Functional Assessment for Clinical Use of Serum-Free Adapted NK-92 Cells
Abstract
1. Introduction
2. Results
2.1. NK-92SF Cells Retained Colony Formation Capacity and High Viability
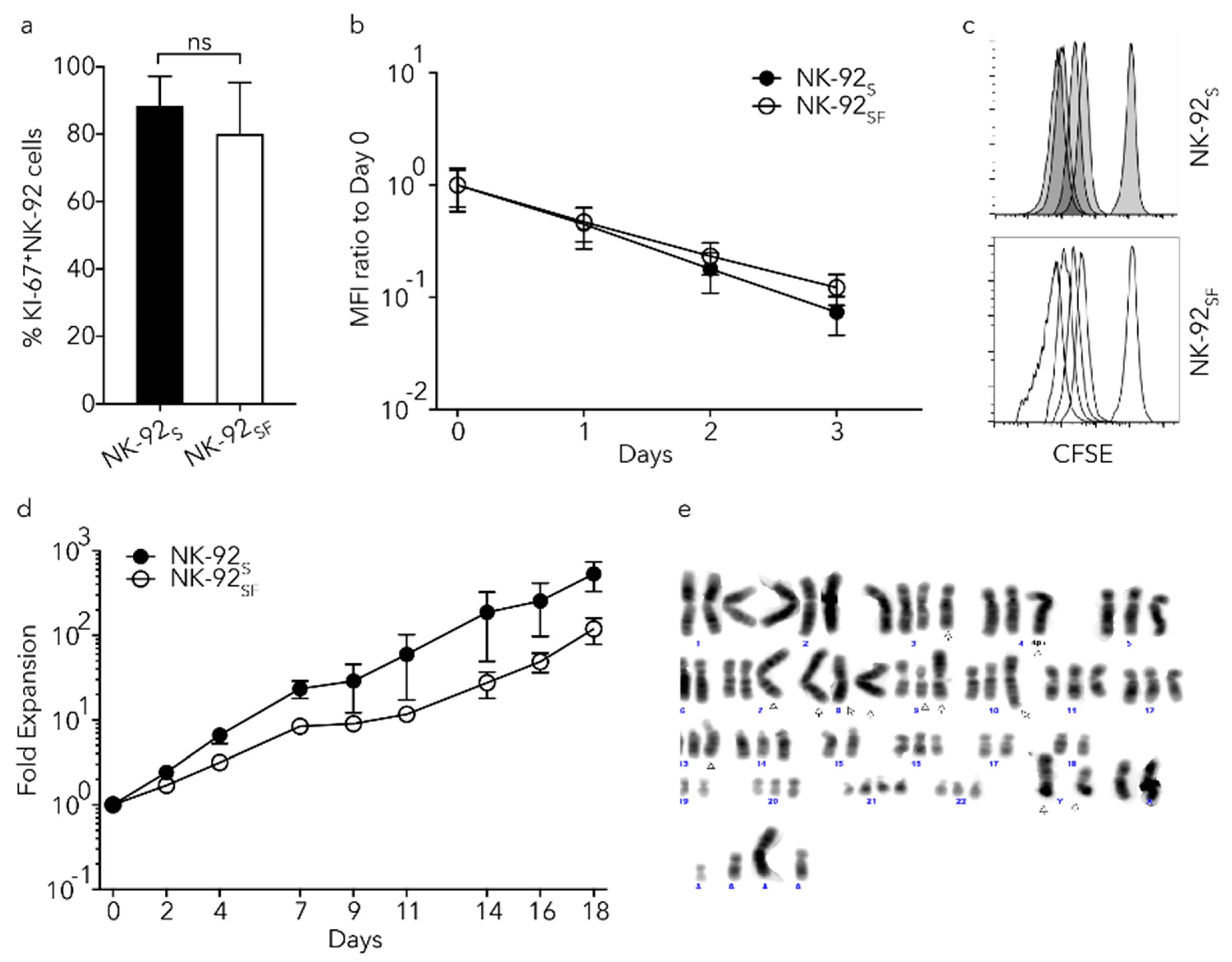
2.2. NK-92 Cells Proliferated in the Absence of Serum
2.3. NK-92SF Cells Showed Varying Karyotypes
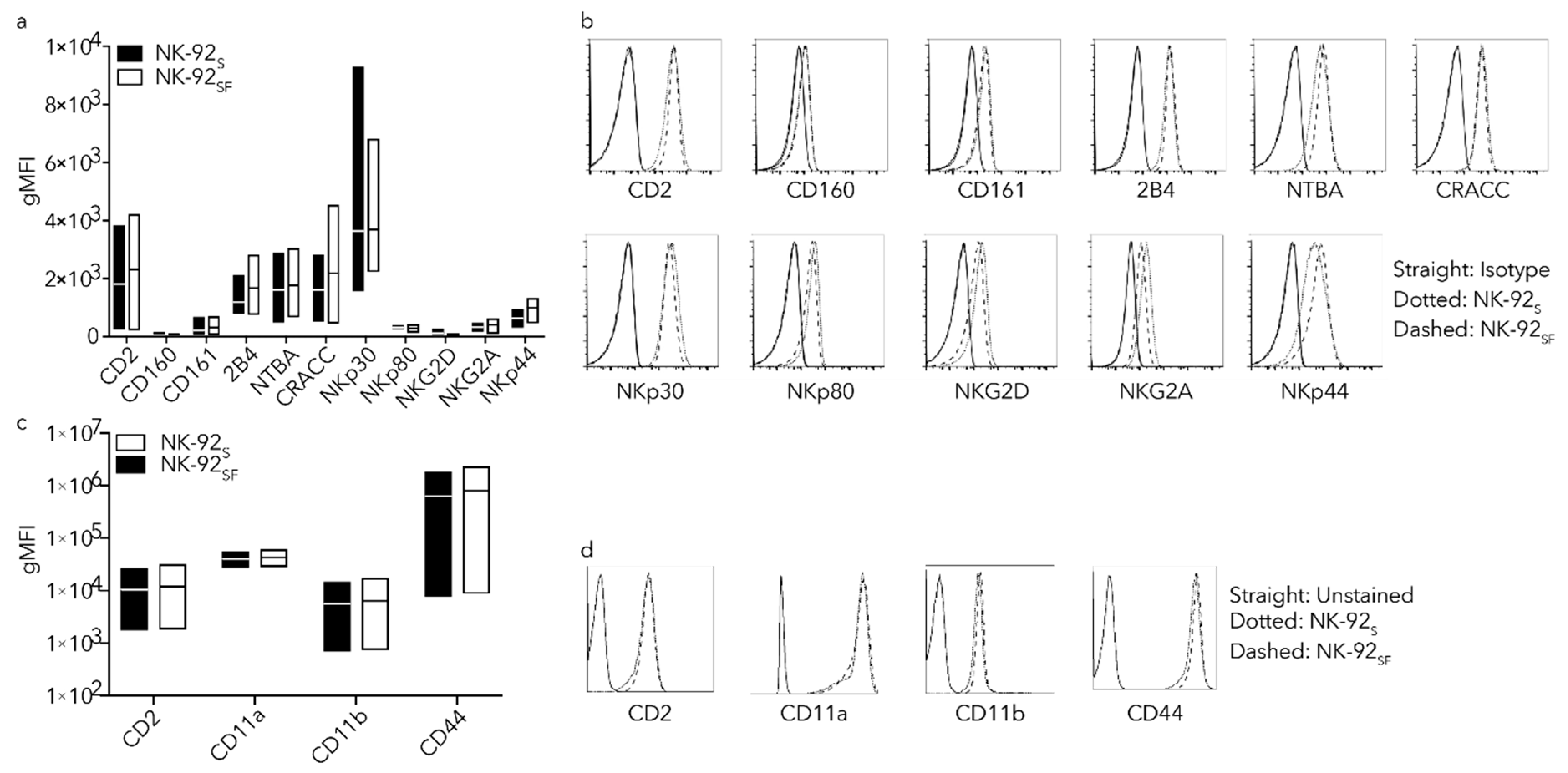
2.4. NK-92SF Cells Showed Similar Receptor Expression Profile as NK-92S Cells
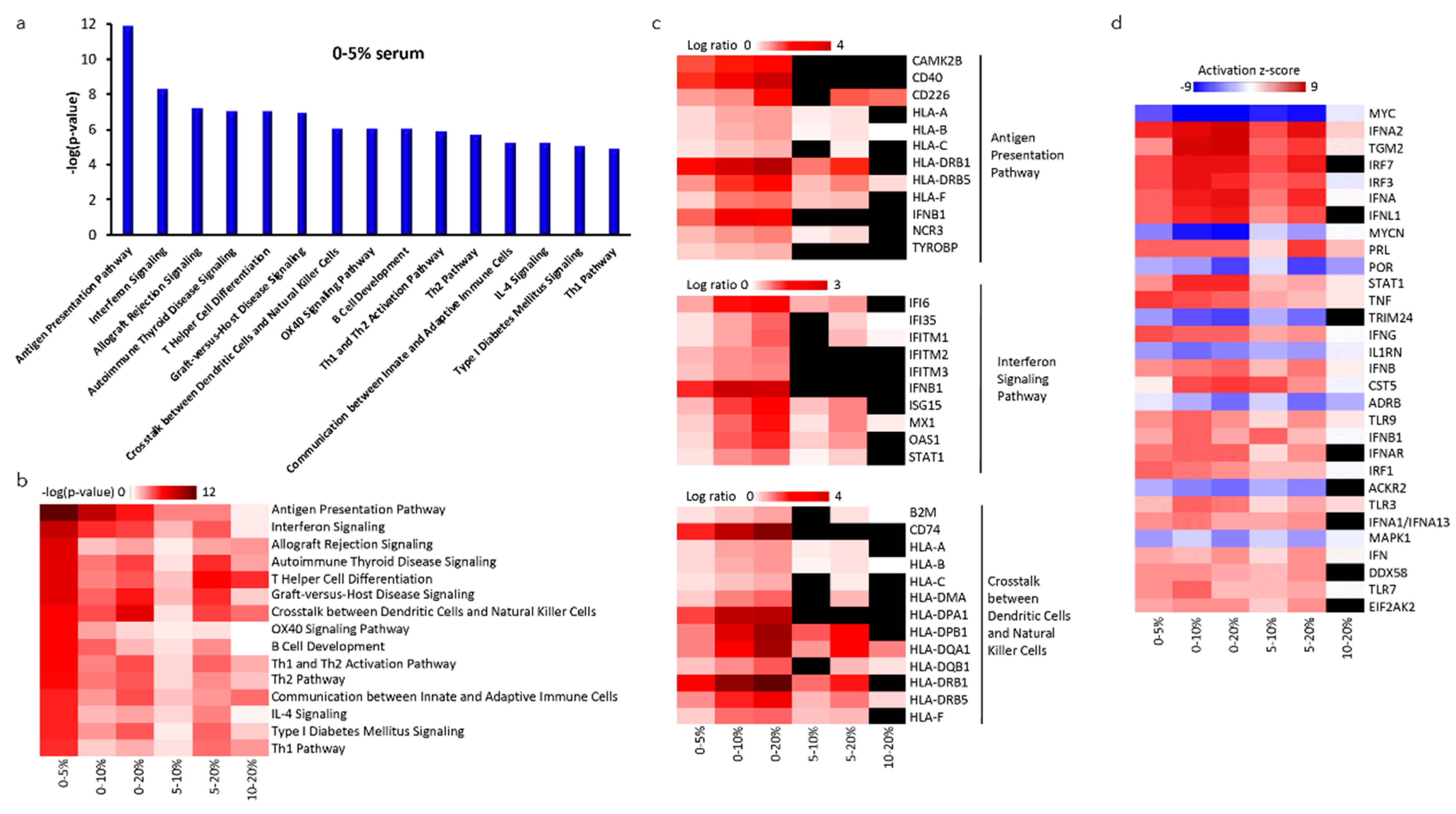
2.5. Altered Gene Expression of NK-92SF Cells
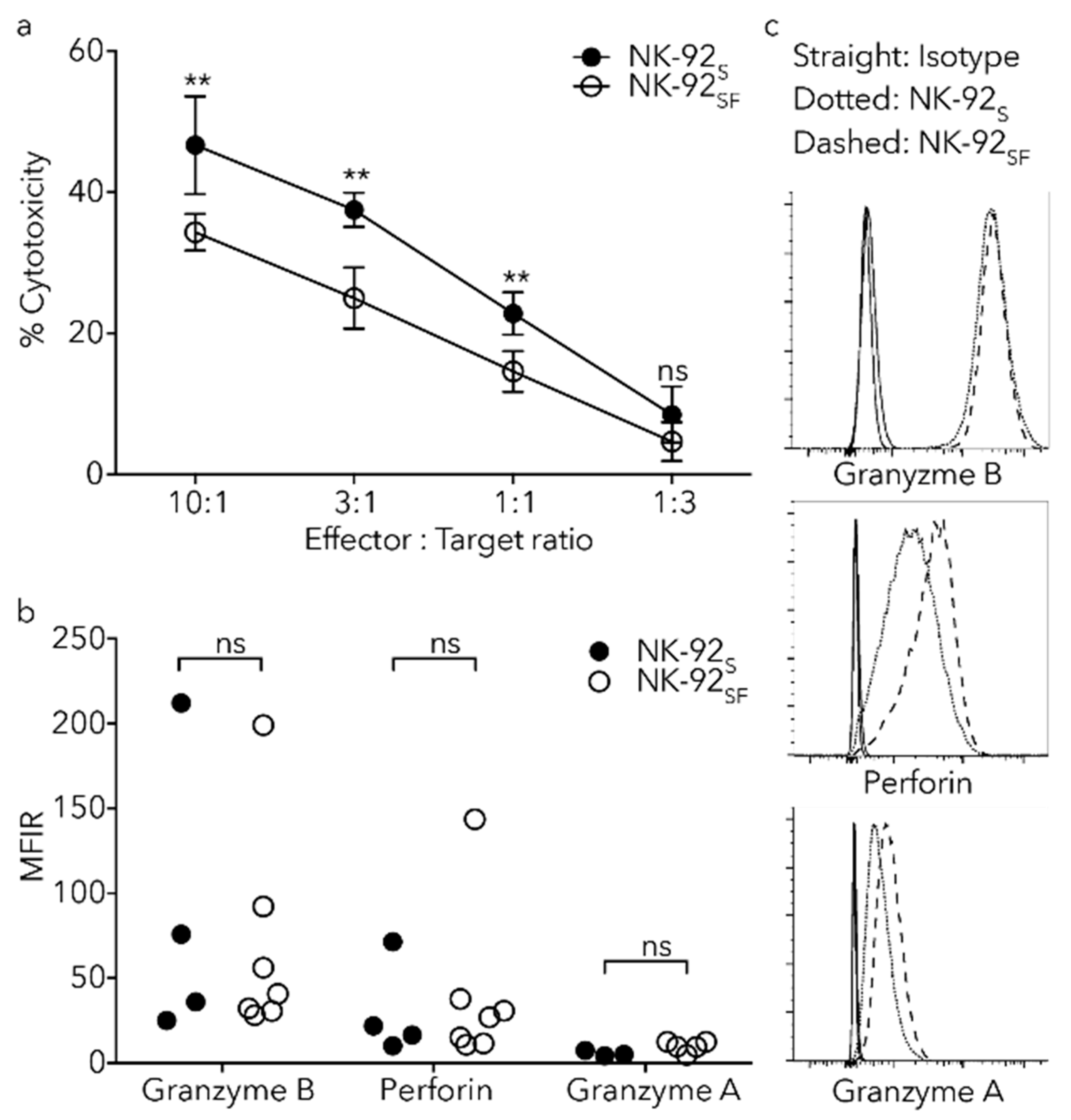
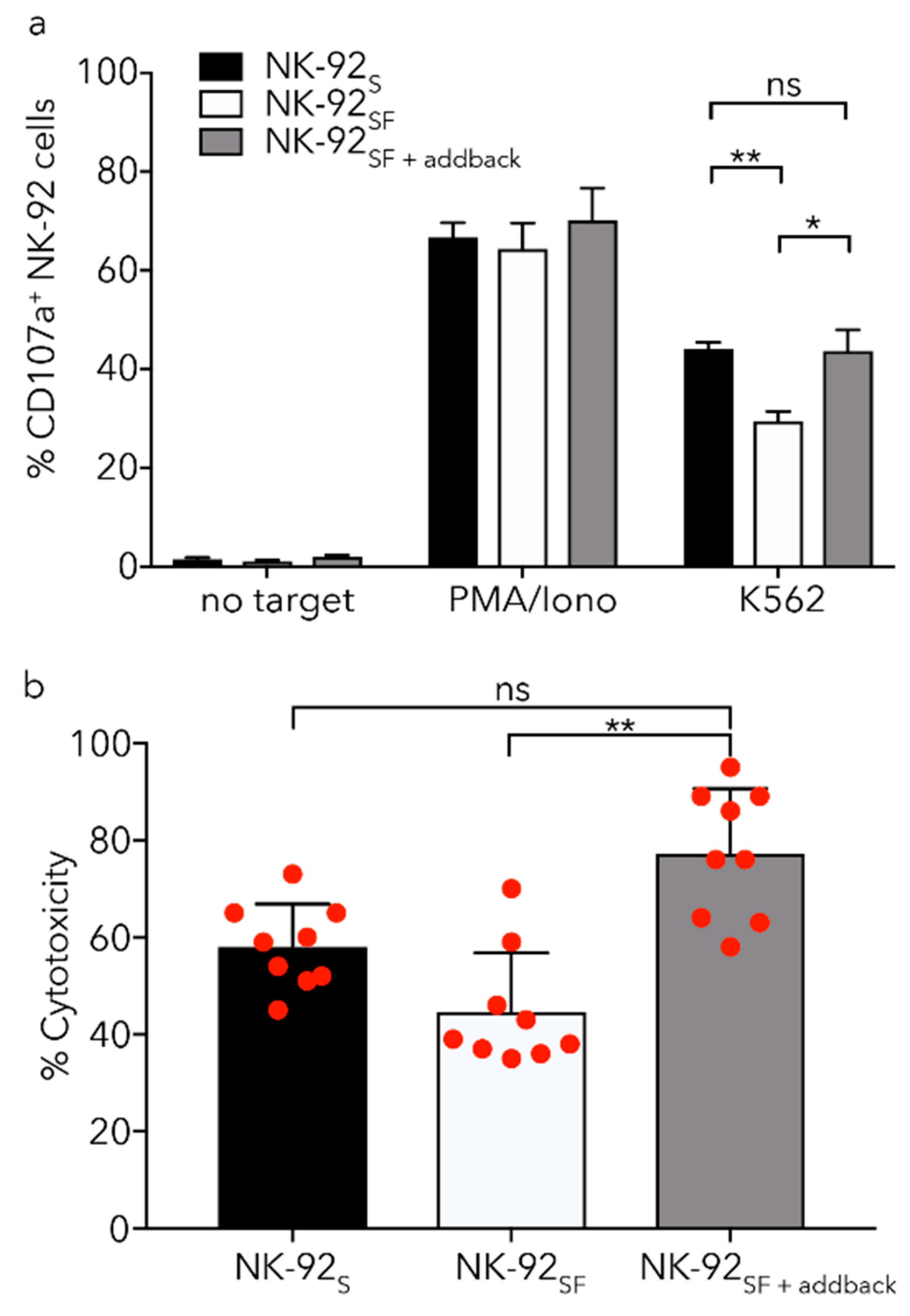
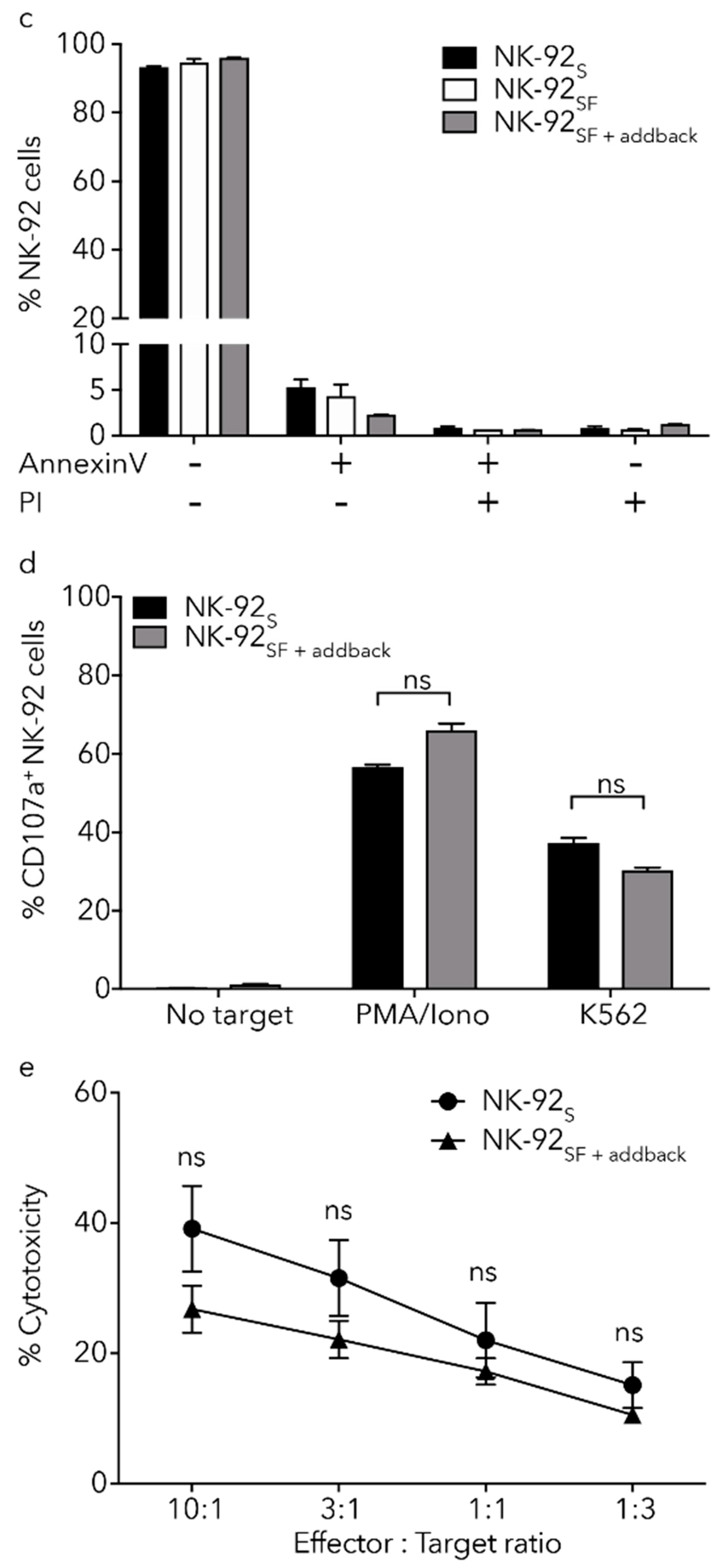
2.6. NK-92SF Cells Displayed Normal Expression of Cytotoxic Molecules but with Reduced Cytotoxic Capacity
2.7. Killing Capacity of NK-92SF Cells Is Recovered after Serum Addback
2.8. Cryoprotection Has Similar Effects on NK-92SF and NK-92S Cells
2.9. Lentiviral Transduction of NK-92 Cells Is Equally Efficient after Serum Reduction
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cell Culture
4.2. Serum Reduction
4.3. Flow Cytometry
4.4. RNA Preparation and Sequencing Data Analysis
4.5. Apoptosis, Cell Cycle Analysis, and Proliferation
4.6. Karyotyping
4.7. Degranulation and Cytotoxicity Assay
4.8. Lentivirus Production
4.9. Lentiviral Transduction
