Poly-ADP-Ribosylation of Estrogen Receptor-Alpha by PARP1 Mediates Antiestrogen Resistance in Human Breast Cancer Cells
Abstract
1. Introduction
2. Results
2.1. Survival of Tamoxifen-Sensitive and -Resistant Breast Cancer Cells Is Inhibited by Tamoxifen-Talazoparib Combination
2.2. Tamoxifen-Talazoparib Combinatorial Efficacy Is ERα-Dependent
2.3. Tamoxifen-Talazoparib Co-Administration Increases Cellular DNA Damage
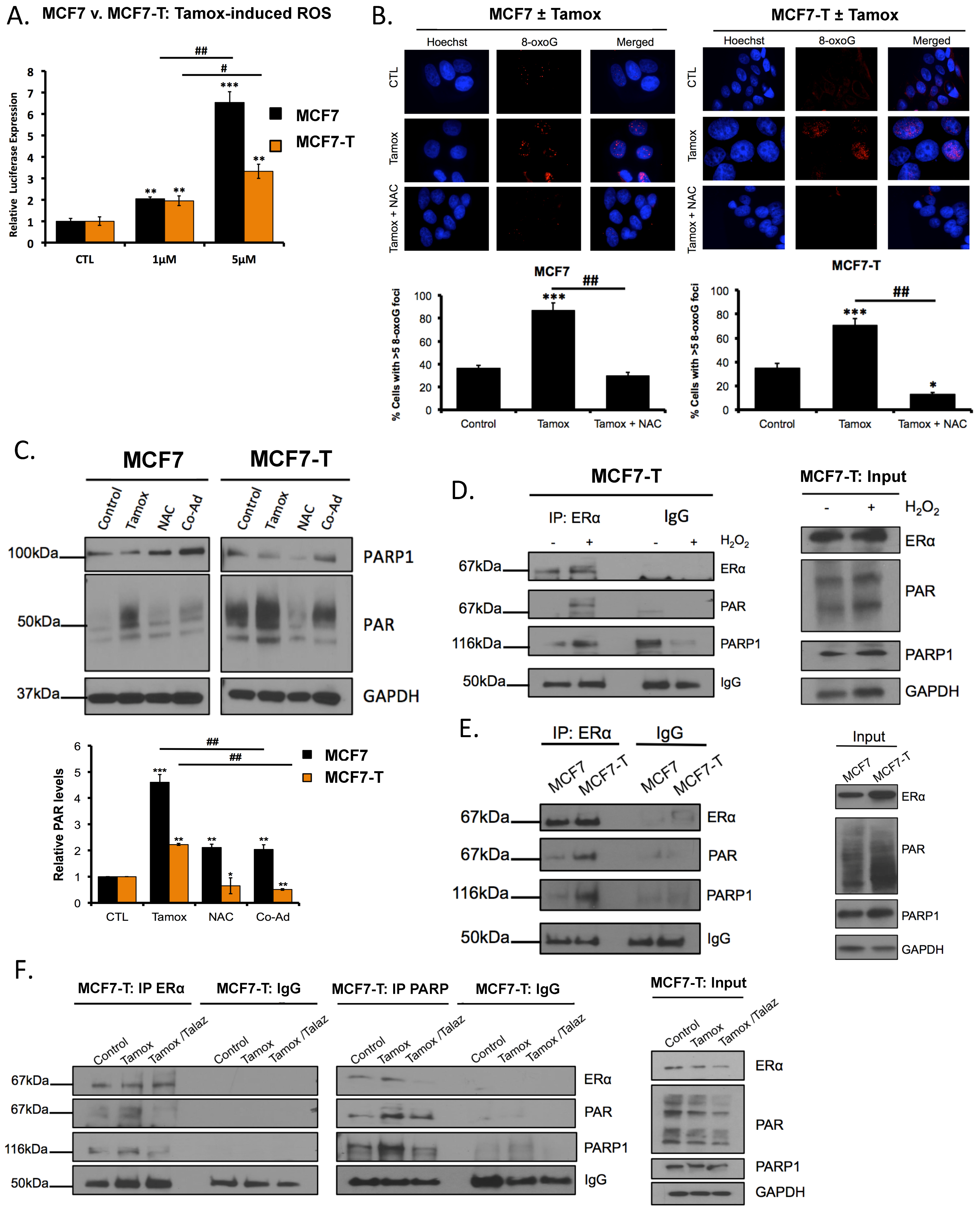
2.4. Tamoxifen Induces ROS, and ROS Promotes PARylaton of ERα
2.5. Tamoxifen Promotes PARylation of ERα and PARylation Mediates ERα Response
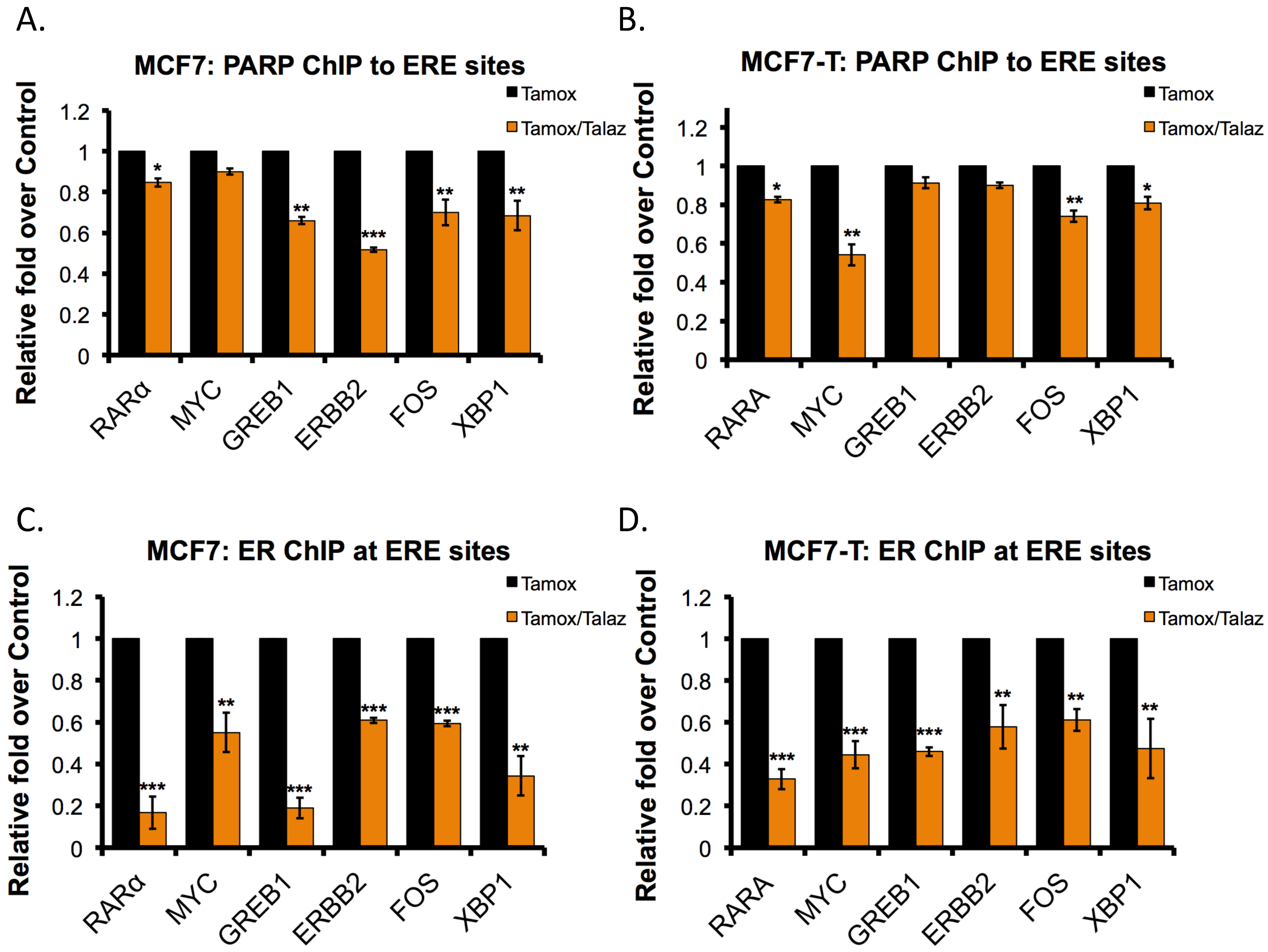
2.6. Talazoparib Reduces PARP1 and ERα Localization to ERα Target Genes
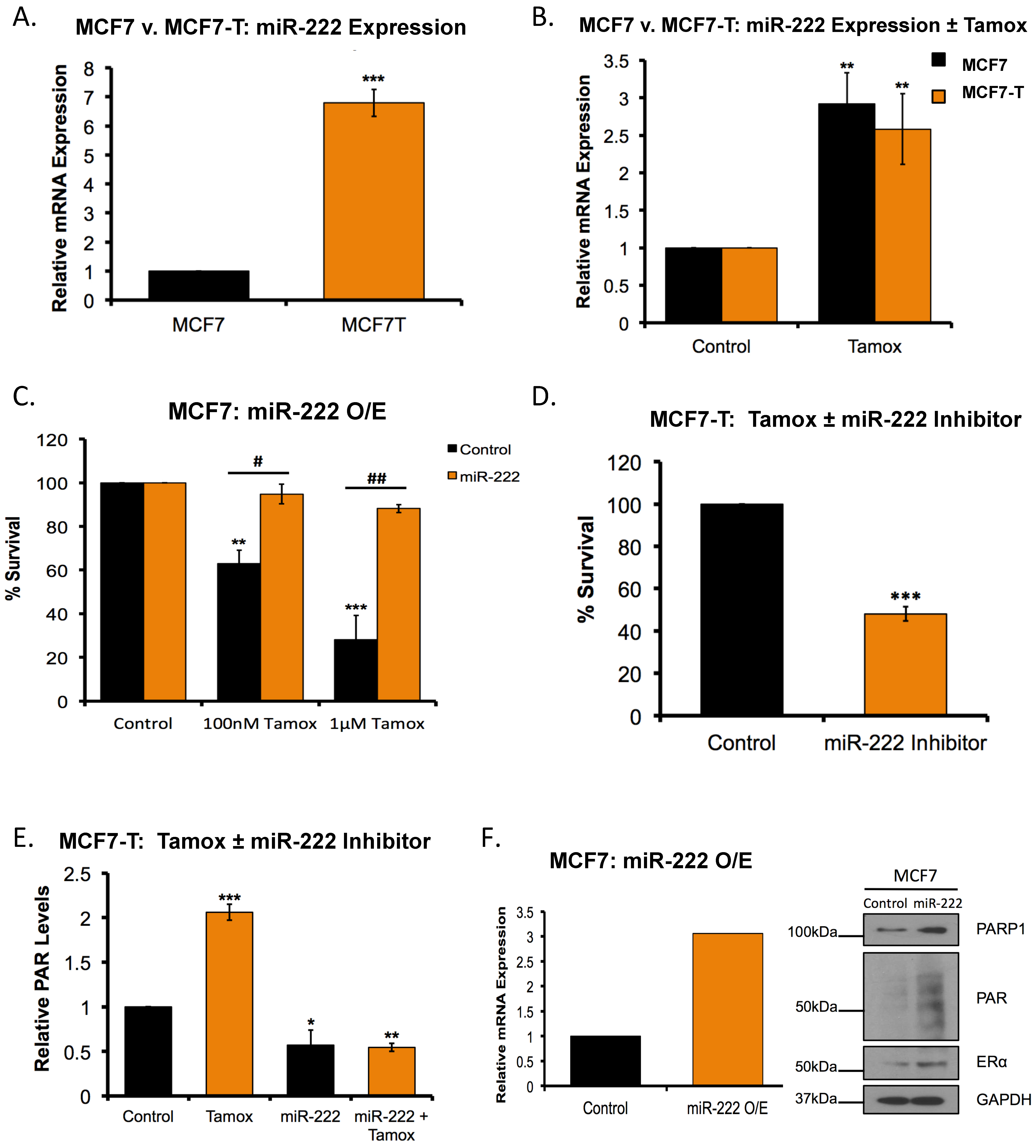
2.7. Overexpression of miR-222 in Tamoxifen Resistant Cells Promotes PARP1 Activation
2.8. miR-222 Regulates the Response to Tamoxifen-PARPi Co-Administration
3. Discussion
4. Materials and Methods
4.1. Cell Lines, Culture Conditions and Reagents
4.2. Clonogenic Survival and MTT Assay
4.3. RNA Extraction, Quantitative RT-PCR and Cell Transfection
4.4. PAR-Capture ELISA
4.5. ROS Assays
4.6. Immunofluorescence
4.7. Western Blot Analysis
4.8. Combination Index and Synergism
4.9. Immunoprecipitation Assays
4.10. Chromatin Immunoprecipitation (ChIP)-qPCR
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Girdler, F.; Brotherick, I. The oestrogen receptors (ER alpha and ER beta) and their role in breast cancer: A review. Breast 2000, 9, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.; Tarulli, G.; Portman, N.; Hickey, T.E.; Tilley, W.D.; Palmieri, C. Pushing estrogen receptor around in breast cancer. Endocr. Relat. Cancer 2016, 23, T227–T241. [Google Scholar] [CrossRef] [PubMed]
- Musgrove, E.A.; Sutherland, R.L. Biological determinants of endocrine resistance in breast cancer. Nat. Rev. Cancer 2009, 9, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.; Park, S.; Teo, W.W.; Korangath, P.; Cho, S.S.; Yoshida, T.; Gyorffy, B.; Goswami, C.P.; Nakshatri, H.; Cruz, L.A.; et al. HOXB7 Is an ERalpha Cofactor in the Activation of HER2 and Multiple ER Target Genes Leading to Endocrine Resistance. Cancer Discov. 2015, 5, 944–959. [Google Scholar] [CrossRef] [PubMed]
