microRNA-23a in Human Cancer: Its Roles, Mechanisms and Therapeutic Relevance
Abstract
1. Introduction
2. Diagnostic and Prognostic Values of miR-23a in Cancer
2.1. miR-23a Expression in Cancer
2.2. Clinical Significance of Tissue Expressing miR-23a in Cancer
2.3. miR-23a as a Non-Invasive Marker in Cancer Diagnosis
2.4. The Prognostic Value of miR-23a in Cancer
3. Biological Role of the miR-23a in Human Cancer
3.1. Carcinogenesis
3.2. Cancer Cell Proliferation and Survival
3.3. Cancer Cell Migration and Invasion
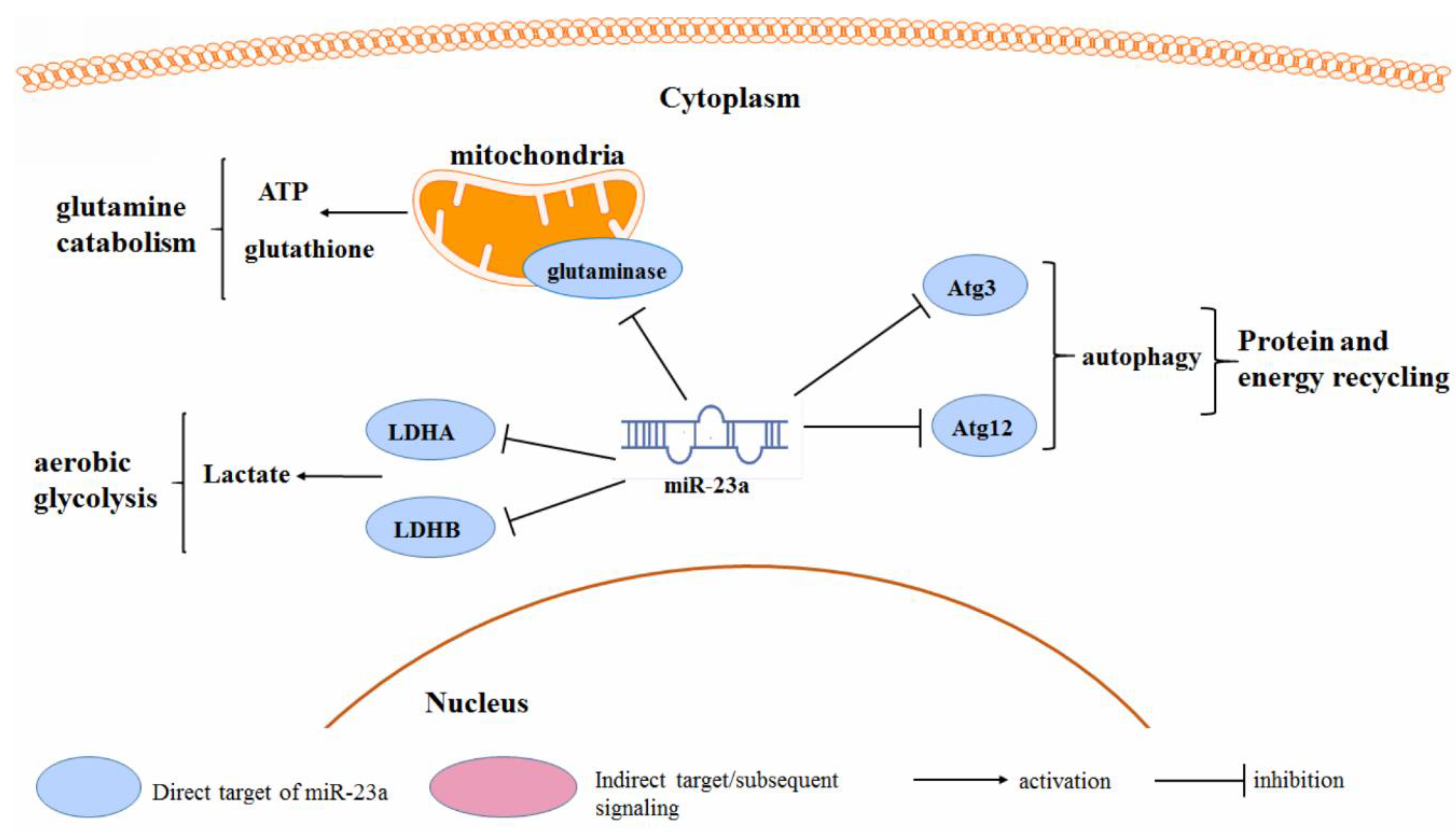
3.4. Cancer Metabolism
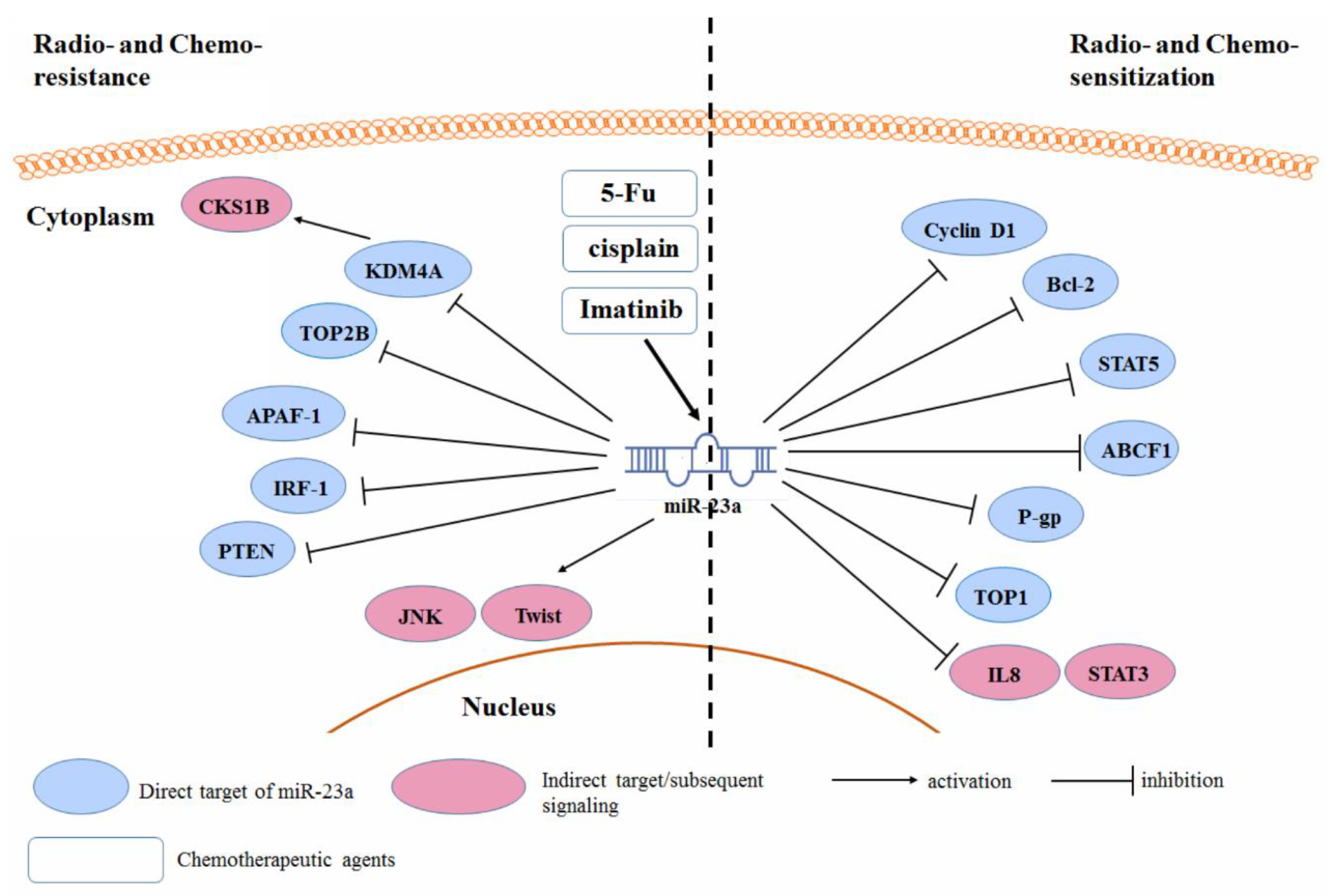
3.5. Radio- and Chemo-Resistance
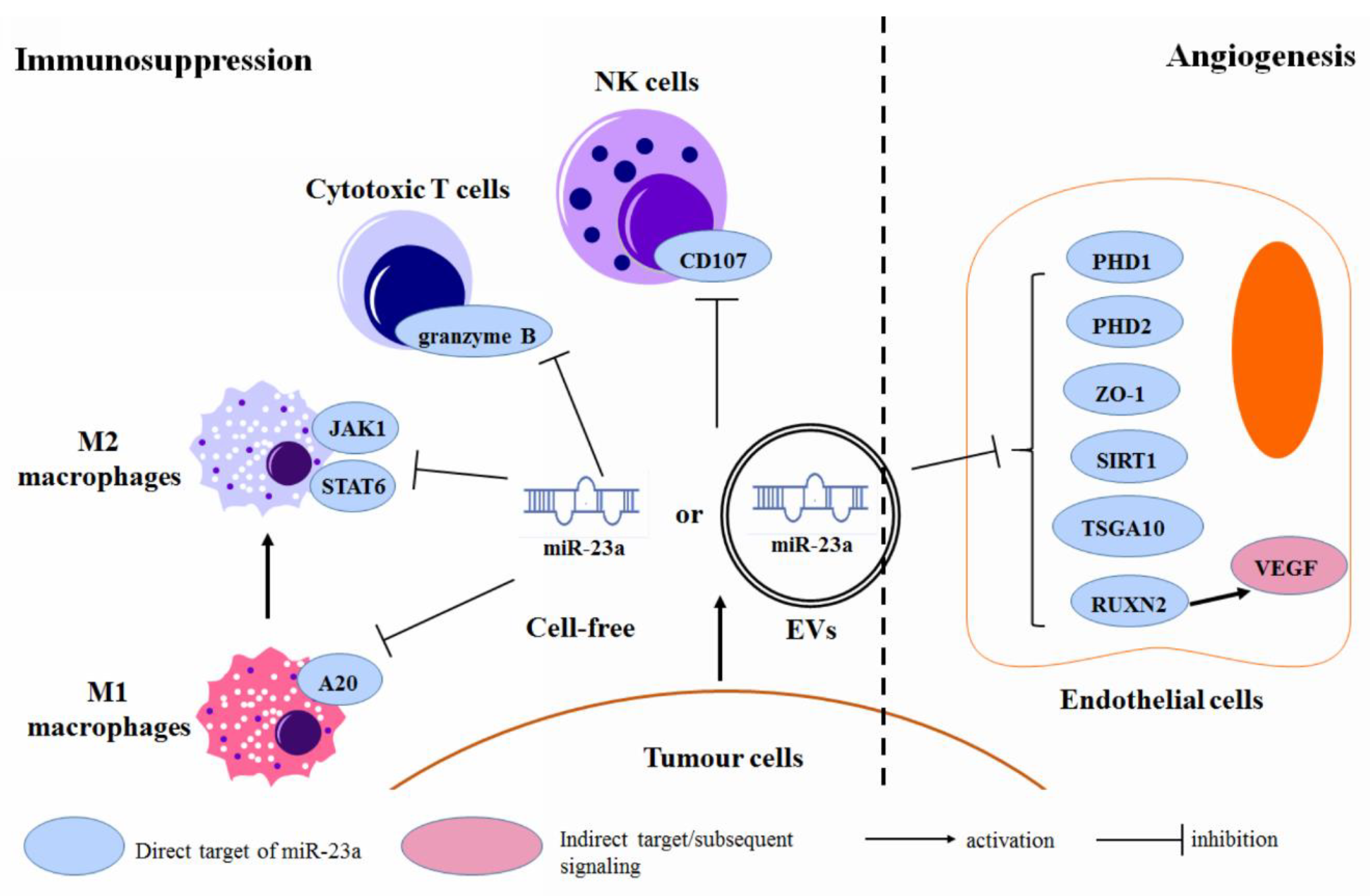
3.6. Regulation of the Tumor Microenvironment
4. Regulation of the miR-23a Expression in Human Cancer
4.1. Transcriptional Factors
4.2. Epigenetic Factors
5. Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pekarsky, Y.; Balatti, V.; Croce, C.M. BCL2 and miR-15/16: From gene discovery to treatment. Cell Death Differ. 2018, 25, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Vidigal, J.A.; Ventura, A. The biological functions of miRNAs: Lessons from in vivo studies. Trends Cell Biol. 2015, 25, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Acunzo, M.; Romano, G.; Wernicke, D.; Croce, C.M. MicroRNA and cancer—A brief overview. Adv. Biol. Regul. 2015, 57, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kurkewich, J.L.; Hansen, J.; Klopfenstein, N.; Zhang, H.; Wood, C.; Boucher, A.; Hickman, J.; Muench, D.E.; Grimes, H.L.; Dahl, R. The miR-23a~27a~24-2 microRNA cluster buffers transcription and signaling pathways during hematopoiesis. PLoS Genet. 2017, 13, e1006887. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Peng, B.; Han, Y. MiR-23a regulates the proliferation and migration of human pulmonary artery smooth muscle cells (HPASMCs) through targeting BMPR2/Smad1 signaling. Biomed. Pharmacother. 2018, 103, 1279–1286. [Google Scholar] [CrossRef]
- Wang, N.; Zhu, M.; Tsao, S.W.; Man, K.; Zhang, Z.; Feng, Y. MiR-23a-mediated inhibition of topoisomerase 1 expression potentiates cell response to etoposide in human hepatocellular carcinoma. Mol. Cancer 2013, 12, 119. [Google Scholar] [CrossRef]
- Hatzl, S.; Geiger, O.; Kuepper, M.K.; Caraffini, V.; Seime, T.; Furlan, T.; Nussbaumer, E.; Wieser, R.; Pichler, M.; Scheideler, M.; et al. Increased Expression of miR-23a Mediates a Loss of Expression in the RAF Kinase Inhibitor Protein RKIP. Cancer Res. 2016, 76, 3644–3654. [Google Scholar] [CrossRef]
- Zhu, X.; Lin, Z.; Du, J.; Zhou, X.; Yang, L.; Liu, G. Studies on microRNAs that are correlated with the cancer stem cells in chronic myeloid leukemia. Mol. Cell. Biochem. 2014, 390, 75–84. [Google Scholar] [CrossRef]
- Xishan, Z.; Xianjun, L.; Ziying, L.; Guangxin, C.; Gang, L. The malignancy suppression role of miR-23a by targeting the BCR/ABL oncogene in chromic myeloid leukemia. Cancer Gene Ther. 2014, 21, 397–404. [Google Scholar] [CrossRef]
- Su, R.; Dong, L.; Zou, D.; Zhao, H.; Ren, Y.; Li, F.; Yi, P.; Li, L.; Zhu, Y.; Ma, Y.; et al. microRNA-23a, -27a and -24 synergistically regulate JAK1/Stat3 cascade and serve as novel therapeutic targets in human acute erythroid leukemia. Oncogene 2016, 35, 6001–6014. [Google Scholar] [CrossRef] [PubMed]
- Aghaee-Bakhtiari, S.H.; Arefian, E.; Naderi, M.; Noorbakhsh, F.; Nodouzi, V.; Asgari, M.; Fard-Esfahani, P.; Mahdian, R.; Soleimani, M. MAPK and JAK/STAT pathways targeted by miR-23a and miR-23b in prostate cancer: Computational and in vitro approaches. Tumour Biol. 2015, 36, 4203–4212. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Chen, R.; Li, X.; Cai, Y.; Ye, Z.; Li, S.; Li, J.; Huang, H.; Peng, S.; Wang, J.; et al. Downregulation of microRNA-23a suppresses prostate cancer metastasis by targeting the PAK6-LIMK1 signaling pathway. Oncotarget 2015, 6, 3904–3917. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Wang, C.; Ma, C.; Wu, Q.; Zhang, W.; Lao, G. MicroRNA-23a regulates epithelial-to-mesenchymal transition in endometrial endometrioid adenocarcinoma by targeting SMAD3. Cancer Cell Int. 2016, 16, 67. [Google Scholar] [CrossRef] [PubMed]
- Koller, K.; Das, S.; Leuschner, I.; Korbelius, M.; Hoefler, G.; Guertl, B. Identification of the transcription factor HOXB4 as a novel target of miR-23a. Genes Chromosom. Cancer 2013, 52, 709–715. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Meng, C.; Shao, Z.; Wang, H.; Yang, S. MiR-23a functions as a tumor suppressor in osteosarcoma. Cell. Physiol. Biochem. 2014, 34, 1485–1496. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, B.; Fu, Y.; He, M.; Wang, J.; Shen, P.; Bai, L. miR-23a suppresses proliferation of osteosarcoma cells by targeting SATB1. Tumour Biol. 2015, 36, 4715–4721. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Wang, H.; Yang, Y.; Guo, S.; Zhang, W.; Liu, Y.; Yi, X.; Ma, J.; Zhao, T.; Liu, L.; et al. Down-regulated miR-23a Contributes to the Metastasis of Cutaneous Melanoma by Promoting Autophagy. Theranostics 2017, 7, 2231–2249. [Google Scholar] [CrossRef]