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dahlberg, C.I.; Sarhan, D.; Chrobok, M.; Duru, A.D.; Alici, E. Natural Killer Cell-Based Therapies Targeting Cancer: Possible Strategies to Gain and Sustain Anti-Tumor Activity. Front. Immunol. 2015, 6, 605. [Google Scholar] [CrossRef] [PubMed]
- Sutlu, T.; Alici, E. Natural killer cell-based immunotherapy in cancer: Current insights and future prospects. J. Intern. Med. 2009, 266, 154–181. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Ma, J.; Chen, Y.; Zhang, J.; Zhao, W.; Zhang, J.; Wei, H.; Ling, B.; Sun, R.; Tian, Z. Establishment, characterization, and successful adaptive therapy against human tumors of NKG cell, a new human NK cell line. Cell Transplant. 2011, 20, 1731–1746. [Google Scholar] [CrossRef] [PubMed]
- Robertson, M.J.; Cochran, K.J.; Cameron, C.; Le, J.M.; Tantravahi, R.; Ritz, J. Characterization of a cell line, NKL, derived from an aggressive human natural killer cell leukemia. Exp. Hematol. 1996, 24, 406–415. [Google Scholar] [PubMed]
- Gong, J.H.; Maki, G.; Klingemann, H.G. Characterization of a human cell line (NK-92) with phenotypical and functional characteristics of activated natural killer cells. Leukemia 1994, 8, 652–658. [Google Scholar]