- Bekele, R.T.; Venkatraman, G.; Liu, R.Z.; Tang, X.; Mi, S.; Benesch, M.G.; Mackey, J.R.; Godbout, R.; Curtis, J.M.; McMullen, T.P.; et al. Oxidative stress contributes to the tamoxifen-induced killing of breast cancer cells: Implications for tamoxifen therapy and resistance. Sci. Rep. 2016, 6, 21164. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Kang, Y.S.; Lee, S.H.; Kim, J.A. Role of NAD(P)H oxidase in the tamoxifen-induced generation of reactive oxygen species and apoptosis in HepG2 human hepatoblastoma cells. Cell Death Differ. 2000, 7, 925–932. [Google Scholar] [CrossRef]
- Kallio, A.; Zheng, A.; Dahllund, J.; Heiskanen, K.M.; Harkonen, P. Role of mitochondria in tamoxifen-induced rapid death of MCF-7 breast cancer cells. Apoptosis 2005, 10, 1395–1410. [Google Scholar] [CrossRef]
- Cook, K.L.; Clarke, P.A.; Parmar, J.; Hu, R.; Schwartz-Roberts, J.L.; Abu-Asab, M.; Warri, A.; Baumann, W.T.; Clarke, R. Knockdown of estrogen receptor-alpha induces autophagy and inhibits antiestrogen-mediated unfolded protein response activation, promoting ROS-induced breast cancer cell death. FASEB J. 2014, 28, 3891–3905. [Google Scholar] [CrossRef]
- Gutierrez, M.C.; Detre, S.; Johnston, S.; Mohsin, S.K.; Shou, J.; Allred, D.C.; Schiff, R.; Osborne, C.K.; Dowsett, M. Molecular changes in tamoxifen-resistant breast cancer: Relationship between estrogen receptor, HER-2, and p38 mitogen-activated protein kinase. J. Clin. Oncol. 2005, 23, 2469–2476. [Google Scholar] [CrossRef]
- Anbalagan, M.; Rowan, B.G. Estrogen receptor alpha phosphorylation and its functional impact in human breast cancer. Mol. Cell. Endocrinol. 2015, 418, 264–272. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, Z.; Cenciarini, M.E.; Proietti, C.J.; Amasino, M.; Hong, T.; Yang, M.; Liao, Y.; Chiang, H.C.; Kaklamani, V.G.; et al. Tamoxifen Resistance in Breast Cancer Is Regulated by the EZH2-ERalpha-GREB1 Transcriptional Axis. Cancer Res. 2018, 78, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Badia, E.; Oliva, J.; Balaguer, P.; Cavailles, V. Tamoxifen resistance and epigenetic modifications in breast cancer cell lines. Curr. Med. Chem. 2007, 14, 3035–3045. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Hafiz, H.A. Epigenetic Mechanisms of Tamoxifen Resistance in Luminal Breast Cancer. Diseases 2017, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Anbalagan, M.; Huderson, B.; Murphy, L.; Rowan, B.G. Post-translational modifications of nuclear receptors and human disease. Nucl. Recept. Signal. 2012, 10, e001. [Google Scholar] [CrossRef] [PubMed]
- Osborne, C.K.; Schiff, R. Mechanisms of endocrine resistance in breast cancer. Annu. Rev. Med. 2011, 62, 233–247. [Google Scholar] [CrossRef]
- Kim, M.Y.; Zhang, T.; Kraus, W.L. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005, 19, 1951–1967. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Kraus, W.L. On PAR with PARP: Cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012, 26, 417–432. [Google Scholar] [CrossRef]
- Wright, R.H.; Castellano, G.; Bonet, J.; Le Dily, F.; Font-Mateu, J.; Ballare, C.; Nacht, A.S.; Soronellas, D.; Oliva, B.; Beato, M. CDK2-dependent activation of PARP-1 is required for hormonal gene regulation in breast cancer cells. Genes Dev. 2012, 26, 1972–1983. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.; Liu, Z.; Xu, X.; Liu, Q.; Zhang, X.; Kong, B.; Wei, J.J.; Gong, Y.; Shao, C. Increased oxidative stress mediates the antitumor effect of PARP inhibition in ovarian cancer. Redox Biol. 2018, 17, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.; Xu, G.; Zhang, C.; Li, B.; Qin, J.; Hao, X.; Liu, Q.; Zhang, X.; Liu, J.; Wei, J.; et al. Berberine induces oxidative DNA damage and impairs homologous recombination repair in ovarian cancer cells to confer increased sensitivity to PARP inhibition. Cell Death Dis. 2017, 8, e3070. [Google Scholar] [CrossRef]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005, 434, 913–917. [Google Scholar] [CrossRef]
