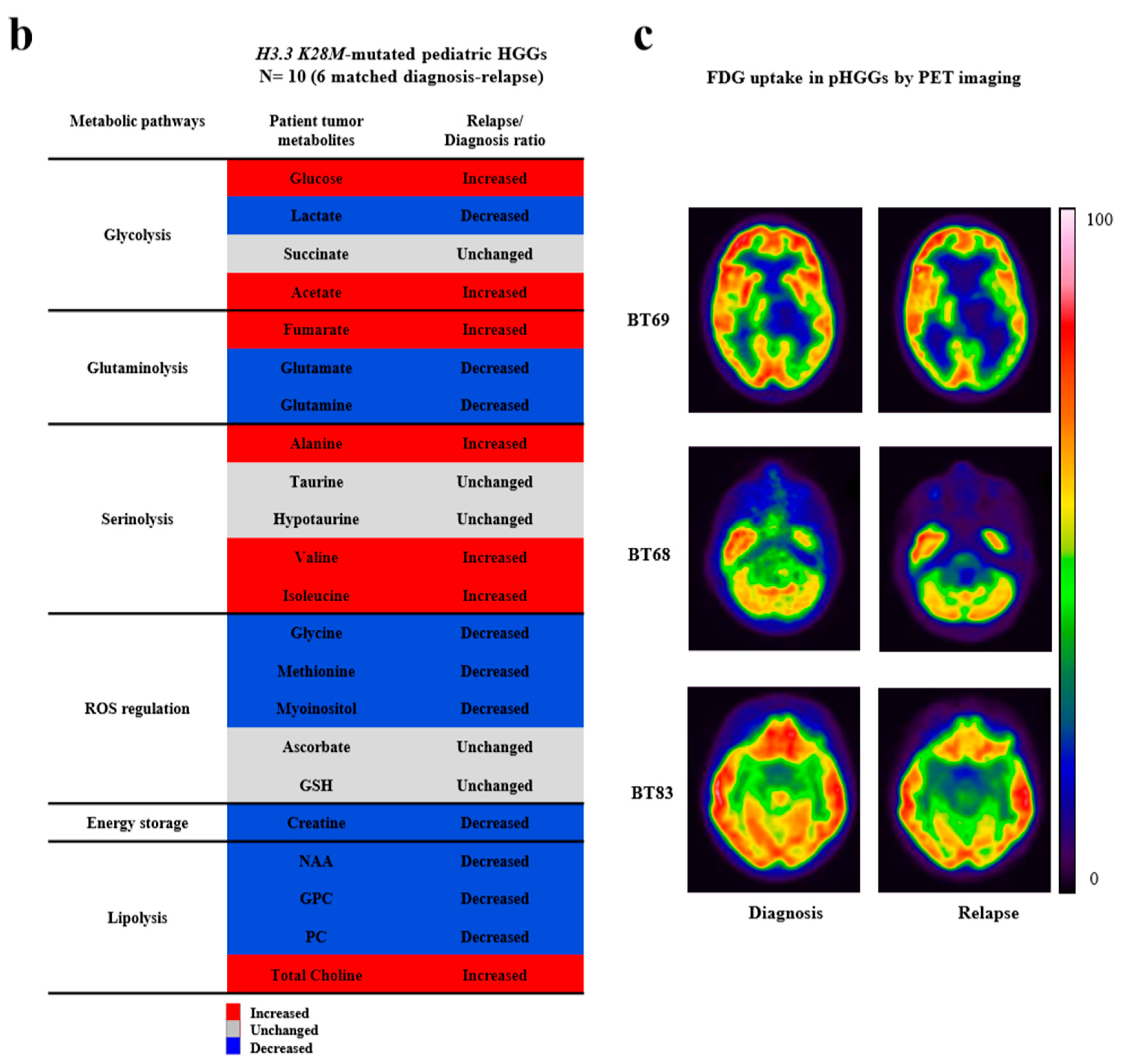
2.3. Importance of In Vitro Hypoxia to Mimic the Metabolic Changes During H3K28M-Mutated pHGG Progression
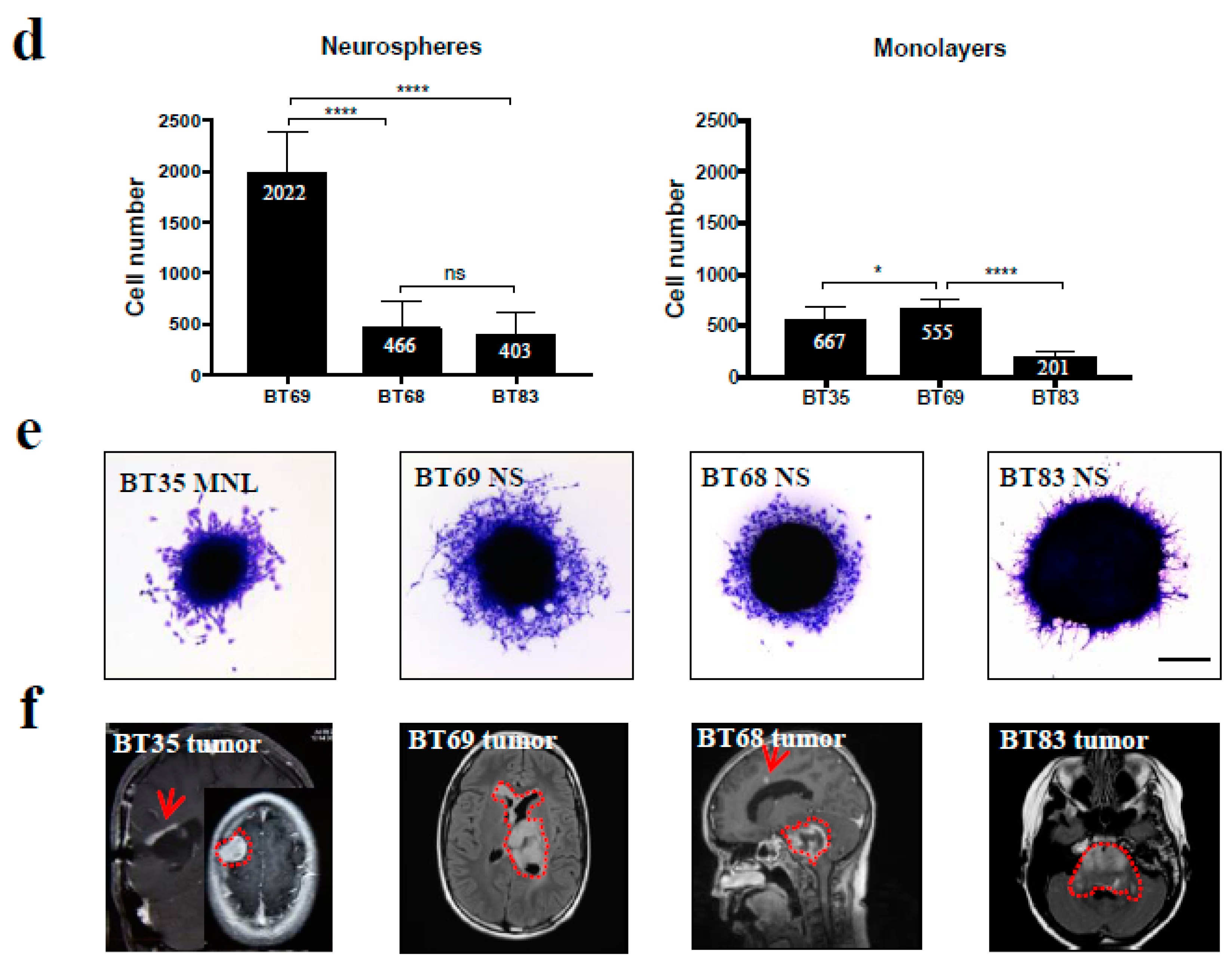
2.4. In Vitro Modeling of Cell Migration from Hypoxic PDCL Spheroids’ Recapitulated Tumor Cell Dissemination in Patients
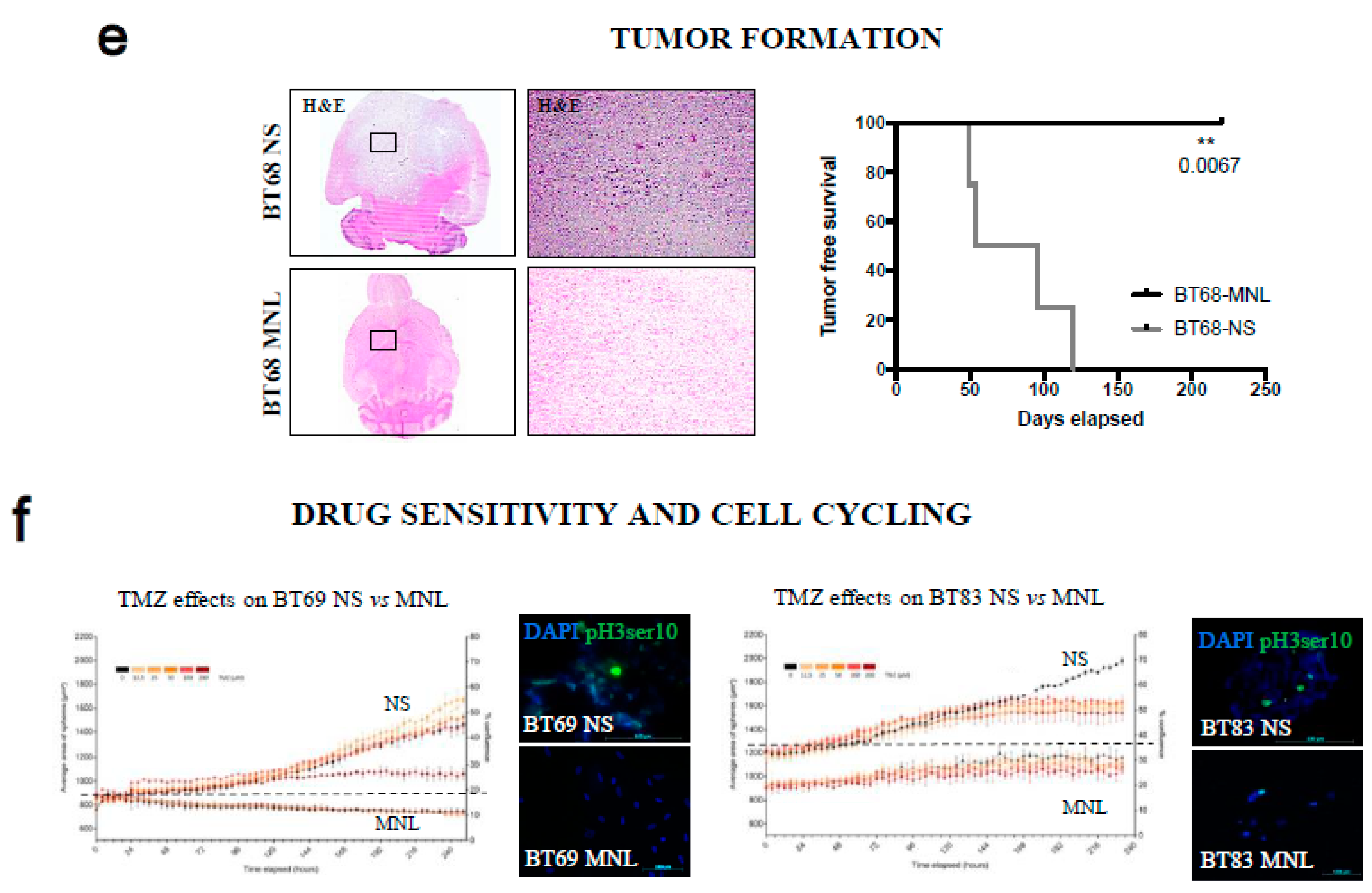
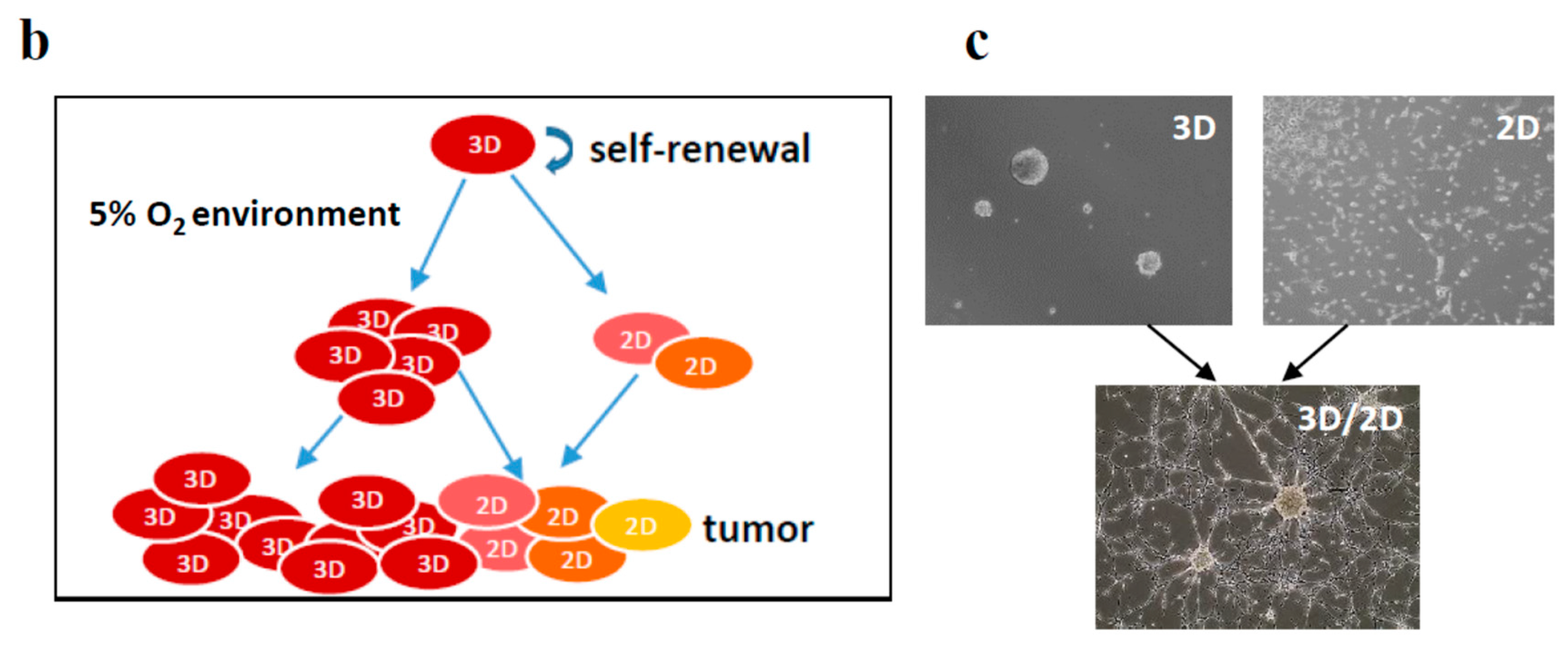
2.5. Both 2D/MNL and 3D/NS Cell Cultures Mimic the Hierarchical pHGG Development and Patient Drug Sensitivity
3. Discussion
4. Methods
4.1. Patient Tumors and Derived-Cell Cultures
4.2. Next Generation Sequencing (NGS) Analyses
4.3. Immunocytochemistry
4.4. Immunohistochemistry (IHC)
4.5. Array Comparative Genomic Hybridization (aCGH) Analyses
4.6. Sample Preparations for NMR (Nuclear Magnetic Resonance) Measurements
4.7. RNA Sequencing Analyses
4.8. Western Blot Analysis
4.9. Spheroid Migration Assay
4.10. Doubling Time
4.11. Clonogenic Assay
4.12. Intracranial Injections
4.13. Quantitative PCR (qPCR) of Human Alu Repeats
4.14. Drug Testing
4.15. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bondy, M.L.; Scheurer, M.E.; Malmer, B.; Barnholtz-Sloan, J.S.; Davis, F.G.; Il’yasova, D.; Kruchko, C.; McCarthy, B.J.; Rajaraman, P.; Schwartzbaum, J.A.; et al. Brain tumor epidemiology: Consensus from the brain tumor epidemiology consortium. Cancer 2008, 113, 1953–1968. [Google Scholar] [CrossRef]