- Gottardo, F.; Liu, C.G.; Ferracin, M.; Calin, G.A.; Fassan, M.; Bassi, P.; Sevignani, C.; Byrne, D.; Negrini, M.; Pagano, F.; et al. Micro-RNA profiling in kidney and bladder cancers. Urol. Oncol. 2007, 25, 387–392. [Google Scholar] [CrossRef]
- Eissa, S.; Matboli, M.; Shehata, H.H. Breast tissue-based microRNA panel highlights microRNA-23a and selected target genes as putative biomarkers for breast cancer. Trans. Res. 2015, 165, 417–427. [Google Scholar] [CrossRef]
- Meng, F.; Henson, R.; Lang, M.; Wehbe, H.; Maheshwari, S.; Mendell, J.T.; Jiang, J.; Schmittgen, T.D.; Patel, T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology 2006, 130, 2113–2129. [Google Scholar] [CrossRef] [PubMed]
- Jahid, S.; Sun, J.; Edwards, R.A.; Dizon, D.; Panarelli, N.C.; Milsom, J.W.; Sikandar, S.S.; Gumus, Z.H.; Lipkin, S.M. miR-23a promotes the transition from indolent to invasive colorectal cancer. Cancer Discov. 2012, 2, 540–553. [Google Scholar] [CrossRef] [PubMed]
- Yong, F.L.; Wang, C.W.; Roslani, A.C.; Law, C.W. The involvement of miR-23a/APAF1 regulation axis in colorectal cancer. Int. J. Mol. Sci. 2014, 15, 11713–11729. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Jin, L.; Jiang, R.; Wang, Q.; Jiang, J.; Mao, C.; Chen, D. Correlations between miRNAs and TGF-β1 in tumor microenvironment of esophageal squamous cell cancer. Chin J. Cell. Mol. Immunol. 2013, 29, 524–528. [Google Scholar]
- Zhu, L.H.; Liu, T.; Tang, H.; Tian, R.Q.; Su, C.; Liu, M.; Li, X. MicroRNA-23a promotes the growth of gastric adenocarcinoma cell line MGC803 and downregulates interleukin-6 receptor. FEBS J. 2010, 277, 3726–3734. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Dai, W.; Sang, A.; Yang, X.; Gao, C. Upregulation of microRNA-23a/b promotes tumor progression and confers poor prognosis in patients with gastric cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 8833–8840. [Google Scholar]
- Hu, X.; Wang, Y.; Liang, H.; Fan, Q.; Zhu, R.; Cui, J.; Zhang, W.; Zen, K.; Zhang, C.Y.; Hou, D.; et al. miR-23a/b promote tumor growth and suppress apoptosis by targeting PDCD4 in gastric cancer. Cell Death Dis. 2017, 8, e3059. [Google Scholar] [CrossRef]
- Hua, K.; Chen, Y.T.; Chen, C.F.; Tang, Y.S.; Huang, T.T.; Lin, Y.C.; Yeh, T.S.; Huang, K.H.; Lee, H.C.; Hsu, M.T.; et al. MicroRNA-23a/27a/24-2 cluster promotes gastric cancer cell proliferation synergistically. Oncol. Lett. 2018, 16, 2319–2325. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Y.; Zhang, H.; Liu, X.; Gong, T.; Li, M.; Sun, L.; Ji, G.; Shi, Y.; Han, Z.; et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol. Cancer Res. 2011, 9, 824–833. [Google Scholar] [CrossRef]
- An, J.; Pan, Y.; Yan, Z.; Li, W.; Cui, J.; Yuan, J.; Tian, L.; Xing, R.; Lu, Y. MiR-23a in amplified 19p13.13 loci targets metallothionein 2A and promotes growth in gastric cancer cells. J. Cell. Biochem. 2013, 114, 2160–2169. [Google Scholar] [CrossRef]
- Li, Y.; Chen, H.; She, P.; Chen, T.; Chen, L.; Yuan, J.; Jiang, B. microRNA-23a promotes cell growth and metastasis in gastric cancer via targeting SPRY2-mediated ERK signaling. Oncol. Lett. 2018, 15, 8433–8441. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.A.; Santosh, V.; Somasundaram, K. Genome-wide expression profiling identifies deregulated miRNAs in malignant astrocytoma. Mod. Pathol. 2010, 23, 1404–1417. [Google Scholar] [CrossRef] [PubMed]
- Koshkin, P.A.; Chistiakov, D.A.; Nikitin, A.G.; Konovalov, A.N.; Potapov, A.A.; Usachev, D.Y.; Pitskhelauri, D.I.; Kobyakov, G.L.; Shishkina, L.V.; Chekhonin, V.P. Analysis of expression of microRNAs and genes involved in the control of key signaling mechanisms that support or inhibit development of brain tumors of different grades. Clin. Chim. Acta 2014, 430, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Lian, S.; Shi, R.; Bai, T.; Liu, Y.; Miao, W.; Wang, H.; Liu, X.; Fan, Y. Anti-miRNA-23a oligonucleotide suppresses glioma cells growth by targeting apoptotic protease activating factor-1. Curr. Pharm. Des. 2013, 19, 6382–6389. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Chen, D.; Cui, Y.; Li, Z.; Huang, J. Targeting microRNA-23a to inhibit glioma cell invasion via HOXD10. Sci. Rep. 2013, 3, 3423. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; He, X.; Ding, J.; Liang, L.; Zhao, Y.; Zhang, Z.; Yao, X.; Pan, Z.; Zhang, P.; Li, J.; et al. Upregulation of miR-23a approximately 27a approximately 24 decreases transforming growth factor-beta-induced tumor-suppressive activities in human hepatocellular carcinoma cells. Int. J. Cancer 2008, 123, 972–978. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Zhao, J.; Dai, X.; Wang, Y.; Ma, R.; Su, Y.; Cui, H.; Niu, J.; Bai, S.; Xiao, Z.; et al. Correlation between miR-23a and onset of hepatocellular carcinoma. Clin. Res. Hepatal. Gastroenterol. 2014, 38, 318–330. [Google Scholar] [CrossRef]