- Yagita, M.; Huang, C.L.; Umehara, H.; Matsuo, Y.; Tabata, R.; Miyake, M.; Konaka, Y.; Takatsuki, K. A novel natural killer cell line (KHYG-1) from a patient with aggressive natural killer cell leukemia carrying a p53 point mutation. Leukemia 2000, 14, 922–930. [Google Scholar] [CrossRef]
- Tsuchiyama, J.; Yoshino, T.; Mori, M.; Kondoh, E.; Oka, T.; Akagi, T.; Hiraki, A.; Nakayama, H.; Shibuya, A.; Ma, Y.; et al. Characterization of a novel human natural killer-cell line (NK-YS) established from natural killer cell lymphoma/leukemia associated with Epstein-Barr virus infection. Blood 1374, 92, 1374–1383. [Google Scholar]
- Arai, S.; Meagher, R.; Swearingen, M.; Myint, H.; Rich, E.; Martinson, J.; Klingemann, H. Infusion of the allogeneic cell line NK-92 in patients with advanced renal cell cancer or melanoma: A phase I trial. Cytotherapy 2008, 10, 625–632. [Google Scholar] [CrossRef]
- Tonn, T.; Schwabe, D.; Klingemann, H.G.; Becker, S.; Esser, R.; Koehl, U.; Suttorp, M.; Seifried, E.; Ottmann, O.G.; Bug, G. Treatment of patients with advanced cancer with the natural killer cell line NK-92. Cytotherapy 2013, 15, 1563–1570. [Google Scholar] [CrossRef]
- Maki, G.; Klingemann, H.G.; Martinson, J.A.; Tam, Y.K. Factors regulating the cytotoxic activity of the human natural killer cell line, NK-92. J. Hematother. Stem Cell Res. 2001, 10, 369–383. [Google Scholar] [CrossRef]
- Klingemann, H.G.; Wong, E.; Maki, G. A cytotoxic NK-cell line (NK-92) for ex vivo purging of leukemia from blood. Biol. Blood Marrow Transplant. 1996, 2, 68–75. [Google Scholar] [PubMed]