- Grill, J.; Massimino, M.; Bouffet, E.; Azizi, A.A.; McCowage, G.; Cañete, A.; Saran, F.; Le Deley, M.C.; Varlet, P.; Morgan, P.S.; et al. Phase II, open-label, randomized, multicenter trial (HERBY) of bevacizumab in pediatric patients with newly diagnosed high-grade glioma. J. Clin. Oncol. 2018, 36, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, L.M.; Veldhuijzen van Zanten, S.E.M.; Colditz, N.; Baugh, J.; Chaney, B.; Hoffmann, M.; Lane, A.; Fuller, C.; Miles, L.; Hawkins, C.; et al. Clinical, radiologic, pathologic, and molecular characteristics of long-term survivors of diffuse intrinsic pontine glioma (DIPG): A collaborative report from the international and european society for pediatric oncology DIPG registries. J. Clin. Oncol. 2018, 36, 1963–1972. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Bax, D.A.; Mackay, A.; Little, S.E.; Carvalho, D.; Viana-Pereira, M.; Tamber, N.; Grigoriadis, A.E.; Ashworth, A.; Reis, R.M.; Ellison, D.W.; et al. A distinct spectrum of copy number aberrations in paediatric high grade gliomas. Clin. Cancer Res. 2010, 16, 3368–3377. [Google Scholar] [CrossRef] [PubMed]
- Harttrampf, A.C.; Lacroix, L.; Deloger, M.; Deschamps, F.; Puget, S.; Auger, N.; Vielh, P.; Varlet, P.; Balogh, Z.; Abbou, S.; et al. Molecular screening for cancer treatment optimization (MOSCATO-01) in pediatric patients: A single-institutional prospective molecular stratification trial. Clin. Cancer Res. 2017, 23, 6101–6112. [Google Scholar] [CrossRef] [PubMed]