- Wang, W.L.; Yang, C.; Han, X.L.; Wang, R.; Huang, Y.; Zi, Y.M.; Li, J.D. MicroRNA-23a expression in paraffin-embedded specimen correlates with overall survival of diffuse large B-cell lymphoma. Med. Oncol. 2014, 31, 919. [Google Scholar] [CrossRef]
- Li, B.; Sun, M.; Gao, F.; Liu, W.; Yang, Y.; Liu, H.; Cheng, Y.; Liu, C.; Cai, J. Up-regulated expression of miR-23a/b targeted the pro-apoptotic Fas in radiation-induced thymic lymphoma. Cell. Physiol. Biochem. 2013, 32, 1729–1740. [Google Scholar] [CrossRef]
- Zhang, X.W.; Liu, N.; Chen, S.; Wang, Y.; Zhang, Z.X.; Sun, Y.Y.; Qiu, G.B.; Fu, W.N. High microRNA-23a expression in laryngeal squamous cell carcinoma is associated with poor patient prognosis. Diagn. Pathol. 2015, 10, 22. [Google Scholar] [CrossRef]
- Cao, M.; Li, Y.; Lu, H.; Meng, Q.; Wang, L.; Cai, L.; Dong, X. MiR-23a-mediated migration/invasion is rescued by its target, IRS-1, in non-small cell lung cancer cells. J. Cancer Res. Clin. Oncol. 2014, 140, 1661–1670. [Google Scholar] [CrossRef] [PubMed]
- Qu, W.Q.; Liu, L.; Yu, Z. Clinical value of microRNA-23a upregulation in non-small cell lung cancer. Int. J. Clin. Exp. Med. 2015, 8, 13598–13603. [Google Scholar] [PubMed]
- Cheng, Y.Y.; Kirschner, M.B.; Cheng, N.C.; Gattani, S.; Klebe, S.; Edelman, J.J.; Vallely, M.P.; McCaughan, B.C.; Jin, H.C.; van Zandwijk, N.; et al. ZIC1 is silenced and has tumor suppressor function in malignant pleural mesothelioma. J. Thorac. Oncol. 2013, 8, 1317–1328. [Google Scholar] [CrossRef] [PubMed]
- Vaksman, O.; Stavnes, H.T.; Kaern, J.; Trope, C.G.; Davidson, B.; Reich, R. miRNA profiling along tumour progression in ovarian carcinoma. J. Cell. Mol. Med. 2011, 15, 1593–1602. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, X.L.; Bai, R.; Liu, W.Y.; Li, X.; Liu, M.; Tang, H. miR-23a promotes IKKalpha expression but suppresses ST7L expression to contribute to the malignancy of epithelial ovarian cancer cells. Br. J. Cancer 2016, 115, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Piepoli, A.; Tavano, F.; Copetti, M.; Mazza, T.; Palumbo, O.; Panza, A.; di Mola, F.F.; Pazienza, V.; Mazzoccoli, G.; Biscaglia, G.; et al. Mirna expression profiles identify drivers in colorectal and pancreatic cancers. PLoS ONE 2012, 7, e33663. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Li, Z.; Jiang, P.; Zhang, X.; Xu, Y.; Chen, K.; Li, X. MicroRNA-23a promotes pancreatic cancer metastasis by targeting epithelial splicing regulator protein 1. Oncotarget 2017, 8, 82854–82871. [Google Scholar] [CrossRef]
- Quan, J.; Jin, L.; Pan, X.; He, T.; Lai, Y.; Chen, P.; Lin, C.; Yang, S.; Zeng, H.; Lai, Y. Oncogenic miR-23a-5p is associated with cellular function in RCC. Mol. Med. Rep. 2017, 16, 2309–2317. [Google Scholar] [CrossRef]
- Tang, H.L.; Deng, M.; Liao, Q.J.; Zeng, X.; Zhou, X.T.; Su, Q. Expression and clinical significance of miR-23a and metastasis suppressor 1 in colon carcinoma. Chin. J. Pathol. 2012, 41, 28–32. [Google Scholar]
- Bao, L.; You, B.; Shi, S.; Shan, Y.; Zhang, Q.; Yue, H.; Zhang, J.; Zhang, W.; Shi, Y.; Liu, Y.; et al. Metastasis-associated miR-23a from nasopharyngeal carcinoma-derived exosomes mediates angiogenesis by repressing a novel target gene TSGA10. Oncogene 2018, 37, 2873–2889. [Google Scholar] [CrossRef]
- Su, L.; Liu, M. Correlation analysis on the expression levels of microRNA-23a and microRNA-23b and the incidence and prognosis of ovarian cancer. Oncol. Lett. 2018, 16, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Li, W.; Liu, C.; Li, W.; Yu, H.; Lei, B.; Ren, Y.; Li, Z.; Pang, D.; Qian, C. MiR-23a promotes TGF-beta1-induced EMT and tumor metastasis in breast cancer cells by directly targeting CDH1 and activating Wnt/beta-catenin signaling. Oncotarget 2017, 8, 69538–69550. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Lu, Z.; Li, H.; Lu, J.; Guo, L.; Ge, Q. Next-generation sequencing of microRNAs for breast cancer detection. J. Biomed. Biotechnol. 2011, 2011, 597145. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Wang, C.; Lu, Z.; Guo, L.; Ge, Q. Analysis of serum genome-wide microRNAs for breast cancer detection. Clin. Chim. Acta 2012, 413, 1058–1065. [Google Scholar] [CrossRef] [PubMed]