- Tam, Y.K.; Maki, G.; Miyagawa, B.; Hennemann, B.; Tonn, T.; Klingemann, H.G. Characterization of genetically altered, interleukin 2-independent natural killer cell lines suitable for adoptive cellular immunotherapy. Hum. Gene Ther. 1999, 10, 1359–1373. [Google Scholar] [CrossRef] [PubMed]
- Tam, Y.K.; Miyagawa, B.; Ho, V.C.; Klingemann, H.G. Immunotherapy of malignant melanoma in a SCID mouse model using the highly cytotoxic natural killer cell line NK-92. J. Hematother. 1999, 8, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Steinherz, P.; Klingemann, H.G.; Dennig, D.; Childs, B.H.; McGuirk, J.; O’Reilly, R.J. Antileukemia activity of a natural killer cell line against human leukemias. Clin. Cancer Res. 1998, 4, 2859–2868. [Google Scholar] [PubMed]
- Karre, K.; Ljunggren, H.G.; Piontek, G.; Kiessling, R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature 1986, 319, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Boyiadzis, M.; Agha, M.; Redner, R.L.; Sehgal, A.; Im, A.; Hou, J.Z.; Farah, R.; Dorritie, K.A.; Raptis, A.; Lim, S.H.; et al. Phase 1 clinical trial of adoptive immunotherapy using “off-the-shelf” activated natural killer cells in patients with refractory and relapsed acute myeloid leukemia. Cytotherapy 2017, 19, 1225–1232. [Google Scholar] [CrossRef]
