Evaluation and Validation of Plasma Proteins Using Two Different Protein Detection Methods for Early Detection of Colorectal Cancer
Abstract
1. Introduction
2. Results
2.1. Characteristics of Study Population
2.2. Individual Markers
2.3. Correlation Analysis
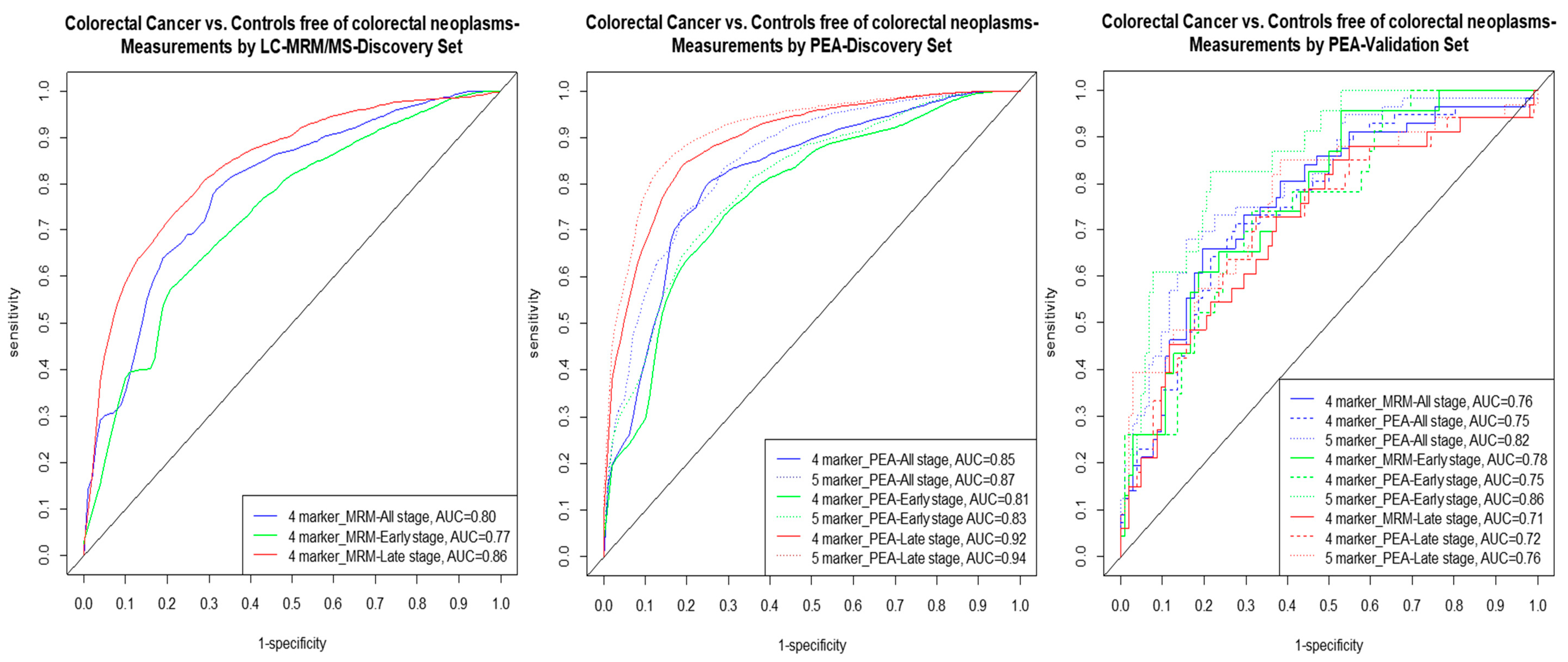
2.4. Multimarker Signatures
3. Discussion
4. Methods
4.1. Study Design
4.2. Study Population: Discovery Set
4.3. Study Population: Validation Set
4.4. Sample Collection and Storage
4.5. Laboratory Assays
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Brenner, H.; Stock, C.; Hoffmeister, M. Effect of screening sigmoidoscopy and screening colonoscopy on colorectal cancer incidence and mortality: Systematic review and meta-analysis of randomised controlled trials and observational studies. BMJ 2014, 348, g2467. [Google Scholar] [CrossRef] [PubMed]
- Zauber, A.G. The impact of screening on colorectal cancer mortality and incidence: Has it really made a difference? Dig. Dis. Sci. 2015, 60, 681–691. [Google Scholar] [CrossRef]
- Atkin, W.; Wooldrage, K.; Parkin, D.M.; Kralj-Hans, I.; MacRae, E.; Shah, U.; Duffy, S.; Cross, A.J. Long term effects of once-only flexible sigmoidoscopy screening after 17 years of follow-up: The uk flexible sigmoidoscopy screening randomised controlled trial. Lancet 2017, 389, 1299–1311. [Google Scholar] [CrossRef]
- Brenner, H.; Chen, C. The colorectal cancer epidemic: Challenges and opportunities for primary, secondary and tertiary prevention. Br. J. Cancer 2018, 119, 785–792. [Google Scholar] [CrossRef] [PubMed]
- Klabunde, C.; Blom, J.; Bulliard, J.L.; Garcia, M.; Hagoel, L.; Mai, V.; Patnick, J.; Rozjabek, H.; Senore, C.; Tornberg, S. Participation rates for organized colorectal cancer screening programmes: An international comparison. J. Med. Screen. 2015, 22, 119–126. [Google Scholar] [CrossRef]
- Bretthauer, M.; Kaminski, M.F.; Løberg, M.; Zauber, A.G.; Regula, J.; Kuipers, E.J.; Påhlman, L.; Hernán, M.A.; McFadden, E.; Sunde, A.; et al. Population-based colonoscopy screening for colorectal cancer: A european randomized trial. JAMA Intern. Med. 2016, 176, 894–902. [Google Scholar] [CrossRef]
- Robertson, D.J.; Lee, J.K.; Boland, C.R.; Dominitz, J.A.; Giardiello, F.M.; Johnson, D.A.; Kaltenbach, T.; Lieberman, D.; Levin, T.R.; Rex, D.K. Recommendations on fecal immunochemical testing to screen for colorectal neoplasia: A consensus statement by the us multi-society task force on colorectal cancer. Gastroenterology 2017, 152, 1217–1237. [Google Scholar] [CrossRef]