- Farmer, H.; McCabe, N.; Lord, C.J.; Tutt, A.N.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.; Knights, C.; et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Bitler, B.G.; Watson, Z.L.; Wheeler, L.J.; Behbakht, K. PARP inhibitors: Clinical utility and possibilities of overcoming resistance. Gynecol. Oncol. 2017, 147, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Schiewer, M.J.; Knudsen, K.E. Transcriptional roles of PARP1 in cancer. Mol. Cancer Res. 2014, 12, 1069–1080. [Google Scholar] [CrossRef] [PubMed]
- Ju, B.G.; Lunyak, V.V.; Perissi, V.; Garcia-Bassets, I.; Rose, D.W.; Glass, C.K.; Rosenfeld, M.G. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science 2006, 312, 1798–1802. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, Y.; Wang, L.; Luo, X.; Huang, K.; Wang, C.; Du, M.; Liu, F.; Luo, T.; Huang, D.; et al. Poly(ADP-ribose) polymerase 1 is a key regulator of estrogen receptor alpha-dependent gene transcription. J. Biol. Chem. 2013, 288, 11348–11357. [Google Scholar] [CrossRef]
- Schiff, R.; Reddy, P.; Ahotupa, M.; Coronado-Heinsohn, E.; Grim, M.; Hilsenbeck, S.G.; Lawrence, R.; Deneke, S.; Herrera, R.; Chamness, G.C.; et al. Oxidative stress and AP-1 activity in tamoxifen-resistant breast tumors in vivo. J. Natl. Cancer Inst. 2000, 92, 1926–1934. [Google Scholar] [CrossRef]
- Du, Y.; Yamaguchi, H.; Wei, Y.; Hsu, J.L.; Wang, H.L.; Hsu, Y.H.; Lin, W.C.; Yu, W.H.; Leonard, P.G.; Lee, G.R.T.; et al. Blocking c-Met-mediated PARP1 phosphorylation enhances anti-tumor effects of PARP inhibitors. Nat. Med. 2016, 22, 194–201. [Google Scholar] [CrossRef]
- Pulliam, N.; Fang, F.; Ozes, A.R.; Tang, J.; Adewuyi, A.; Keer, H.N.; Lyons, J.F.; Baylin, S.B.; Matei, D.; Nakshatri, H.; et al. An Effective Epigenetic-PARP inhibitor Combination Therapy for Breast and Ovarian Cancers Independent of BRCA-mutations. Clin. Cancer Res. 2018. [Google Scholar] [CrossRef]