- Sahm, F.; Schrimpf, D.; Jones, D.T.W.; Meyer, J.; Kratz, A.; Reuss, D.; Capper, D.; Koelsche, C.; Korshunov, A.; Wiestler, B.; et al. Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol. 2016, 131, 903–910. [Google Scholar] [CrossRef]
- Mount, C.W.; Majzner, R.G.; Sundaresh, S.; Arnold, E.P.; Kadapakkam, M.; Haile, S.; Hulleman, E.; Woo, P.J.; Rietberg, S.P.; Vogel, H.; et al. Potent antitumor efficacy of anti-GD2 CAR T-cells in H3K27M+ diffuse midline gliomas. Nat. Med. 2018, 24, 572–579. [Google Scholar] [CrossRef]
- Plessier, A.; Le Dret, L.; Varlet, P.; Beccaria, K.; Lacombe, J.; Mériaux, S.; Geffroy, F.; Fiette, L.; Flamant, P.; Chrétien, F.; et al. New in vivo avatars of diffuse intrinsic pontine gliomas (DIPG) from stereotactic biopsies performed at diagnosis. Oncotarget 2017, 8, 52543. [Google Scholar] [CrossRef]
- Blandin, A.F.; Noulet, F.; Renner, G.; Mercier, M.C.; Choulier, L.; Vauchelles, R.; Ronde, P.; Carreiras, F.; Etienne-Selloum, N.; Vereb, G.; et al. Glioma cell dispersion is driven by α5 integrin-mediated cell–matrix and cell–cell interactions. Cancer Lett. 2016, 376, 328–338. [Google Scholar] [CrossRef]
- Gencoglu, M.F.; Barney, L.E.; Hall, C.L.; Brooks, E.A.; Schwartz, A.D.; Corbett, D.C.; Stevens, K.R.; Peyton, S.R. Comparative study of multicellular tumor spheroid formation methods and implications for drug screening. ACS Biomater. Sci. Eng. 2018, 4, 410–420. [Google Scholar] [CrossRef] [PubMed]