- Adler, A.; Geiger, S.; Keil, A.; Bias, H.; Schatz, P.; deVos, T.; Dhein, J.; Zimmermann, M.; Tauber, R.; Wiedenmann, B. Improving compliance to colorectal cancer screening using blood and stool based tests in patients refusing screening colonoscopy in germany. BMC Gastroenterol. 2014, 14, 183. [Google Scholar] [CrossRef]
- Ponomarenko, E.A.; Poverennaya, E.V.; Ilgisonis, E.V.; Pyatnitskiy, M.A.; Kopylov, A.T.; Zgoda, V.G.; Lisitsa, A.V.; Archakov, A.I. The size of the human proteome: The width and depth. Int. J. Anal. Chem. 2016, 2016, 7436849. [Google Scholar] [CrossRef]
- Kuzyk, M.A.; Smith, D.; Yang, J.; Cross, T.J.; Jackson, A.M.; Hardie, D.B.; Anderson, N.L.; Borchers, C.H. Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol. Cell. Proteomics 2009, 8, 1860–1877. [Google Scholar] [CrossRef] [PubMed]
- Domanski, D.; Percy, A.J.; Yang, J.; Chambers, A.G.; Hill, J.S.; Freue, G.V.; Borchers, C.H. Mrm-based multiplexed quantitation of 67 putative cardiovascular disease biomarkers in human plasma. Proteomics 2012, 12, 1222–1243. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zucknick, M.; Werner, S.; Knebel, P.; Brenner, H. Head-to-head comparison and evaluation of 92 plasma protein biomarkers for early detection of colorectal cancer in a true screening setting. Clin. Cancer Res. 2015, 21, 3318–3326. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Qian, J.; Werner, S.; Cuk, K.; Knebel, P.; Brenner, H. Development and validation of a panel of five proteins as blood biomarkers for early detection of colorectal cancer. Clin. Epidemiol. 2017, 9, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, M.; Erben, V.; Schrotz-King, P.; Brenner, H. Cell line secretome and tumor tissue proteome markers for early detection of colorectal cancer: A systematic review. Cancers 2017, 9, 156. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, M.; Gies, A.; Werner, S.; Schrotz-King, P.; Brenner, H. Blood-based protein signatures for early detection of colorectal cancer: A systematic review. Clin. Transl. Gastroenterol. 2017, 8, e128. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.G.; Gerszten, R.E. Emerging affinity-based proteomic technologies for large-scale plasma profiling in cardiovascular disease. Circulation 2017, 135, 1651–1664. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Kim, J.W.; Jeon, S.Y.; Park, B.K.; Han, B.G. Proteomic analysis of the effect of storage temperature on human serum. Ann. Clin. Lab. Sci. 2010, 40, 61–70. [Google Scholar]