- Williams, B.A.; Law, A.D.; Routy, B.; denHollander, N.; Gupta, V.; Wang, X.H.; Chaboureau, A.; Viswanathan, S.; Keating, A. A phase I trial of NK-92 cells for refractory hematological malignancies relapsing after autologous hematopoietic cell transplantation shows safety and evidence of efficacy. Oncotarget 2017, 8, 89256–89268. [Google Scholar] [CrossRef]
- Tang, X.; Yang, L.; Li, Z.; Nalin, A.P.; Dai, H.; Xu, T.; Yin, J.; You, F.; Zhu, M.; Shen, W.; et al. First-in-man clinical trial of CAR NK-92 cells: Safety test of CD33-CAR NK-92 cells in patients with relapsed and refractory acute myeloid leukemia. Am. J. Cancer Res. 2018, 8, 1083–1089. [Google Scholar] [PubMed]
- Tam, Y.K.; Martinson, J.A.; Doligosa, K.; Klingemann, H.G. Ex vivo expansion of the highly cytotoxic human natural killer-92 cell-line under current good manufacturing practice conditions for clinical adoptive cellular immunotherapy. Cytotherapy 2003, 5, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Gstraunthaler, G. Alternatives to the use of fetal bovine serum: Serum-free cell culture. Altex 2003, 20, 275–281. [Google Scholar]
- MacLeod, R.A.; Nagel, S.; Kaufmann, M.; Greulich-Bode, K.; Drexler, H.G. Multicolor-FISH analysis of a natural killer cell line (NK-92). Leuk. Res. 2002, 26, 1027–1033. [Google Scholar] [CrossRef]