- Hubert, C.G.; Rivera, M.; Spangler, L.C.; Wu, Q.; Mack, S.C.; Prager, B.C.; Couce, M.; McLendon, R.E.; Sloan, A.E.; Rich, J.N. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 2016, 76, 2465–2477. [Google Scholar] [CrossRef] [PubMed]
- Grasso, C.S.; Tang, Y.; Truffaux, N.; Berlow, N.E.; Liu, L.; Debily, M.A.; Quist, M.J.; Davis, L.E.; Huang, E.C.; Woo, P.J.; et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat. Med. 2015, 21, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Hennika, T.; Hu, G.; Olaciregui, N.G.; Barton, K.L.; Ehteda, A.; Chitranjan, A.; Gifford, A.J.; Tsoli, M.; Ziegler, D.S.; Carcaboso, A.M.; et al. Pre-clinical study of panobinostat in xenograft and genetically engineered murine diffuse intrinsic pontine glioma models. PLoS ONE 2017, 12, e0169485. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.R.; Mackay, A.; Truffaux, N.; Butterfield, Y.; Morozova, O.; Philippe, C.; Castel, D.; Grasso, C.S.; Vinci, M.; Carvalho, D.; et al. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat. Genet. 2014, 46, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Filbin, M.G.; Tirosh, I.; Hovestadt, V.; Shaw, M.L.; Escalante, L.E.; Mathewson, N.D.; Neftel, C.; Frank, N.; Pelton, K.; Hebert, C.M.; et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 2018, 360, 331–335. [Google Scholar] [CrossRef]