- Enroth, S.; Hallmans, G.; Grankvist, K.; Gyllensten, U. Effects of long-term storage time and original sampling month on biobank plasma protein concentrations. EBioMedicine 2016, 12, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Bjorkesten, J.; Galli, J.; Ekman, D.; Broberg, J.; Nordberg, N.; Tillander, A.; Kamali-Moghaddam, M.; Tybring, G.; Landegren, U. Strong impact on plasma protein profiles by precentrifugation delay but not by repeated freeze-thaw cycles, as analyzed using multiplex proximity extension assays. Clin. Chem. Lab. Med. 2018, 56, 582–594. [Google Scholar] [CrossRef]
- Alnabulsi, A.; Murray, G.I. Proteomics for early detection of colorectal cancer: Recent updates. Expert Rev. Proteomics 2018, 15, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Surinova, S.; Choi, M.; Tao, S.; Schueffler, P.J.; Chang, C.-Y.; Clough, T.; Vyslouzil, K.; Khoylou, M.; Srovnal, J.; Liu, Y.; et al. Prediction of colorectal cancer diagnosis based oncirculating plasma proteins. EMBO Mol. Med. 2015, 7, 1166–1178. [Google Scholar] [CrossRef] [PubMed]
- Ciarloni, L.; Ehrensberger, S.H.; Imaizumi, N.; Monnier-Benoit, S.; Nichita, C.; Myung, S.J.; Kim, J.S.; Song, S.Y.; Kim, T.I.; van der Weg, B.; et al. Development and clinical validation of a blood test based on 29-gene expression for early detection of colorectal cancer. Clin. Cancer Res. 2016, 22, 4604–4611. [Google Scholar] [CrossRef] [PubMed]
- Barderas, R.; Mendes, M.; Torres, S.; Bartolome, R.A.; Lopez-Lucendo, M.; Villar-Vazquez, R.; Pelaez-Garcia, A.; Fuente, E.; Bonilla, F.; Casal, J.I. In-depth characterization of the secretome of colorectal cancer metastatic cells identifies key proteins in cell adhesion, migration, and invasion. Mol. Cell. Proteomics 2013, 12, 1602–1620. [Google Scholar] [CrossRef] [PubMed]
- Rodia, M.T.; Solmi, R.; Pasini, F.; Nardi, E.; Mattei, G.; Ugolini, G.; Ricciardiello, L.; Strippoli, P.; Miglio, R.; Lauriola, M. Lgals4, ceacam6, tspan8, and col1a2: Blood markers for colorectal cancer-validation in a cohort of subjects with positive fecal immunochemical test result. Clin. Colorectal Cancer 2018, 17, e217–e228. [Google Scholar] [CrossRef]