- Alici, E.; Chrobok, M.; Lund, J.; Ahmadi, T.; Khan, I.; Duru, A.D.; Nahi, H. Re-challenging with anti-CD38 monotherapy in triple-refractory multiple myeloma patients is a feasible and safe approach. Br. J. Haematol. 2016, 174, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Voskoboinik, I.; Whisstock, J.C.; Trapani, J.A. Perforin and granzymes: Function, dysfunction and human pathology. Nat. Rev. Immunol. 2015, 15, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Price, P.J.; Gregory, E.A. Relationship between in vitro growth promotion and biophysical and biochemical properties of the serum supplement. In Vitro 1982, 18, 576–584. [Google Scholar] [CrossRef]
- Bjare, U. Serum-free cell culture. Pharmacol. Ther. 1992, 53, 355–374. [Google Scholar] [CrossRef]
- Francis, G.L. Albumin and mammalian cell culture: Implications for biotechnology applications. Cytotechnology 2010, 62, 1–16. [Google Scholar] [CrossRef]
- Muller, L.; Aigner, P.; Stoiber, D. Type I Interferons and Natural Killer Cell Regulation in Cancer. Front. Immunol. 2017, 8, 304. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.; Gonen-Gross, T.; Fitchett, J.; Rowe, T.; Daniels, M.; Arnon, T.I.; Gazit, R.; Joseph, A.; Schjetne, K.W.; Steinle, A.; et al. Novel APC-like properties of human NK cells directly regulate T cell activation. J. Clin. Investig. 2004, 114, 1612–1623. [Google Scholar] [CrossRef] [PubMed]