- Fan, M.; Yan, P.S.; Hartman-Frey, C.; Chen, L.; Paik, H.; Oyer, S.L.; Salisbury, J.D.; Cheng, A.S.; Li, L.; Abbosh, P.H.; et al. Diverse gene expression and DNA methylation profiles correlate with differential adaptation of breast cancer cells to the antiestrogens tamoxifen and fulvestrant. Cancer Res 2006, 66, 11954–11966. [Google Scholar] [CrossRef]
- Chou, T.C.; Talalay, P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 1984, 22, 27–55. [Google Scholar] [CrossRef]
- Lord, C.J.; Ashworth, A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017, 355, 1152–1158. [Google Scholar] [CrossRef] [PubMed]
- Brunner, N.; Boysen, B.; Jirus, S.; Skaar, T.C.; Holst-Hansen, C.; Lippman, J.; Frandsen, T.; Spang-Thomsen, M.; Fuqua, S.A.; Clarke, R. MCF7/LCC9: An antiestrogen-resistant MCF-7 variant in which acquired resistance to the steroidal antiestrogen ICI 182,780 confers an early cross-resistance to the nonsteroidal antiestrogen tamoxifen. Cancer Res. 1997, 57, 3486–3493. [Google Scholar] [PubMed]
- Kocanova, S.; Mazaheri, M.; Caze-Subra, S.; Bystricky, K. Ligands specify estrogen receptor alpha nuclear localization and degradation. BMC Cell Biol. 2010, 11, 98. [Google Scholar] [CrossRef] [PubMed]
- Razandi, M.; Oh, P.; Pedram, A.; Schnitzer, J.; Levin, E.R. ERs associate with and regulate the production of caveolin: Implications for signaling and cellular actions. Mol. Endocrinol. 2002, 16, 100–115. [Google Scholar] [CrossRef] [PubMed]
- Sakai, W.; Swisher, E.M.; Karlan, B.Y.; Agarwal, M.K.; Higgins, J.; Friedman, C.; Villegas, E.; Jacquemont, C.; Farrugia, D.J.; Couch, F.J.; et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 2008, 451, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Jiang, Z.; Wang, C.; Yang, Y.; Li, L.; He, H.; Yu, Z. Sensitization of tamoxifen-resistant breast cancer cells by Z-ligustilide through inhibiting autophagy and accumulating DNA damages. Oncotarget 2017, 8, 29300–29317. [Google Scholar] [CrossRef] [PubMed]
- Neijenhuis, S.; Bajrami, I.; Miller, R.; Lord, C.J.; Ashworth, A. Identification of miRNA modulators to PARP inhibitor response. DNA Repair 2013, 12, 394–402. [Google Scholar] [CrossRef]