- Gomez-Roman, N.; Stevenson, K.; Gilmour, L.; Hamilton, G.; Chalmers, A.J. A novel 3D human glioblastoma cell culture system for modeling drug and radiation responses. Neuro-Oncology 2017, 19, 229–241. [Google Scholar] [CrossRef]
- Meel, M.H.; Sewing, A.C.P.; Waranecki, P.; Metselaar, D.S.; Wedekind, L.E.; Koster, J.; van Vuurden, D.G.; Kaspers, G.J.L.; Hulleman, E. Culture methods of diffuse intrinsic pontine glioma cells determine response to targeted therapies. Exp. Cell Res. 2017, 360, 397–403. [Google Scholar] [CrossRef]
- Vinci, M.; Burford, A.; Molinari, V.; Kessler, K.; Popov, S.; Clarke, M.; Taylor, K.R.; Pemberton, H.N.; Lord, C.J.; Gutteridge, A.; et al. Functional diversity and cooperativity between subclonal populations of pediatric glioblastoma and diffuse intrinsic pontine glioma cells. Nat. Med. 2018, 24, 1204–1215. [Google Scholar] [CrossRef]
- Wang, X.; Prager, B.C.; Wu, Q.; Kim, L.J.Y.; Gimple, R.C.; Shi, Y.; Yang, K.; Morton, A.R.; Zhou, W.; Zhu, Z.; et al. Reciprocal signaling between glioblastoma stem cells and differentiated tumor cells promotes malignant progression. Cell Stem Cell 2018, 22, 514–528. [Google Scholar] [CrossRef]
- Brahimi-Horn, M.C.; Pouysségur, J. Oxygen, a source of life and stress. FEBS Lett. 2007, 581, 3582–3591. [Google Scholar] [CrossRef] [PubMed]