- Nowakowska, P.; Romanski, A.; Miller, N.; Odendahl, M.; Bonig, H.; Zhang, C.; Seifried, E.; Wels, W.S.; Tonn, T. Clinical grade manufacturing of genetically modified, CAR-expressing NK-92 cells for the treatment of ErbB2-positive malignancies. Cancer Immunol. Immunother. 2018, 67, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Tomalka, A.G.; Resto-Garay, I.; Campbell, K.S.; Popkin, D.L. In vitro Evidence That Combination Therapy With CD16-Bearing NK-92 Cells and FDA-Approved Alefacept Can Selectively Target the Latent HIV Reservoir in CD4+ CD2hi Memory T Cells. Front. Immunol. 2018, 9, 2552. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, H.; Ding, J.; Liu, H.; Li, H.; Li, H.; Lu, M.; Miao, Y.; Li, L.; Zheng, J. Combination Therapy with EpCAM-CAR-NK-92 Cells and Regorafenib against Human Colorectal Cancer Models. J. Immunol. Res. 2018, 4263520. [Google Scholar] [CrossRef] [PubMed]
- Mickel, R.A.; Kessler, D.J.; Taylor, J.M.; Lichtenstein, A. Natural killer cell cytotoxicity in the peripheral blood, cervical lymph nodes, and tumor of head and neck cancer patients. Cancer Res. 1988, 48, 5017–5022. [Google Scholar] [PubMed]