- Rodia, M.T.; Ugolini, G.; Mattei, G.; Montroni, I.; Zattoni, D.; Ghignone, F.; Veronese, G.; Marisi, G.; Lauriola, M.; Strippoli, P.; et al. Systematic large-scale meta-analysis identifies a panel of two mrnas as blood biomarkers for colorectal cancer detection. Oncotarget 2016, 7, 30295–30306. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Kohn, M.A.; Carpenter, C.R.; Newman, T.B. Understanding the direction of bias in studies of diagnostic test accuracy. Acad. Emerg. Med. 2013, 20, 1194–1206. [Google Scholar] [CrossRef]
- Whiting, P.; Rutjes, A.W.; Reitsma, J.B.; Glas, A.S.; Bossuyt, P.M.; Kleijnen, J. Sources of variation and bias in studies of diagnostic accuracy: A systematic review. Ann. Intern. Med. 2004, 140, 189–202. [Google Scholar] [CrossRef]
- Potter, N.T.; Hurban, P.; White, M.N.; Whitlock, K.D.; Lofton-Day, C.E.; Tetzner, R.; Koenig, T.; Quigley, N.B.; Weiss, G. Validation of a real-time pcr-based qualitative assay for the detection of methylated sept9 DNA in human plasma. Clin. Chem. 2014, 60, 1183–1191. [Google Scholar] [CrossRef]
- Church, T.R.; Wandell, M.; Lofton-Day, C.; Mongin, S.J.; Burger, M.; Payne, S.R.; Castaños-Vélez, E.; Blumenstein, B.A.; Rösch, T.; Osborn, N.; et al. Prospective evaluation of methylated sept9 in plasma for detection of asymptomatic colorectal cancer. Gut 2014, 63, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Wild, N.; Andres, H.; Rollinger, W.; Krause, F.; Dilba, P.; Tacke, M.; Karl, J. A combination of serum markers for the early detection of colorectal cancer. Clin. Cancer Res. 2010, 16, 6111–6121. [Google Scholar] [CrossRef]
- Lumachi, F.; Marino, F.; Orlando, R.; Chiara, G.B.; Basso, S.M. Simultaneous multianalyte immunoassay measurement of five serum tumor markers in the detection of colorectal cancer. Anticancer Res. 2012, 32, 985–988. [Google Scholar] [PubMed]
- Werner, S.; Krause, F.; Rolny, V.; Strobl, M.; Morgenstern, D.; Datz, C.; Chen, H.; Brenner, H. Evaluation of a 5-marker blood test for colorectal cancer early detection in a colorectal cancer screening setting. Clin. Cancer Res. 2016, 22, 1725–1733. [Google Scholar] [CrossRef] [PubMed]
- Brenner, H.; Calderazzo, S.; Seufferlein, T.; Ludwig, L.; Dikopoulos, N.; Mangold, J.; Böck, W.; Stolz, T.; Eisenbach, T.; Block, T.; et al. Effect of a single aspirin dose prior to fecal immunochemical testing on test sensitivity for detecting advanced colorectal neoplasms: A randomized clinical trialeffect of single-dose aspirin prior to fit on colorectal cancer detectioneffect of single-dose aspirin prior to fit on colorectal cancer detection. JAMA 2019, 321, 1686–1692. [Google Scholar] [PubMed]