- Lal, A.; Lash, A.E.; Altschul, S.F.; Velculescu, V.; Zhang, L.; McLendon, R.E.; Marra, M.A.; Prange, C.; Morin, P.J.; Polyak, K.; et al. A public database for gene expression in human cancers. Cancer Res. 1999, 59, 5403–5407. [Google Scholar] [PubMed]
- Schneider, T.; Osl, F.; Friess, T.; Stockinger, H.; Scheuer, W.V. Quantification of human Alu sequences by real-time PCR--an improved method to measure therapeutic efficacy of anti-metastatic drugs in human xenotransplants. Clin. Exp. Metastasis 2002, 19, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-H.; Liu, W.-T.; Hung, H.-C.; Gean, C.-Y.; Tsai, H.-M.; Su, C.-L.; Gean, P.W. Synergistic inhibition of tumor growth by combination treatment with drugs against different subpopulations of glioblastoma cells. BMC Cancer 2017, 17, 905. [Google Scholar] [CrossRef] [PubMed]
- Carreau, A.; Hafny-Rahbi, B.E.; Matejuk, A.; Grillon, C.; Kieda, C. Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J. Cell Mol. Med. 2011, 15, 1239–1253. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.; Moussalieh, F.M.; Mackay, A.; Cicek, A.E.; Coca, A.; Chenard, M.P.; Weingertner, N.; Lhermitte, B.; Letouzé, E.; Guérin, E.; et al. Characterization of the Transcriptional and Metabolic Responses of Pediatric High Grade Gliomas to mTOR-HIF-1a Axis Inhibition. Available online: http://www.impactjournals.com/oncotarget/index.php?journal=oncotarget&.page=article&op=view&path[]=16500&pubmed-linkout=1 (accessed on 19 June 2017).