- Rao, X.; Di Leva, G.; Li, M.; Fang, F.; Devlin, C.; Hartman-Frey, C.; Burow, M.E.; Ivan, M.; Croce, C.M.; Nephew, K.P. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene 2011, 30, 1082–1097. [Google Scholar] [CrossRef]
- Di Leva, G.; Gasparini, P.; Piovan, C.; Ngankeu, A.; Garofalo, M.; Taccioli, C.; Iorio, M.V.; Li, M.; Volinia, S.; Alder, H.; et al. MicroRNA cluster 221-222 and estrogen receptor alpha interactions in breast cancer. J. Natl. Cancer Inst. 2010, 102, 706–721. [Google Scholar] [CrossRef]
- Manavalan, T.T.; Teng, Y.; Appana, S.N.; Datta, S.; Kalbfleisch, T.S.; Li, Y.; Klinge, C.M. Differential expression of microRNA expression in tamoxifen-sensitive MCF-7 versus tamoxifen-resistant LY2 human breast cancer cells. Cancer Lett. 2011, 313, 26–43. [Google Scholar] [CrossRef] [PubMed]
- Early Breast Cancer Trialists’ Collaborative Group. Long-term outcomes for neoadjuvant versus adjuvant chemotherapy in early breast cancer: Meta-analysis of individual patient data from ten randomised trials. Lancet Oncol. 2018, 19, 27–39. [Google Scholar] [CrossRef]
- Dowsett, M.; Nicholson, R.I.; Pietras, R.J. Biological characteristics of the pure antiestrogen fulvestrant: Overcoming endocrine resistance. Breast Cancer Res. Treat. 2005, 93, S11–S18. [Google Scholar] [CrossRef] [PubMed]
- Jordan, V.C. Tamoxifen Resistance Trumped and Oral Selective Estrogen Receptor Degraders Arrive. Clin. Cancer Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Trachootham, D.; Alexandre, J.; Huang, P. Targeting cancer cells by ROS-mediated mechanisms: A radical therapeutic approach? Nat. Rev. Drug Discov. 2009, 8, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Muvarak, N.E.; Chowdhury, K.; Xia, L.; Robert, C.; Choi, E.Y.; Cai, Y.; Bellani, M.; Zou, Y.; Singh, Z.N.; Duong, V.H.; et al. Enhancing the cytotoxic effects of PARP inhibitors with DNA demethylating agents—A potential therapy for cancer. Cancer Cell 2016, 30, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Ozes, A.R.; Miller, D.F.; Ozes, O.N.; Fang, F.; Liu, Y.; Matei, D.; Huang, T.; Nephew, K.P. NF-kappaB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene 2016, 35, 5350–5361. [Google Scholar] [CrossRef] [PubMed]