- Sutlu, T.; Nystrom, S.; Gilljam, M.; Stellan, B.; Applequist, S.E.; Alici, E. Inhibition of intracellular antiviral defense mechanisms augments lentiviral transduction of human natural killer cells: Implications for gene therapy. Hum. Gene Ther. 2012, 23, 1090–1100. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chrobok, M.; Dahlberg, C.I.M.; Sayitoglu, E.C.; Beljanski, V.; Nahi, H.; Gilljam, M.; Stellan, B.; Sutlu, T.; Duru, A.D.; Alici, E. Functional Assessment for Clinical Use of Serum-Free Adapted NK-92 Cells. Cancers 2019, 11, 69. https://doi.org/10.3390/cancers11010069
Chrobok M, Dahlberg CIM, Sayitoglu EC, Beljanski V, Nahi H, Gilljam M, Stellan B, Sutlu T, Duru AD, Alici E. Functional Assessment for Clinical Use of Serum-Free Adapted NK-92 Cells. Cancers. 2019; 11(1):69. https://doi.org/10.3390/cancers11010069
Chicago/Turabian StyleChrobok, Michael, Carin I. M. Dahlberg, Ece Canan Sayitoglu, Vladimir Beljanski, Hareth Nahi, Mari Gilljam, Birgitta Stellan, Tolga Sutlu, Adil Doganay Duru, and Evren Alici. 2019. "Functional Assessment for Clinical Use of Serum-Free Adapted NK-92 Cells" Cancers 11, no. 1: 69. https://doi.org/10.3390/cancers11010069
APA StyleChrobok, M., Dahlberg, C. I. M., Sayitoglu, E. C., Beljanski, V., Nahi, H., Gilljam, M., Stellan, B., Sutlu, T., Duru, A. D., & Alici, E. (2019). Functional Assessment for Clinical Use of Serum-Free Adapted NK-92 Cells. Cancers, 11(1), 69. https://doi.org/10.3390/cancers11010069