- Hundt, S.; Haug, U.; Brenner, H. Comparative evaluation of immunochemical fecal occult blood tests for colorectal adenoma detection. Ann. Intern. Med. 2009, 150, 162–169. [Google Scholar] [CrossRef]
- Brenner, H.; Tao, S.; Haug, U. Low-dose aspirin use and performance of immunochemical fecal occult blood tests. JAMA 2010, 304, 2513–2520. [Google Scholar] [CrossRef] [PubMed]
- Gies, A.; Cuk, K.; Schrotz-King, P.; Brenner, H. Direct comparison of diagnostic performance of 9 quantitative fecal immunochemical tests for colorectal cancer screening. Gastroenterology 2018, 154, 93–104. [Google Scholar] [CrossRef]
- Brenner, H.; Hoffmeister, M.; Arndt, V.; Stegmaier, C.; Altenhofen, L.; Haug, U. Protection from right- and left-sided colorectal neoplasms after colonoscopy: Population-based study. J. Natl. Cancer Inst. 2010, 102, 89–95. [Google Scholar] [CrossRef]
- Percy, A.J.; Chambers, A.G.; Parker, C.E.; Borchers, C.H. Absolute quantitation of proteins in human blood by multiplexed multiple reaction monitoring mass spectrometry. Methods Mol. Biol. 2013, 1000, 167–189. [Google Scholar]
- Assarsson, E.; Lundberg, M.; Holmquist, G.; Björkesten, J.; Bucht Thorsen, S.; Ekman, D.; Eriksson, A.; Rennel Dickens, E.; Ohlsson, S.; Edfeldt, G.; et al. Homogenous 96-plex pea immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE 2014, 9, e95192. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B-Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Efron, B.; Tibshirani, R. Improvements on cross-validation: The 632+ bootstrap method. J. Am. Stat. Assoc. 1997, 92, 548–560. [Google Scholar]
- The R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Gies, A.; Bhardwaj, M.; Stock, C.; Schrotz-King, P.; Brenner, H. Quantitative fecal immunochemical tests for colorectal cancer screening. Int. J. Cancer. 2018, 143, 234–244. [Google Scholar] [CrossRef] [PubMed]
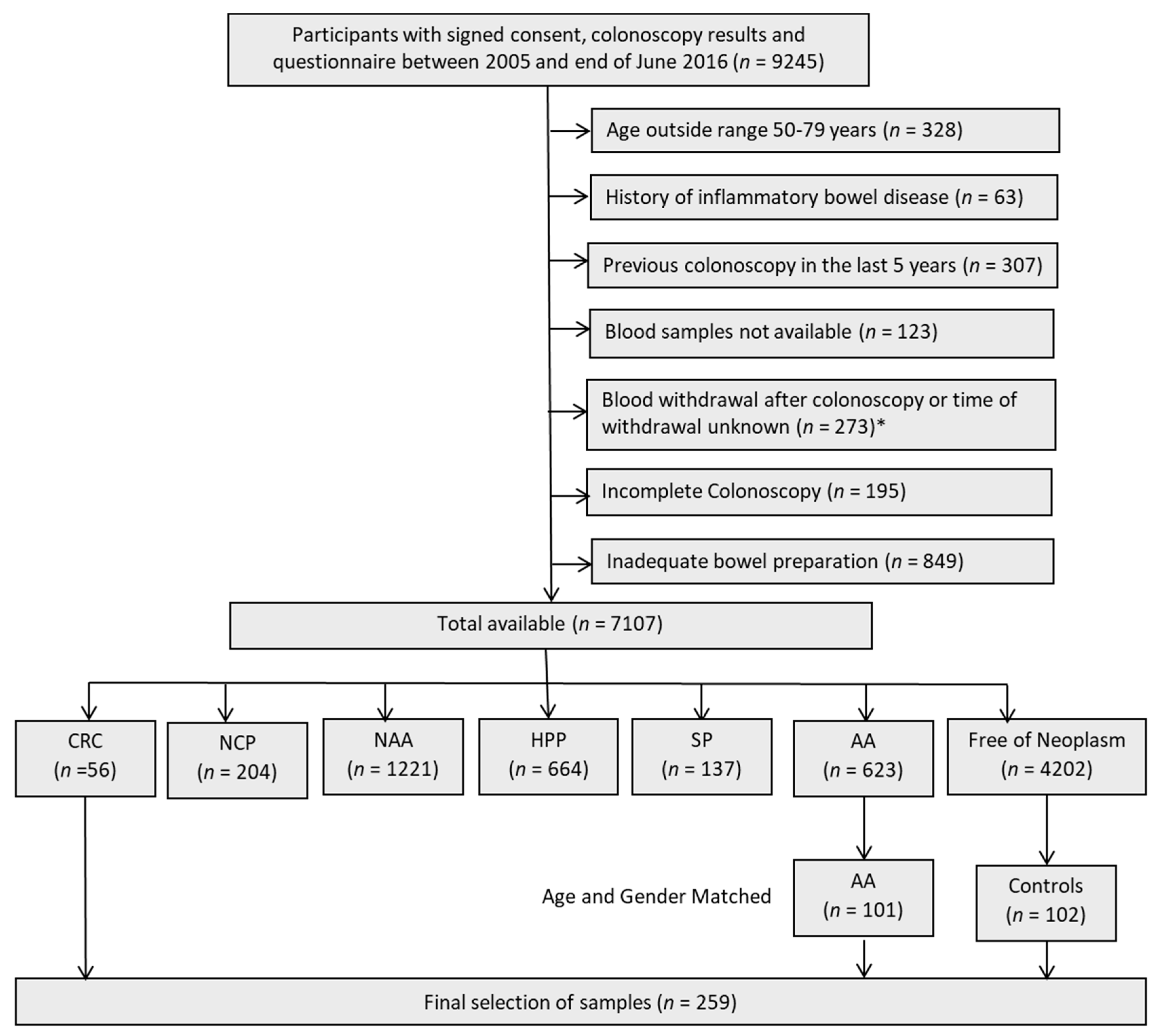
| Group | Discovery Set | Validation Set | Participants of Screening Colonoscopy | ||||
|---|---|---|---|---|---|---|---|
| iDa (Clinical) CRC | ASTER (Mostly Screening) Controls | BLITZ Matched Set (Screening) | BLITZ (Screening) | ||||
| CRC | AA | Controls | AA | Controls | |||