- Nakatani, F.; Ferracin, M.; Manara, M.C.; Ventura, S.; Del Monaco, V.; Ferrari, S.; Alberghini, M.; Grilli, A.; Knuutila, S.; Schaefer, K.L.; et al. miR-34a predicts survival of Ewing’s sarcoma patients and directly influences cell chemo-sensitivity and malignancy. J. Pathol. 2012, 226, 796–805. [Google Scholar] [CrossRef] [PubMed]
- Frampton, A.E.; Castellano, L.; Colombo, T.; Giovannetti, E.; Krell, J.; Jacob, J.; Pellegrino, L.; Roca-Alonso, L.; Funel, N.; Gall, T.M.; et al. Integrated molecular analysis to investigate the role of microRNAs in pancreatic tumour growth and progression. Lancet 2015, 385 (Suppl. 1), S37. [Google Scholar] [CrossRef]
- Komatsu, S.; Ichikawa, D.; Kawaguchi, T.; Takeshita, H.; Miyamae, M.; Ohashi, T.; Okajima, W.; Imamura, T.; Kiuchi, J.; Arita, T.; et al. Plasma microRNA profiles: Identification of miR-23a as a novel biomarker for chemoresistance in esophageal squamous cell carcinoma. Oncotarget 2016, 7, 62034–62048. [Google Scholar] [CrossRef]
- Yang, C.; Wang, C.; Chen, X.; Chen, S.; Zhang, Y.; Zhi, F.; Wang, J.; Li, L.; Zhou, X.; Li, N.; et al. Identification of seven serum microRNAs from a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for malignant astrocytomas. Int. J. Cancer 2013, 132, 116–127. [Google Scholar] [CrossRef]
- Cinpolat, O.; Unal, Z.N.; Ismi, O.; Gorur, A.; Unal, M. Comparison of microRNA profiles between benign and malignant salivary gland tumors in tissue, blood and saliva samples: A prospective, case-control study. Braz. J. Otorhinolaryngol. 2017, 83, 276–284. [Google Scholar] [CrossRef]
- Humeau, M.; Vignolle-Vidoni, A.; Sicard, F.; Martins, F.; Bournet, B.; Buscail, L.; Torrisani, J.; Cordelier, P. Salivary MicroRNA in Pancreatic Cancer Patients. PLoS ONE 2015, 10, e0130996. [Google Scholar] [CrossRef]
- Khare, D.; Goldschmidt, N.; Bardugo, A.; Gur-Wahnon, D.; Ben-Dov, I.Z.; Avni, B. Plasma microRNA profiling: Exploring better biomarkers for lymphoma surveillance. PLoS ONE 2017, 12, e0187722. [Google Scholar] [CrossRef] [PubMed]
- Yong, F.L.; Law, C.W.; Wang, C.W. Potentiality of a triple microRNA classifier: MiR-193a-3p, miR-23a and miR-338-5p for early detection of colorectal cancer. BMC Cancer 2013, 13, 280. [Google Scholar] [CrossRef] [PubMed]
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H.; et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 2014, 9, e92921. [Google Scholar] [CrossRef] [PubMed]
- Ostenfeld, M.S.; Jensen, S.G.; Jeppesen, D.K.; Christensen, L.L.; Thorsen, S.B.; Stenvang, J.; Hvam, M.L.; Thomsen, A.; Mouritzen, P.; Rasmussen, M.H.; et al. miRNA profiling of circulating EpCAM(+) extracellular vesicles: Promising biomarkers of colorectal cancer. J. Extracell. Vesicles 2016, 5, 31488. [Google Scholar] [CrossRef] [PubMed]
- Vychytilova-Faltejskova, P.; Radova, L.; Sachlova, M.; Kosarova, Z.; Slaba, K.; Fabian, P.; Grolich, T.; Prochazka, V.; Kala, Z.; Svoboda, M.; et al. Serum-based microRNA signatures in early diagnosis and prognosis prediction of colon cancer. Carcinogenesis 2016, 37, 941–950. [Google Scholar] [CrossRef]
- Quan, J.; Liu, S.; Dai, K.; Jin, L.; He, T.; Pan, X.; Lai, Y. MicroRNA-23a/24-2/27a as a potential diagnostic biomarker for cancer: A systematic review and meta-analysis. Mol. Clin. Oncol. 2018, 8, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Vasaikar, S.V.; Straub, P.; Wang, J.; Zhang, B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018, 46, D956–D963. [Google Scholar] [CrossRef]
- El-Halawany, M.S.; Ismail, H.M.; Zeeneldin, A.A.; Elfiky, A.; Tantawy, M.; Kobaisi, M.H.; Hamed, I.; Abdel Wahab, A.H. Investigating the pretreatment miRNA expression patterns of advanced hepatocellular carcinoma patients in association with response to TACE treatment. Biomed Res. Int. 2015, 2015, 649750. [Google Scholar] [CrossRef]
- Qu, J.Q.; Yi, H.M.; Ye, X.; Li, L.N.; Zhu, J.F.; Xiao, T.; Yuan, L.; Li, J.Y.; Wang, Y.Y.; Feng, J.; et al. MiR-23a sensitizes nasopharyngeal carcinoma to irradiation by targeting IL-8/Stat3 pathway. Oncotarget 2015, 6, 28341–28356. [Google Scholar] [CrossRef]
- Ramassone, A.; Pagotto, S.; Veronese, A.; Visone, R. Epigenetics and MicroRNAs in Cancer. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef]