- Ozes, A.R.; Pulliam, N.; Ertosun, M.G.; Yilmaz, O.; Tang, J.; Copuroglu, E.; Matei, D.; Ozes, O.N.; Nephew, K.P. Protein kinase A-mediated phosphorylation regulates STAT3 activation and oncogenic EZH2 activity. Oncogene 2018. [Google Scholar] [CrossRef]
- Sakai, W.; Swisher, E.M.; Jacquemont, C.; Chandramohan, K.V.; Couch, F.J.; Langdon, S.P.; Wurz, K.; Higgins, J.; Villegas, E.; Taniguchi, T. Functional restoration of BRCA2 protein by secondary BRCA2 mutations in BRCA2-mutated ovarian carcinoma. Cancer Res. 2009, 69, 6381–6386. [Google Scholar] [CrossRef]
- Available online: http://www.combosyn.com/feature.html (accessed on 4 January 2019).



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pulliam, N.; Tang, J.; Wang, W.; Fang, F.; Sood, R.; O’Hagan, H.M.; Miller, K.D.; Clarke, R.; Nephew, K.P. Poly-ADP-Ribosylation of Estrogen Receptor-Alpha by PARP1 Mediates Antiestrogen Resistance in Human Breast Cancer Cells. Cancers 2019, 11, 43. https://doi.org/10.3390/cancers11010043
Pulliam N, Tang J, Wang W, Fang F, Sood R, O’Hagan HM, Miller KD, Clarke R, Nephew KP. Poly-ADP-Ribosylation of Estrogen Receptor-Alpha by PARP1 Mediates Antiestrogen Resistance in Human Breast Cancer Cells. Cancers. 2019; 11(1):43. https://doi.org/10.3390/cancers11010043
Chicago/Turabian StylePulliam, Nicholas, Jessica Tang, Weini Wang, Fang Fang, Riddhi Sood, Heather M. O’Hagan, Kathy D. Miller, Robert Clarke, and Kenneth P. Nephew. 2019. "Poly-ADP-Ribosylation of Estrogen Receptor-Alpha by PARP1 Mediates Antiestrogen Resistance in Human Breast Cancer Cells" Cancers 11, no. 1: 43. https://doi.org/10.3390/cancers11010043
APA StylePulliam, N., Tang, J., Wang, W., Fang, F., Sood, R., O’Hagan, H. M., Miller, K. D., Clarke, R., & Nephew, K. P. (2019). Poly-ADP-Ribosylation of Estrogen Receptor-Alpha by PARP1 Mediates Antiestrogen Resistance in Human Breast Cancer Cells. Cancers, 11(1), 43. https://doi.org/10.3390/cancers11010043