| Total | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) |
| 96 | 94 | 56 | 101 | 102 | 623 | 4202 | |
| Age in years | |||||||
| 50–59 | 22 (23) | 25 (27) | 10 (18) | 22 (21) | 21 (21) | 237 (38) | 1916 (46) |
| 60–69 | 41 (43) | 44 (46) | 28 (50) | 49 (49) | 50 (49) | 247 (40) | 1614 (38) |
| 70–79 | 33 (34) | 25 (27) | 18 (32) | 30 (30) | 31 (30) | 139 (22) | 672 (16) |
| Mean | 64.8 | 64.1 | 66.0 | 65.5 | 65.4 | 63.3 | 61.9 |
| Median | 65.0 | 66.0 | 65.0 | 65.0 | 65.5 | 62.0 | 60.0 |
| SD | 7.0 | 7.4 | 5.8 | 6.6 | 6.9 | 5.9 | 6.5 |
| Gender distribution | |||||||
| Male | 59 (61) | 55 (59) | 36 (64) | 65 (64) | 66 (65) | 393 (63) | 1808 (43) |
| Female | 37 (39) | 39 (41) | 20 (36) | 36 (36) | 36 (35) | 230 (37) | 2394 (57) |
| Stage distribution | |||||||
| Stage I | 17 (18) | - | 17 (30) | - | - | - | - |
| Stage II | 31 (32) | - | 6 (11) | - | - | - | - |
| Stage III | 22 (23) | - | 26 (46) | - | - | - | - |
| Stage IV | 26 (27) | - | (13) | - | - | - | - |
| Early stage (I/II) | 48 (50) | - | 23 (41) | - | - | - | - |
| Late stage (III/IV) | 48 (50) | - | 33 (59) | - | - | - | - |
| Protein Biomarkers | Discovery Set (LC/MRM-MS Measurements) | Discovery Set (PEA Measurements) | Validation Set (PEA Measurements) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p-val | p-valadj | AUC (95% CI) | AUCBS (95% CI) | Se% at 90% Sp | p-val | p-valadj | AUC (95% CI) | AUCBS (95% CI) | Se% at 90% Sp | p-val | p-valadj | AUC (95% CI) | AUCBS (95% CI) | Se% at 90% Sp | |
| CDH5 | 0.35 | 0.38 | 0.54 (0.46–0.62) | 0.48 (0.36–0.61) | 8 | 0.20 | 0.25 | 0.55 (0.47–0.64) | 0.50 (0.36–0.64) | 10 | <0.05 | <0.05 | 0.62 (0.54–0.71) | 0.59 (0.50–0.73) | 12 |
| Gal 3 | 0.51 | 0.51 | 0.53 (0.45–0.61) | 0.47 (0.36–0.59) | 7 | 0.84 | 0.84 | 0.51 (0.43–0.59) | 0.46 (0.37–0.55) | 7 | 0.13 | 0.16 | 0.57 (0.48–0.67) | 0.51 (0.33–0.67) | 11 |
| IGFBP2 | <0.05 | <0.05 | 0.61 (0.53–0.69) | 0.58 (0.50–0.71) | 24 | <0.05 | <0.05 | 0.61 (0.53–0.69) | 0.58 (0.50–0.72) | 21 | 0.43 | 0.46 | 0.54 (0.44–0.63) | 0.48 (0.34–0.62) | 10 |
| MASP1 | <0.001 | <0.001 | 0.68 (0.61–0.76) | 0.67 (0.58–0.78) | 27 | <0.001 | <0.001 | 0.65 (0.57–0.72) | 0.63 (0.53–0.75) | 20 | 0.08 | 0.11 | 0.58 (0.49–0.68) | 0.53 (0.37–0.68) | 13 |
| MMP9 | 0.24 | 0.33 | 0.55 (0.47–0.63) | 0.50 (0.37–0.62) | 10 | 0.32 | 0.36 | 0.55 (0.38–0.54) | 0.50 (0.38–0.62) | 10 | 0.05 | 0.08 | 0.59 (0.50–0.69) | 0.55 (0.46–0.71) | 16 |