- Tsai, Y.S.; Lin, C.S.; Chiang, S.L.; Lee, C.H.; Lee, K.W.; Ko, Y.C. Areca nut induces miR-23a and inhibits repair of DNA double-strand breaks by targeting FANCG. Toxicol. Sci. 2011, 123, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Hsu, S.H.; Frankel, W.; Ghoshal, K.; Jacob, S.T. Stat3-mediated activation of microRNA-23a suppresses gluconeogenesis in hepatocellular carcinoma by down-regulating glucose-6-phosphatase and peroxisome proliferator-activated receptor gamma, coactivator 1 α. Hepatology 2012, 56, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Ebert, M.S.; Sharp, P.A. Roles for microRNAs in conferring robustness to biological processes. Cell 2012, 149, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zhu, A.; Tian, L.; Xin, Y.; Liu, X.; Peng, Y.; Zhang, J.; Miao, Y.; Wei, J. miR23a suppresses pancreatic cancer cell progression by inhibiting PLK1 expression. Mol. Med. Rep. 2018, 18, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, Q.; Fan, Y.; Wang, S.; Liu, X.; Zhu, L.; Liu, M.; Tang, H. Downregulation of PPP2R5E expression by miR-23a suppresses apoptosis to facilitate the growth of gastric cancer cells. FEBS Lett. 2014, 588, 3160–3169. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jiao, T.; Wang, Y.; Su, W.; Tang, Z.; Han, C. Long non-coding RNA GAS5 acts as a molecular sponge to regulate miR-23a in gastric cancer. Int. J. Clin. Exp. Pathol. 2016, 9, 11412–11419. [Google Scholar]
- Liu, N.; Sun, Y.Y.; Zhang, X.W.; Chen, S.; Wang, Y.; Zhang, Z.X.; Song, S.W.; Qiu, G.B.; Fu, W.N. Oncogenic miR-23a in Pancreatic Ductal Adenocarcinogenesis Via Inhibiting APAF1. Dig. Dis. Sci. 2015, 60, 2000–2008. [Google Scholar] [CrossRef]
- Zhang, X.W.; Liu, N.; Chen, S.; Wang, Y.E.; Sun, K.L.; Xu, Z.M.; Fu, W.N. Upregulation of microRNA-23a regulates proliferation and apoptosis by targeting APAF-1 in laryngeal carcinoma. Oncology Lett. 2015, 10, 410–416. [Google Scholar] [CrossRef]
- Yan, Y.; Liang, Z.; Du, Q.; Yang, M.; Geller, D.A. MicroRNA-23a downregulates the expression of interferon regulatory factor-1 in hepatocellular carcinoma cells. Oncol. Rep. 2016, 36, 633–640. [Google Scholar] [CrossRef]
- Huang, F.Y.; Wong, D.K.; Seto, W.K.; Lai, C.L.; Yuen, M.F. Estradiol induces apoptosis via activation of miRNA-23a and p53: Implication for gender difference in liver cancer development. Oncotarget 2015, 6, 34941–34952. [Google Scholar] [CrossRef]
- Wang, H.; Shen, L.; Li, X.; Sun, M. MicroRNAs contribute to the anticancer effect of 1′-acetoxychavicol acetate in human head and neck squamous cell carcinoma cell line HN4. Biosci. Biotechnol. Biochem. 2013, 77, 2348–2355. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Seike, M.; Soeno, C.; Mizutani, H.; Kitamura, K.; Minegishi, Y.; Noro, R.; Yoshimura, A.; Cai, L.; Gemma, A. MiR-23a regulates TGF-β-induced epithelial-mesenchymal transition by targeting E-cadherin in lung cancer cells. Int. J. Oncol. 2012, 41, 869–875. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Li, W.; Wang, Y.; Xie, T.; Cai, Y.; Wang, Z.; Jiang, B. miR-23a inhibits E-cadherin expression and is regulated by AP-1 and NFAT4 complex during Fas-induced EMT in gastrointestinal cancer. Carcinogenesis 2014, 35, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Yang, T.; Kuang, Y.; Kong, B.; Yu, S.; Shu, H.; Zhou, H.; Gu, J. MicroRNA-23a promotes neuroblastoma cell metastasis by targeting CDH1. Oncology Lett. 2014, 7, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Tian, K.; Di, R.; Wang, L. MicroRNA-23a enhances migration and invasion through PTEN in osteosarcoma. Cancer Gene Ther. 2015, 22, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; He, Y.H.; Huang, X.; Tao, S.Q.; Wang, X.N.; Yan, H.; Ding, K.S.; Lobie, P.E.; Wu, W.Y.; Wu, Z.S. MiR-23a modulates X-linked inhibitor of apoptosis-mediated autophagy in human luminal breast cancer cell lines. Oncotarget 2017, 8, 80709–80721. [Google Scholar] [CrossRef]
- Li, X.; Liu, X.; Xu, W.; Zhou, P.; Gao, P.; Jiang, S.; Lobie, P.E.; Zhu, T. c-MYC-regulated miR-23a/24-2/27a cluster promotes mammary carcinoma cell invasion and hepatic metastasis by targeting Sprouty2. J. Biol. Chem. 2013, 288, 18121–18133. [Google Scholar] [CrossRef]
- Chien, M.H.; Lee, W.J.; Yang, Y.C.; Li, Y.L.; Chen, B.R.; Cheng, T.Y.; Yang, P.W.; Wang, M.Y.; Jan, Y.H.; Lin, Y.K.; et al. KSRP suppresses cell invasion and metastasis through miR-23a-mediated EGR3 mRNA degradation in non-small cell lung cancer. Biochim. Biophys. Acta Gene Requl. Mech. 2017, 1860, 1013–1024. [Google Scholar] [CrossRef]
- Saumet, A.; Vetter, G.; Bouttier, M.; Antoine, E.; Roubert, C.; Orsetti, B.; Theillet, C.; Lecellier, C.H. Estrogen and retinoic acid antagonistically regulate several microRNA genes to control aerobic glycolysis in breast cancer cells. Mol. Biosyst. 2012, 8, 3242–3253. [Google Scholar] [CrossRef]