- Grande, S.; Palma, A.; Ricci-Vitiani, L.; Luciani, A.M.; Buccarelli, M.; Biffoni, M.; Molinari, A.; Calcabrini, A.; D’Amore, E.; Guidoni, L.; et al. Metabolic Heterogeneity Evidenced by MRS Among Patient-Derived Glioblastoma Multiforme Stem-Like Cells Accounts for Cell Clustering and Different Responses to Drugs. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5835274/ (accessed on 4 April 2018).
- Zhao, H.; Heimberger, A.B.; Lu, Z.; Wu, X.; Hodges, T.R.; Song, R.; Shen, J. Metabolomics profiling in plasma samples from glioma patients correlates with tumor phenotypes. Oncotarget 2016, 7, 20486–20495. [Google Scholar] [CrossRef]
- Maudsley, A.A.; Gupta, R.K.; Stoyanova, R.; Parra, N.A.; Roy, B.; Sheriff, S.; Hussain, N.; Behari, S. Mapping of glycine distributions in Gliomas. Am. J. Neuroradiol. 2014, 35, S31–S36. [Google Scholar] [CrossRef]
- Chinnaiyan, P.; Kensicki, E.; Bloom, G.; Prabhu, A.; Sarcar, B.; Kahali, S.; Eschrich, S.; Qu, X.; Forsyth, P.; Gillies, R. The metabolomic signature of malignant glioma reflects accelerated anabolic metabolism. Cancer Res. 2012, 72, 5878–5888. [Google Scholar] [CrossRef]
- Wenger, A.; Larsson, S.; Danielsson, A.; Elbæk, K.J.; Kettunen, P.; Tisell, M.; Sabel, M.; Lannering, B.; Nordborg, C.; Schepke, E.; et al. Stem cell cultures derived from pediatric brain tumors accurately model the originating tumors. Oncotarget 2017, 8, 18626–18639. [Google Scholar] [CrossRef]
- Rahman, M.; Reyner, K.; Deleyrolle, L.; Millette, S.; Azari, H.; Day, B.W.; Stringer, B.W.; Boyd, A.W.; Johns, T.G.; Blot, V.; et al. Neurosphere and adherent culture conditions are equivalent for malignant glioma stem cell lines. Anat. Cell Biol. 2015, 48, 25–35. [Google Scholar] [CrossRef]
- Bradshaw, A.; Wickremsekera, A.; Tan, S.T.; Peng, L.; Davis, P.F.; Itinteang, T. Cancer stem cell hierarchy in glioblastoma multiforme. Front. Surg. 2016, 3, 21. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhao, Y.-J.; Lin, Q.; Yu, L.; Liu, Z.; Lindsay, H.; Kogiso, M.; Rao, P.; Li, X.N.; Lu, X. Cytogenetic landscape of paired neurospheres and traditional monolayer cultures in pediatric malignant brain tumors. Neuro-Oncology 2015, 17, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Mack, S.C.; Hubert, C.G.; Miller, T.E.; Taylor, M.D.; Rich, J.N. An epigenetic gateway to brain tumor cell identity. Nat. Neurosci. 2016, 19, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Ströbele, S.; Nonnenmacher, L.; Siegelin, M.D.; Tepper, M.; Stroh, S.; Hasslacher, S.; Enzenmüller, S.; Strauss, G.; Baumann, B.; et al. A paired comparison between glioblastoma “stem cells” and differentiated cells. Int. J. Cancer 2016, 138, 1709–1718. [Google Scholar] [CrossRef]
- Chassot, A.; Canale, S.; Varlet, P.; Puget, S.; Roujeau, T.; Negretti, L.; Dhermain, F.; Rialland, X.; Raquin, M.A.; Grill, J.; et al. Radiotherapy with concurrent and adjuvant temozolomide in children with newly diagnosed diffuse intrinsic pontine glioma. J. Neuro Oncol. 2012, 106, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Moussallieh, F.-M.; Elbayed, K.; Chanson, J.; Rudolf, G.; Piotto, M.; De Seze, J.; Namer, I.J. Serum analysis by 1H nuclear magnetic resonance spectroscopy: A new tool for distinguishing neuromyelitis optica from multiple sclerosis. Mult. Scler. J. 2014, 20, 558–565. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Achuthan, P.; Akanni, W.; Amode, M.R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; Irón, C.G.; et al. Ensembl 2018. Nucleic Acids Res. 2018, 46, D754–D761. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq - A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-seq Data with DESeq2. Available online: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4302049/ (accessed on 5 December 2014).