| MPO | 0.06 | 0.11 | 0.58 (0.50–0.66) | 0.47 (0.36–0.59) | 8 | <0.005 | <0.005 | 0.63 (0.55–0.71) | 0.60 (0.52–0.73) | 17 | 0.53 | 0.53 | 0.53 (0.44–0.62) | 0.46 (0.35–0.57) | 6 |
| OPN | <0.001 | <0.001 | 0.64 (0.57–0.72) | 0.62 (0.54–0.75) | 26 | <0.001 | <0.001 | 0.75 (0.68–0.82) | 0.73 (0.65–0.84) | 35 | <0.05 | <0.05 | 0.62 (0.53–0.71) | 0.59 (0.49–0.73) | 18 |
| PON3 | <0.001 | <0.001 | 0.73 (0.66–0.80) | 0.72 (0.63–0.82) | 32 | <0.001 | <0.001 | 0.75 (0.68–0.82) | 0.74 (0.66–0.84) | 43 | 0.05 | 0.08 | 0.59 (0.51–0.68) | 0.54 (0.37–0.69) | 11 |
| PRTN3 | 0.13 | 0.21 | 0.56 (0.48–0.64) | 0.51 (0.36–0.65) | 10 | <0.001 | <0.001 | 0.64 (0.56–0.72) | 0.61 (0.54–0.74) | 18 | 0.06 | 0.09 | 0.59 (0.50–0.68) | 0.49 (0.31–0.65) | 5 |
| SPARC | 0.30 | 0.37 | 0.54 (0.46–0.63) | 0.49 (0.37–0.61) | 10 | 0.33 | 0.36 | 0.54 (0.46–0.62) | 0.49 (0.36–0.62) | 9 | 0.41 | 0.46 | 0.54 (0.45–0.63) | 0.48 (0.36–0.61) | 5 |
| TR | <0.001 | <0.001 | 0.67 (0.60–0.75) | 0.66 (0.57–0.77) | 35 | <0.001 | <0.001 | 0.70 (0.63–0.77) | 0.69 (0.60–0.79) | 36 | <0.001 | <0.001 | 0.74 (0.66–0.82) | 0.72 (0.64–0.85) | 33 |
| AREG | - | - | - | - | - | <0.001 | <0.001 | 0.79 (0.73–0.86) | 0.78 (0.70–0.87) | 54 | <0.001 | <0.001 | 0.72 (0.64–0.80) | 0.70 (0.61–0.83) | 35 |
| Protein Biomarkers Discovered in the Signatures | Discovery Set (LC/MRM-MS Measurements) | Discovery Set (PEA Measurements) | Validation Set (PEA Measurements) as in Screening Population | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AUCBS | AUC | Se % at 80% Sp | Se % at 90% Sp | AUCBS | AUC | Se % at 80% Sp | Se % at 90% Sp | Weighted AUC (95% CI) | Se % at 80% Sp | Se % at 90% Sp | |
| All stages CRC | |||||||||||
| MASP1 + OPN + PON3 + TR # | 0.80 | 0.80 | 65 | 35 | - | - | - | - | 0.76 (0.67–0.85) | 46 | 36 |
| - | - | - | - | 0.84 | 0.85 | 73 | 42 | 0.75 (0.65–0.84) | 46 | 36 | |
| AREG + MASP1 + OPN+ PON3 + TR § | - | - | - | - | 0.87 | 0.87 | 74 | 57 | 0.82 (0.74–0.89) | 71 | 50 |
| Early stages CRC | |||||||||||
| MASP1 + OPN + PON3 + TR # | 0.75 | 0.77 | 56 | 38 | - | - | - | - | 0.78 (0.66–0.88) | 43 | 30 |
| - | - | - | - | 0.79 | 0.81 | 64 | 30 | 0.75 (0.63–0.86) | 52 | 35 | |
| AREG + MASP1 + OPN + PON3 + TR § | - | - | - | - | 0.81 | 0.83 | 69 | 42 | 0.86 (0.77–0.92) | 83 | 43 |
| Late stages CRC | |||||||||||
| MASP1 + OPN+ PON3 + TR # | 0.84 | 0.86 | 72 | 59 | - | - | - | - | 0.71 (0.59–0.83) | 48 | 21 |