- Poyyakkara, A.; Raji, G.R.; Kunhiraman, H.; Edatt, L.; Kumar, S.V.B. ER stress mediated regulation of miR23a confer Hela cells better adaptability to utilize glycolytic pathway. J. Cell. Biochem. 2018, 119, 4907–4917. [Google Scholar] [CrossRef]
- Gao, P.; Tchernyshyov, I.; Chang, T.C.; Lee, Y.S.; Kita, K.; Ochi, T.; Zeller, K.I.; De Marzo, A.M.; Van Eyk, J.E.; Mendell, J.T.; et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009, 458, 762–765. [Google Scholar] [CrossRef] [PubMed]
- Rathore, M.G.; Saumet, A.; Rossi, J.F.; de Bettignies, C.; Tempe, D.; Lecellier, C.H.; Villalba, M. The NF-kappaB member p65 controls glutamine metabolism through miR-23a. Int. J. Biochem. Cell Biol. 2012, 44, 1448–1456. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Wang, Y.; Wang, X.; Zhou, D.; Wang, X.; Zhou, M.; He, Z. Effect of the LncRNA GAS5-MiR-23a-ATG3 Axis in Regulating Autophagy in Patients with Breast Cancer. Cell. Physiol. Biochem. 2018, 48, 194–207. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, S.; Cangemi, A.; Galvano, A.; Fanale, D.; Buscemi, S.; Ciaccio, M.; Russo, A.; Castorina, S.; Bazan, V. Analysis of miRNA expression profile induced by short term starvation in breast cancer cells treated with doxorubicin. Oncotarget 2017, 8, 71924–71932. [Google Scholar] [CrossRef] [PubMed]
- Farhadi, E.; Zaker, F.; Safa, M.; Rezvani, M.R. miR-101 sensitizes K562 cell line to imatinib through Jak2 downregulation and inhibition of NF-kappaB target genes. Tumour Biol. 2016, 37, 14117–14128. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Yang, F.; Wang, Y.; Wang, Y.; Xue, G.; Mei, Q.; Wang, F.; Sun, S. MicroRNA-23a antisense enhances 5-fluorouracil chemosensitivity through APAF-1/caspase-9 apoptotic pathway in colorectal cancer cells. J. Cell. Biochem. 2014, 115, 772–784. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, X.; Liao, D.; Wang, X.; Wu, Z.; Nie, J.; Bai, M.; Fu, X.; Mei, Q.; Han, W. Elevated microRNA-23a Expression Enhances the Chemoresistance of Colorectal Cancer Cells with Microsatellite Instability to 5-Fluorouracil by Directly Targeting ABCF1. Curr. Protein Pept. Sci. 2015, 16, 301–309. [Google Scholar] [CrossRef]
- Yu, Z.W.; Zhong, L.P.; Ji, T.; Zhang, P.; Chen, W.T.; Zhang, C.P. MicroRNAs contribute to the chemoresistance of cisplatin in tongue squamous cell carcinoma lines. Oral Oncol. 2010, 46, 317–322. [Google Scholar] [CrossRef]
- Peng, F.; Zhang, H.; Du, Y.; Tan, P. miR-23a promotes cisplatin chemoresistance and protects against cisplatin-induced apoptosis in tongue squamous cell carcinoma cells through Twist. Oncol. Rep. 2015, 33, 942–950. [Google Scholar] [CrossRef]
- Black, J.C.; Zhang, H.; Kim, J.; Getz, G.; Whetstine, J.R. Regulation of Transient Site-specific Copy Gain by MicroRNA. J. Biol. Chem. 2016, 291, 4862–4871. [Google Scholar] [CrossRef]
- Liu, X.; Ru, J.; Zhang, J.; Zhu, L.H.; Liu, M.; Li, X.; Tang, H. miR-23a targets interferon regulatory factor 1 and modulates cellular proliferation and paclitaxel-induced apoptosis in gastric adenocarcinoma cells. PLoS ONE 2013, 8, e64707. [Google Scholar] [CrossRef]
- Han, Z.; Zhou, X.; Li, S.; Qin, Y.; Chen, Y.; Liu, H. Inhibition of miR-23a increases the sensitivity of lung cancer stem cells to erlotinib through PTEN/PI3K/Akt pathway. Oncol. Rep. 2017, 38, 3064–3070. [Google Scholar] [CrossRef] [PubMed]
- Jin, A.H.; Zhou, X.P.; Zhou, F.Z. Inhibition of microRNA-23a increases cisplatin sensitivity of ovarian cancer cells: The possible molecular mechanisms. J. South. Med. Univ. 2015, 35, 125–128. [Google Scholar]
- Jin, A.H.; Wei, Z.L. Molecular mechanism of increased sensitivity of cisplatin to ovarian cancer by inhibition of microRNA-23a expression. Int. J. Clin. Exp. Med. 2015, 8, 13329–13334. [Google Scholar] [PubMed]
- Tan, H.Y.; Wang, N.; Lam, W.; Guo, W.; Feng, Y.; Cheng, Y.C. Targeting tumour microenvironment by tyrosine kinase inhibitor. Mol. Cancer 2018, 17, 43. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef]
- Endzelins, E.; Berger, A.; Melne, V.; Bajo-Santos, C.; Sobolevska, K.; Abols, A.; Rodriguez, M.; Santare, D.; Rudnickiha, A.; Lietuvietis, V.; et al. Detection of circulating miRNAs: Comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer 2017, 17, 730. [Google Scholar] [CrossRef]
- Ma, S.; Liu, M.; Xu, Z.; Li, Y.; Guo, H.; Ge, Y.; Liu, Y.; Zheng, D.; Shi, J. A double feedback loop mediated by microRNA-23a/27a/24-2 regulates M1 versus M2 macrophage polarization and thus regulates cancer progression. Oncotarget 2016, 7, 13502–13519. [Google Scholar] [CrossRef]