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014, 42, D199–D205. [Google Scholar] [CrossRef] [PubMed]
- Selway, J.G. Metabolism at a Glance, 3rd ed.; Blackwell Publishing: Malden, MI, USA, 2014. [Google Scholar]



| Patients | BT35 | BT69 | BT68 | BT83 |
|---|---|---|---|---|
| Sample | Relapse | Relapse | Diagnosis | Diagnosis |
| Location | Thalamic | Thalamic | Pons | Pons |
| Age (years) | 18 | 14 | 9 | 13 |
| Diagnosis | HGG | HGG | DIPG | DIPG |
| 1st line therapy | Stupp protocol | Stupp protocol | RT + cilengitide | RT + dasatinib |
| 2nd line therapy | RT + Iri/Beva | RAPIRI protocol | No treatment | RAPIRI protocol |
| Survival (months) | 12 | 17 | 10 | 9 |
| Imaging | ||||
| Contrast-enhanced T1-weighted MRI |  | 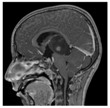 | 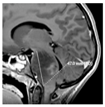 | 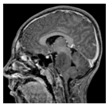 |
| ADC (mm(2)/s) | 1047 | 1029 | 945 | 1068 |
| T2-weighted FLAIR MRI |  | 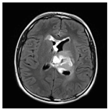 | 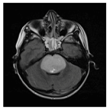 | 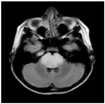 |
| Histology (diagnosis) | ||||
| H&E | 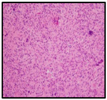 |  | 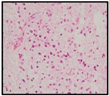 | 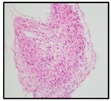 |
| IHC | ||||
| Ki67 (%) | 50–80 | 20–25 | 15 | 10–15 |
| GFAP | pos | pos | pos | pos |
| p53 | pos | neg | pos | pos |
| EGFR | neg | pos | pos | pos |
| PTEN | pos | pos | NA | pos |
| HIF-1α (%) | 20 | 50 | 50 | 20 |
| Olig2 (%) | 70 | 100 | 10 | 50 |
| Olig2 (%) at relapse | 30 | 50 | NA | NA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blandin, A.-F.; Durand, A.; Litzler, M.; Tripp, A.; Guérin, É.; Ruhland, E.; Obrecht, A.; Keime, C.; Fuchs, Q.; Reita, D.; et al. Hypoxic Environment and Paired Hierarchical 3D and 2D Models of Pediatric H3.3-Mutated Gliomas Recreate the Patient Tumor Complexity. Cancers 2019, 11, 1875. https://doi.org/10.3390/cancers11121875
Blandin A-F, Durand A, Litzler M, Tripp A, Guérin É, Ruhland E, Obrecht A, Keime C, Fuchs Q, Reita D, et al. Hypoxic Environment and Paired Hierarchical 3D and 2D Models of Pediatric H3.3-Mutated Gliomas Recreate the Patient Tumor Complexity. Cancers. 2019; 11(12):1875. https://doi.org/10.3390/cancers11121875
Chicago/Turabian StyleBlandin, Anne-Florence, Aurélie Durand, Marie Litzler, Aurélien Tripp, Éric Guérin, Elisa Ruhland, Adeline Obrecht, Céline Keime, Quentin Fuchs, Damien Reita, and et al. 2019. "Hypoxic Environment and Paired Hierarchical 3D and 2D Models of Pediatric H3.3-Mutated Gliomas Recreate the Patient Tumor Complexity" Cancers 11, no. 12: 1875. https://doi.org/10.3390/cancers11121875
APA StyleBlandin, A.-F., Durand, A., Litzler, M., Tripp, A., Guérin, É., Ruhland, E., Obrecht, A., Keime, C., Fuchs, Q., Reita, D., Lhermitte, B., Coca, A., Jones, C., Rebel, I. L., Villa, P., Namer, I. J., Dontenwill, M., Guenot, D., & Entz-Werle, N. (2019). Hypoxic Environment and Paired Hierarchical 3D and 2D Models of Pediatric H3.3-Mutated Gliomas Recreate the Patient Tumor Complexity. Cancers, 11(12), 1875. https://doi.org/10.3390/cancers11121875