| - | - | - | - | 0.90 | 0.92 | 85 | 67 | 0.72 (0.59–0.83) | 55 | 33 | |
| AREG + MASP1 + OPN+ PON3 + TR § | - | - | - | - | 0.92 | 0.94 | 88 | 78 | 0.76 (0.64–0.86) | 58 | 45 |
| AA | |||||||||||
| MASP1 + OPN+ PON3 + TR # | - | - | - | - | - | - | - | - | 0.58 (0.48–0.68) | 28 | 19 |
| - | - | - | - | - | - | - | - | 0.59 (0.49–0.68) | 32 | 21 | |
| AREG + MASP1 + OPN + PON3 + TR § | - | - | - | - | - | - | - | - | 0.60 (0.51–0.69) | 36 | 23 |
| Biomarkers | Name | Uniprot ID | Molecular Function | Biological Process |
|---|---|---|---|---|
| AREG | amphiregulin | P15514 | cytokine, growth factor | Cell–cell signaling, cell proliferation |
| MASP1 | mannan-binding lectin serine protease 1 | P48740 | hydrolase, protease, serine protease | Complement activation lectin pathway, Immunity, Innate immunity |
| OPN/SPP1 | osteopontin | P10451 | cytokine | Biomineralization, Cell adhesion |
| PON3 | serum paraoxonase lactonase 3 | Q15166 | hydrolase | Calcium, Metal-binding |
| TR/TFRC | transferrin receptor protein 1 | P02786 | host cell receptor for virus entry, receptor | Endocytosis, Host–virus interaction |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhardwaj, M.; Gies, A.; Weigl, K.; Tikk, K.; Benner, A.; Schrotz-King, P.; Borchers, C.H.; Brenner, H. Evaluation and Validation of Plasma Proteins Using Two Different Protein Detection Methods for Early Detection of Colorectal Cancer. Cancers 2019, 11, 1426. https://doi.org/10.3390/cancers11101426
Bhardwaj M, Gies A, Weigl K, Tikk K, Benner A, Schrotz-King P, Borchers CH, Brenner H. Evaluation and Validation of Plasma Proteins Using Two Different Protein Detection Methods for Early Detection of Colorectal Cancer. Cancers. 2019; 11(10):1426. https://doi.org/10.3390/cancers11101426
Chicago/Turabian StyleBhardwaj, Megha, Anton Gies, Korbinian Weigl, Kaja Tikk, Axel Benner, Petra Schrotz-King, Christoph H. Borchers, and Hermann Brenner. 2019. "Evaluation and Validation of Plasma Proteins Using Two Different Protein Detection Methods for Early Detection of Colorectal Cancer" Cancers 11, no. 10: 1426. https://doi.org/10.3390/cancers11101426
APA StyleBhardwaj, M., Gies, A., Weigl, K., Tikk, K., Benner, A., Schrotz-King, P., Borchers, C. H., & Brenner, H. (2019). Evaluation and Validation of Plasma Proteins Using Two Different Protein Detection Methods for Early Detection of Colorectal Cancer. Cancers, 11(10), 1426. https://doi.org/10.3390/cancers11101426