- Lin, R.; Chen, L.; Chen, G.; Hu, C.; Jiang, S.; Sevilla, J.; Wan, Y.; Sampson, J.H.; Zhu, B.; Li, Q.J. Targeting miR-23a in CD8+ cytotoxic T lymphocytes prevents tumor-dependent immunosuppression. J. Clin. Investig. 2014, 124, 5352–5367. [Google Scholar] [CrossRef]
- Berchem, G.; Noman, M.Z.; Bosseler, M.; Paggetti, J.; Baconnais, S.; Le Cam, E.; Nanbakhsh, A.; Moussay, E.; Mami-Chouaib, F.; Janji, B.; et al. Hypoxic tumor-derived microvesicles negatively regulate NK cell function by a mechanism involving TGF-β and miR23a transfer. Oncoimmunology 2016, 5, e1062968. [Google Scholar] [CrossRef]
- Lu, L.; Chen, X.M.; Tao, H.M.; Xiong, W.; Jie, S.H.; Li, H.Y. Regulation of the expression of zinc finger protein genes by microRNAs enriched within acute lymphoblastic leukemia-derived microvesicles. Genet. Mol. Res. 2015, 14, 11884–11895. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, T.Y.; Lee, M.S.; Mun, J.Y.; Ihm, C.; Kim, S.A. Exosome cargo reflects TGF-β1-mediated epithelial-to-mesenchymal transition (EMT) status in A549 human lung adenocarcinoma cells. Biochem. Biophys. Res. Commun. 2016, 478, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.L.; Hung, J.Y.; Chang, W.A.; Lin, Y.S.; Pan, Y.C.; Tsai, P.H.; Wu, C.Y.; Kuo, P.L. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene 2017, 36, 4929–4942. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Liu, L.; Chen, C.; Ming, P.; Huang, Q.; Li, C.; Cao, D.; Xu, X.; Ge, W. The extracellular vesicles secreted by lung cancer cells in radiation therapy promote endothelial cell angiogenesis by transferring miR-23a. PeerJ 2017, 5, e3627. [Google Scholar] [CrossRef] [PubMed]
- Sruthi, T.V.; Edatt, L.; Raji, G.R.; Kunhiraman, H.; Shankar, S.S.; Shankar, V.; Ramachandran, V.; Poyyakkara, A.; Kumar, S.V.B. Horizontal transfer of miR-23a from hypoxic tumor cell colonies can induce angiogenesis. J. Cell. Phys. 2018, 233, 3498–3514. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.D.; Guo, T.; Liu, L.; Wang, C.; Zhang, K.; Liu, H.Q.; Wang, F.; Bai, W.D.; Zhang, M.Y. MiR-23a targets RUNX2 and suppresses ginsenoside Rg1-induced angiogenesis in endothelial cells. Oncotarget 2017, 8, 58072–58085. [Google Scholar] [CrossRef] [PubMed]
- Cameron, J.E.; Fewell, C.; Yin, Q.; McBride, J.; Wang, X.; Lin, Z.; Flemington, E.K. Epstein-Barr virus growth/latency III program alters cellular microRNA expression. Virology 2008, 382, 257–266. [Google Scholar] [CrossRef]
- Honegger, A.; Schilling, D.; Bastian, S.; Sponagel, J.; Kuryshev, V.; Sultmann, H.; Scheffner, M.; Hoppe-Seyler, K.; Hoppe-Seyler, F. Dependence of intracellular and exosomal microRNAs on viral E6/E7 oncogene expression in HPV-positive tumor cells. PLoS Pathog. 2015, 11, e1004712. [Google Scholar] [CrossRef] [PubMed]
- Saito, A.; Suzuki, H.I.; Horie, M.; Ohshima, M.; Morishita, Y.; Abiko, Y.; Nagase, T. An integrated expression profiling reveals target genes of TGF-beta and TNF-alpha possibly mediated by microRNAs in lung cancer cells. PLoS ONE 2013, 8, e56587. [Google Scholar] [CrossRef]
- Alarmo, E.L.; Havunen, R.; Hayrynen, S.; Penkki, S.; Ketolainen, J.; Nykter, M.; Kallioniemi, A. Bone morphogenetic protein 4 regulates microRNA expression in breast cancer cell lines in diverse fashion. Genes Chromosom. Cancer 2016, 55, 227–236. [Google Scholar] [CrossRef]
- Wen, Y.C.; Lee, W.J.; Tan, P.; Yang, S.F.; Hsiao, M.; Lee, L.M.; Chien, M.H. By inhibiting snail signaling and miR-23a-3p, osthole suppresses the EMT-mediated metastatic ability in prostate cancer. Oncotarget 2015, 6, 21120–21136. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Sheng, Y.; Zhang, J.; Zheng, Z.; Ji, L. The altered microRNA profile in andrographolide-induced inhibition of hepatoma tumor growth. Gene 2016, 588, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Wang, N.; Tsao, S.W.; Yuen, M.F.; Feng, Y.; Wan, T.S.; Man, K.; Feng, Y. Up-regulation of microRNAs, miR21 and miR23a in human liver cancer cells treated with Coptidis rhizoma aqueous extract. Exp. Ther. Med. 2011, 2, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhu, M.; Wang, X.; Tan, H.Y.; Tsao, S.W.; Feng, Y. Berberine-induced tumor suppressor p53 up-regulation gets involved in the regulatory network of MIR-23a in hepatocellular carcinoma. Biochim. Biophys. Acta 2014, 1839, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.C.; Ye, H.; Zeng, Z.; Chin, Y.E.; Huang, Y.N.; Fu, G.H. The NF-kappaB p65/miR-23a-27a-24 cluster is a target for leukemia treatment. Oncotarget 2015, 6, 33554–33567. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.U.H.; Rathore, M.G.; Allende-Vega, N.; Vo, D.N.; Belkhala, S.; Orecchioni, S.; Talarico, G.; Bertolini, F.; Cartron, G.; Lecellier, C.H.; et al. Human Leukemic Cells performing Oxidative Phosphorylation (OXPHOS) Generate an Antioxidant Response Independently of Reactive Oxygen species (ROS) Production. EBioMedicine 2016, 3, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Wang, S.; Zhu, L.; Wu, C.; Yin, B.; Zhao, J.; Yuan, J.; Qiang, B.; Peng, X. cAMP response element-binding protein promotes gliomagenesis by modulating the expression of oncogenic microRNA-23a. Proc. Natl. Acad. Sci. USA 2012, 109, 15805–15810. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Ishii, H.; Nishida, N.; Takemasa, I.; Mizushima, T.; Ikeda, M.; Yokobori, T.; Mimori, K.; Yamamoto, H.; Sekimoto, M.; et al. Significance of Lgr5(+ve) cancer stem cells in the colon and rectum. Ann. Surg. Oncol. 2011, 18, 1166–1174. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, L.; Kandpal, R.P. EphB6 receptor modulates micro RNA profile of breast carcinoma cells. PLoS ONE 2011, 6, e22484. [Google Scholar] [CrossRef]
- He, X.X.; Kuang, S.Z.; Liao, J.Z.; Xu, C.R.; Chang, Y.; Wu, Y.L.; Gong, J.; Tian, D.A.; Guo, A.Y.; Lin, J.S. The regulation of microRNA expression by DNA methylation in hepatocellular carcinoma. Mol. Biosyst. 2015, 11, 532–539. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Z.X.; Chen, S.; Qiu, G.B.; Xu, Z.M.; Fu, W.N. Methylation Status of SP1 Sites within miR-23a-27a-24-2 Promoter Region Influences Laryngeal Cancer Cell Proliferation and Apoptosis. Biomed Res. Int. 2016, 2016, 2061248. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Si, J.; Wang, Y.; Huang, Z.; Zhu, H.; Feng, S.; Wu, X.; Wu, L. Long Noncoding RNA GAS5 Suppresses Tumorigenesis by Inhibiting miR-23a Expression in Non-Small Cell Lung Cancer. Oncol. Res. 2017, 25, 1027–1037. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Weng, X.D.; Wang, L.; Liu, X.H.; Zhu, H.C.; Guo, J.; Ning, J.Z.; Xiao, C.C. LncRNA XIST acts as a tumor suppressor in prostate cancer through sponging miR-23a to modulate RKIP expression. Oncotarget 2017, 8, 94358–94370. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z. The guideline of the design and validation of MiRNA mimics. Methods Mol. Biol. 2011, 676, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.Y.; Chang, D.C.; Miller, J.D.; Lin, S.L. The microRNA: Overview of the RNA gene that modulates gene functions. Methods Mol. Biol. 2006, 342, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Ortega, M.M.; Bouamar, H. Guidelines on Designing MicroRNA Sponges: From Construction to Stable Cell Line. Methods Mol. Biol. 2017, 1509, 221–233. [Google Scholar] [CrossRef] [PubMed]
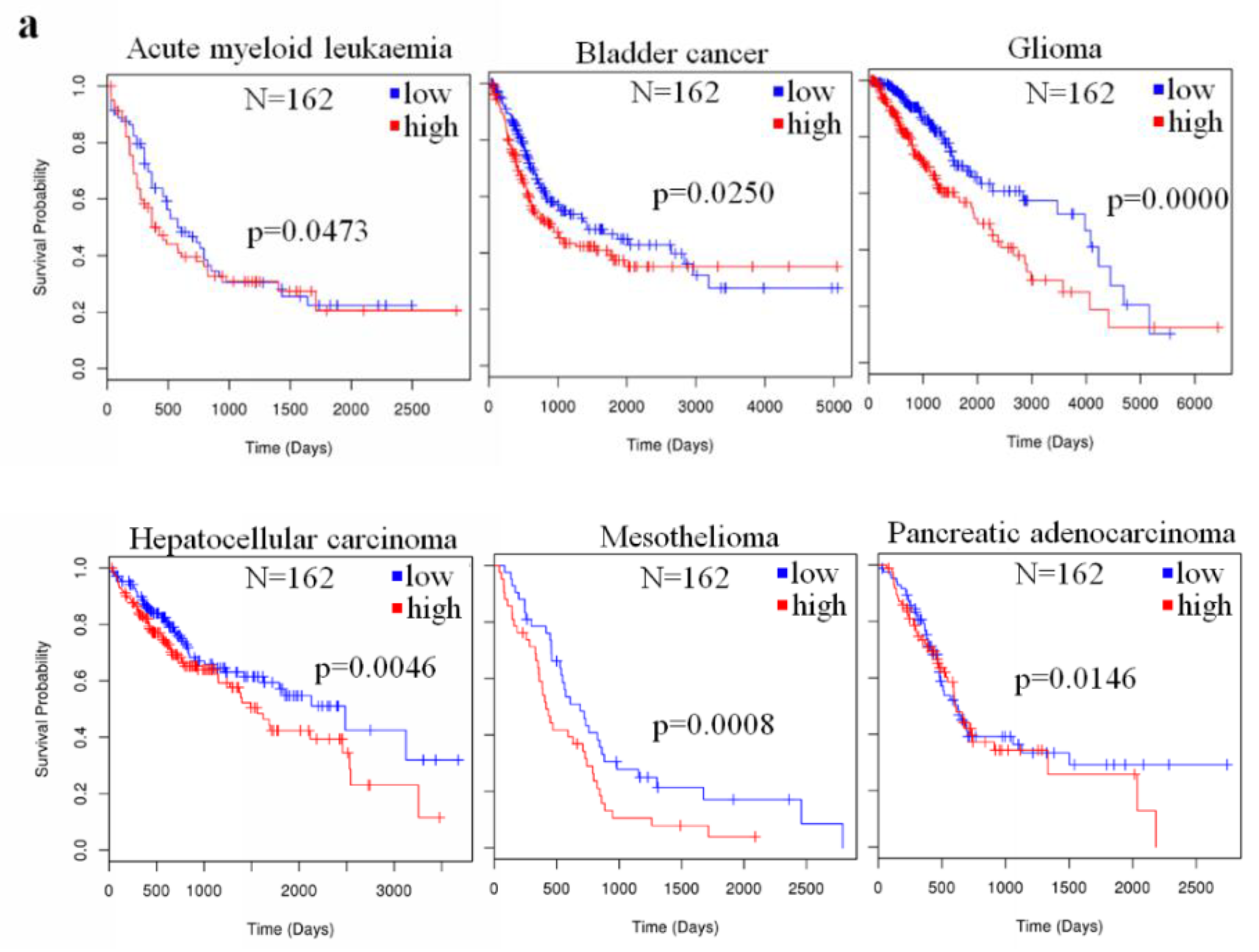
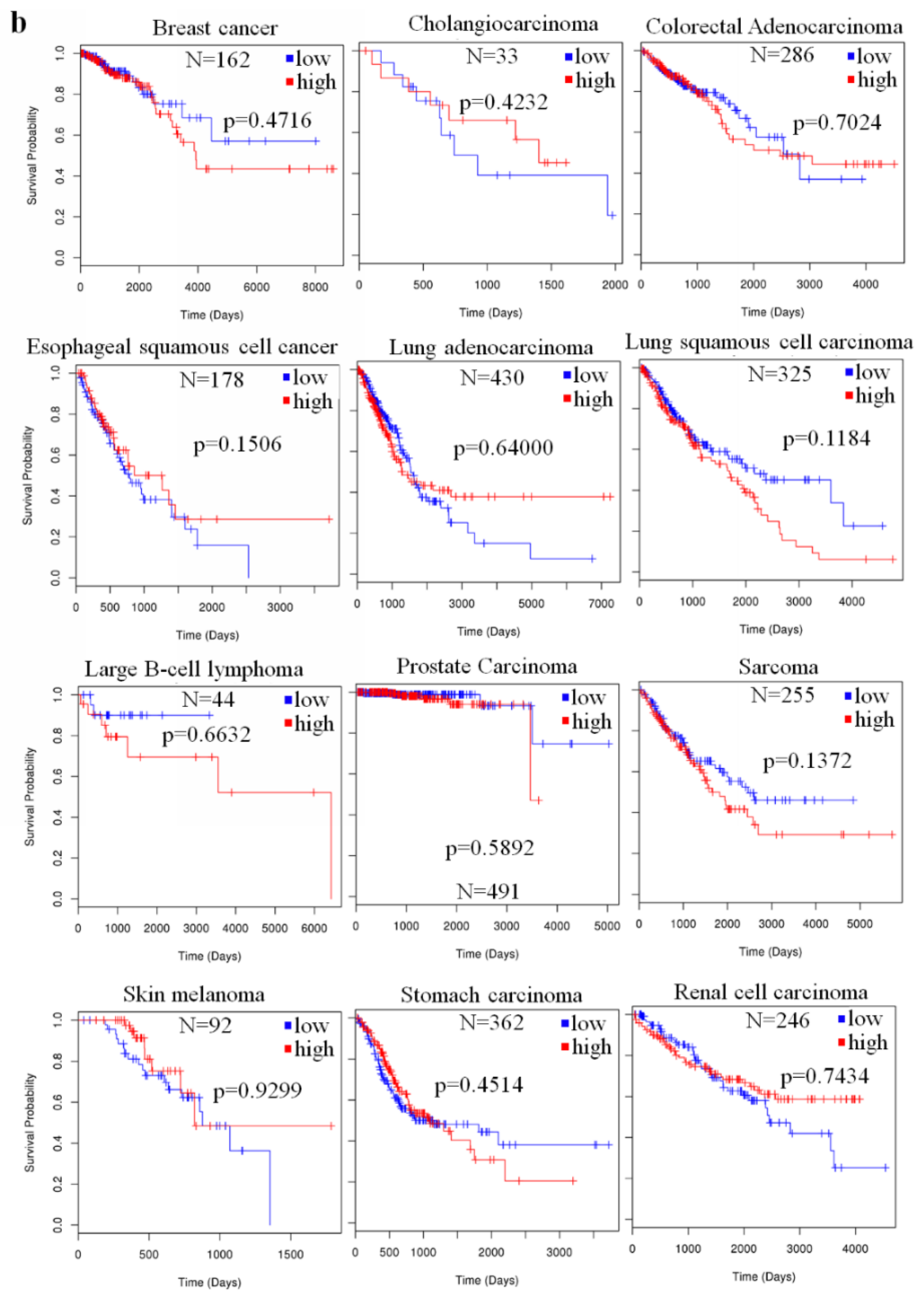
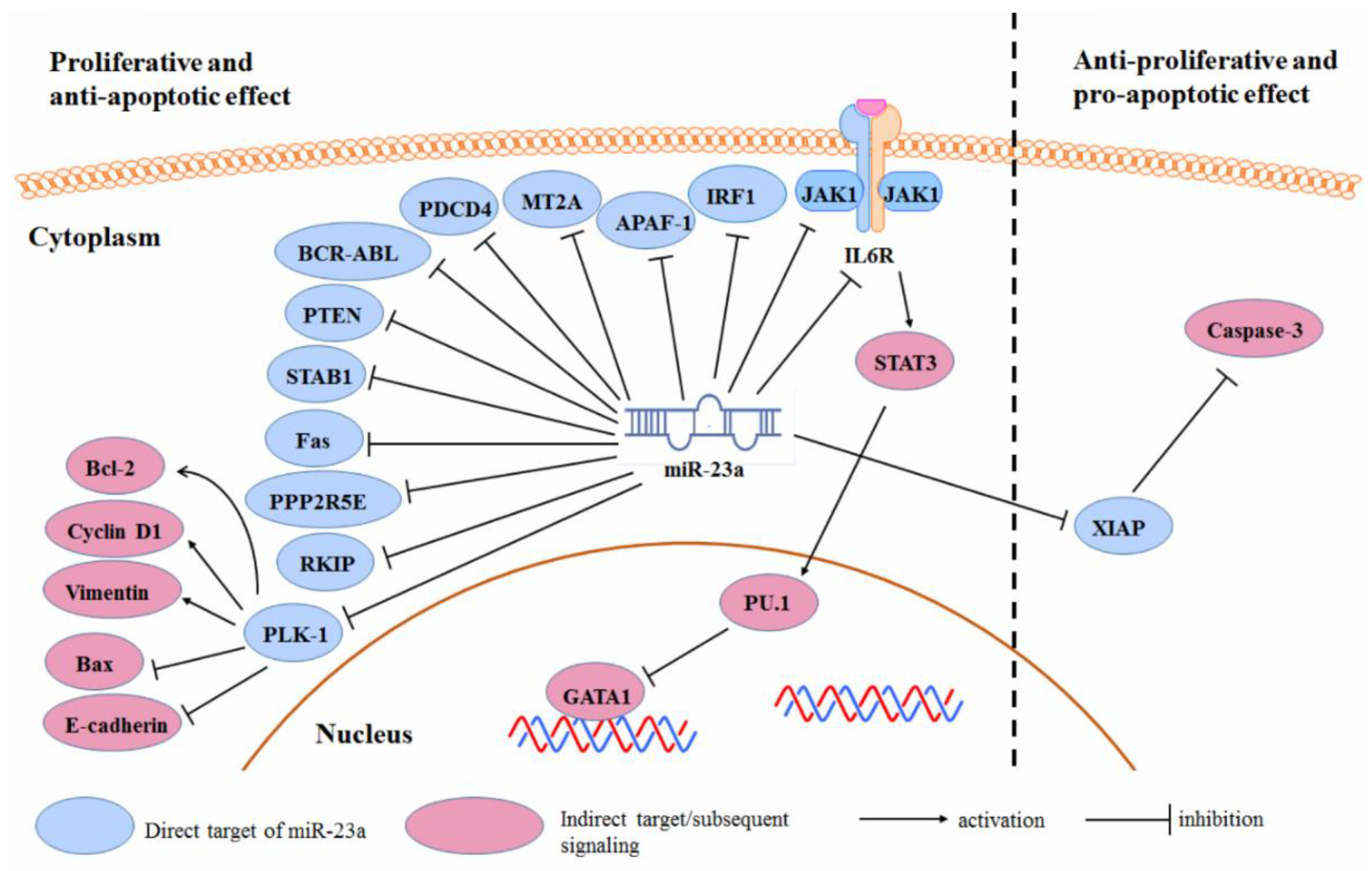
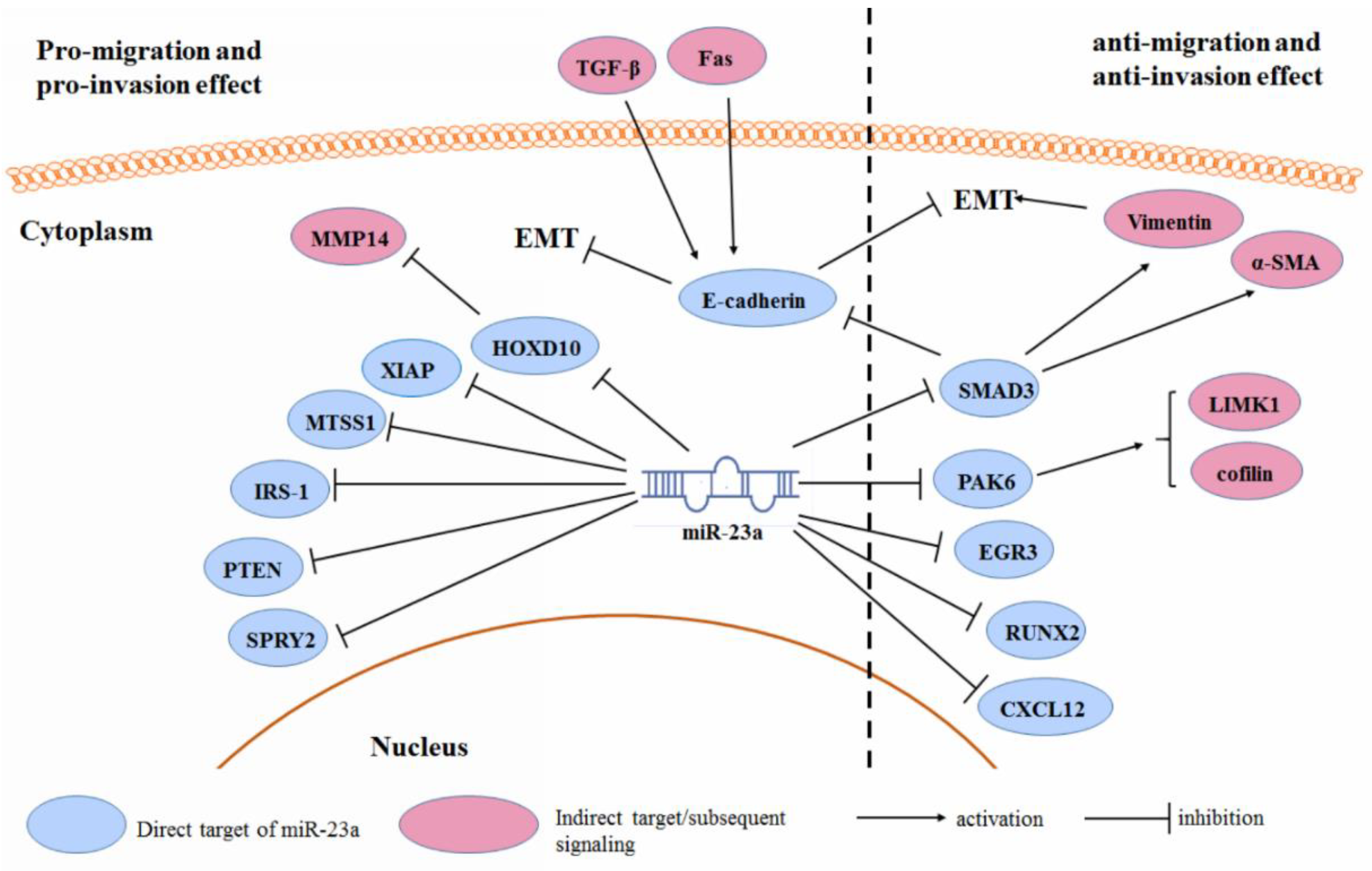
| Type of Cancer | References |
|---|---|
| Down-Regulated | |
| Acute myelogenous leukemia | [8] |
| Chronic myelogenous leukemia | [9,10] |
| Endometrial endometrioid adenocarcinoma | [14] |
| Melanoma | [18] |
| Nephroblastoma | [15] |
| Osteosarcoma | [16,17] |
| Prostate carcinoma | [12,13] |
| Up-Regulated | |
| Bladder cancer | [19] |
| Breast cancer | [20] |
| Cholangiocarcinoma | [21] |
| Colorectal cancer | [22,23] |
| Esophageal squamous cell cancer (ESCC) | [24] |
| Gastric carcinoma | [25,26,27,28,29,30,31] |
| Glioblastoma & glioma | [32,33,34,35] |
| Hepatocellular carcinoma | [36,37] |
| Large B-cell lymphoma | [38,39] |
| Laryngeal cancer | [40] |
| Lung carcinoma | [41,42] |
| Malignant pleural mesothelioma (MPM) | [43] |
| Ovarian carcinoma | [44,45] |
| Pancreatic cancer | [46,47] |
| Renal cell carcinoma (RCC) | [48] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, N.; Tan, H.-Y.; Feng, Y.-G.; Zhang, C.; Chen, F.; Feng, Y. microRNA-23a in Human Cancer: Its Roles, Mechanisms and Therapeutic Relevance. Cancers 2019, 11, 7. https://doi.org/10.3390/cancers11010007
Wang N, Tan H-Y, Feng Y-G, Zhang C, Chen F, Feng Y. microRNA-23a in Human Cancer: Its Roles, Mechanisms and Therapeutic Relevance. Cancers. 2019; 11(1):7. https://doi.org/10.3390/cancers11010007
Chicago/Turabian StyleWang, Ning, Hor-Yue Tan, Yi-Gang Feng, Cheng Zhang, Feiyu Chen, and Yibin Feng. 2019. "microRNA-23a in Human Cancer: Its Roles, Mechanisms and Therapeutic Relevance" Cancers 11, no. 1: 7. https://doi.org/10.3390/cancers11010007
APA StyleWang, N., Tan, H.-Y., Feng, Y.-G., Zhang, C., Chen, F., & Feng, Y. (2019). microRNA-23a in Human Cancer: Its Roles, Mechanisms and Therapeutic Relevance. Cancers, 11(1), 7. https://doi.org/10.3390/cancers11010007





