Detection of TP53 Mutations in Tissue or Liquid Rebiopsies at Progression Identifies ALK+ Lung Cancer Patients with Poor Survival
Abstract
:1. Introduction
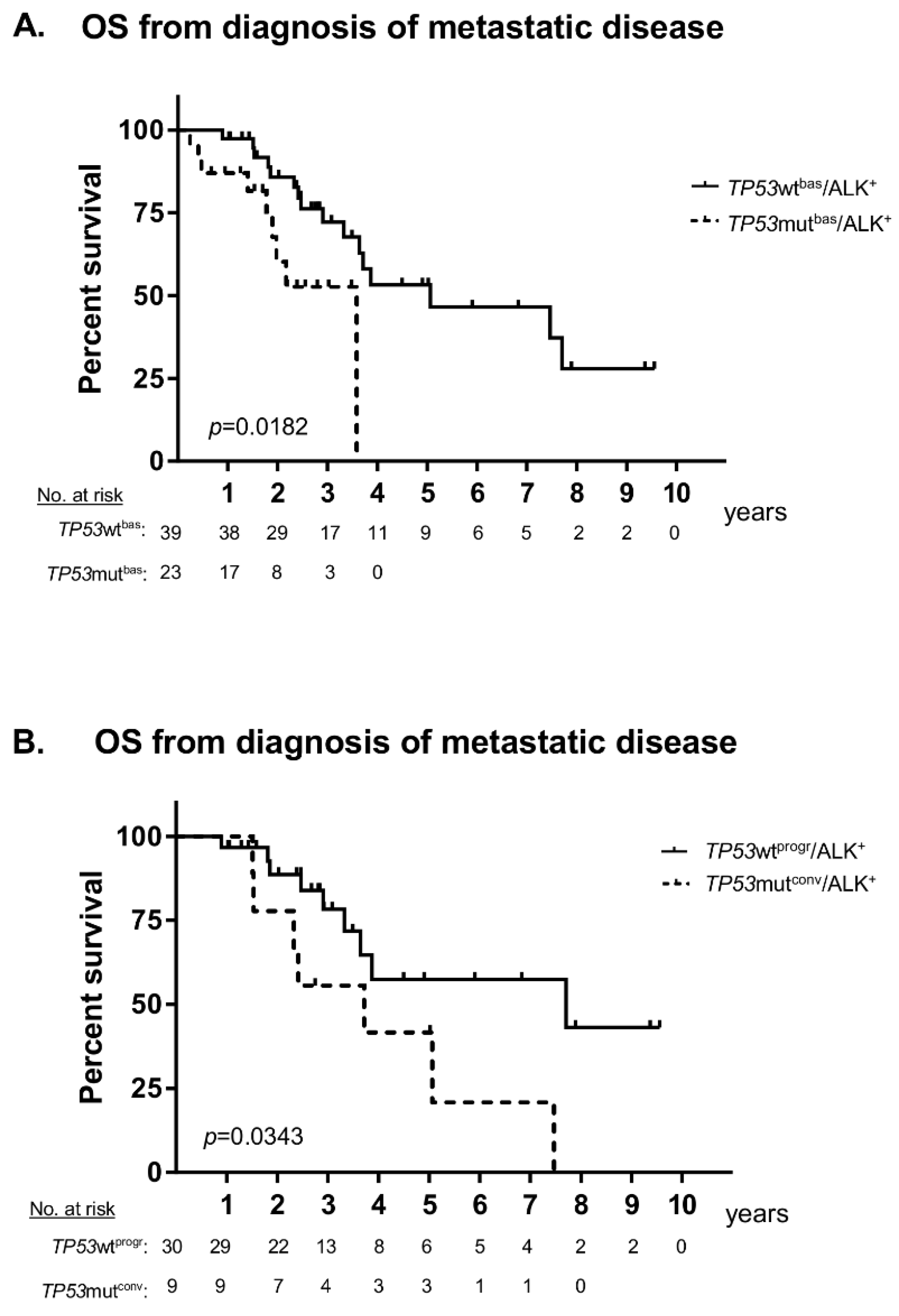
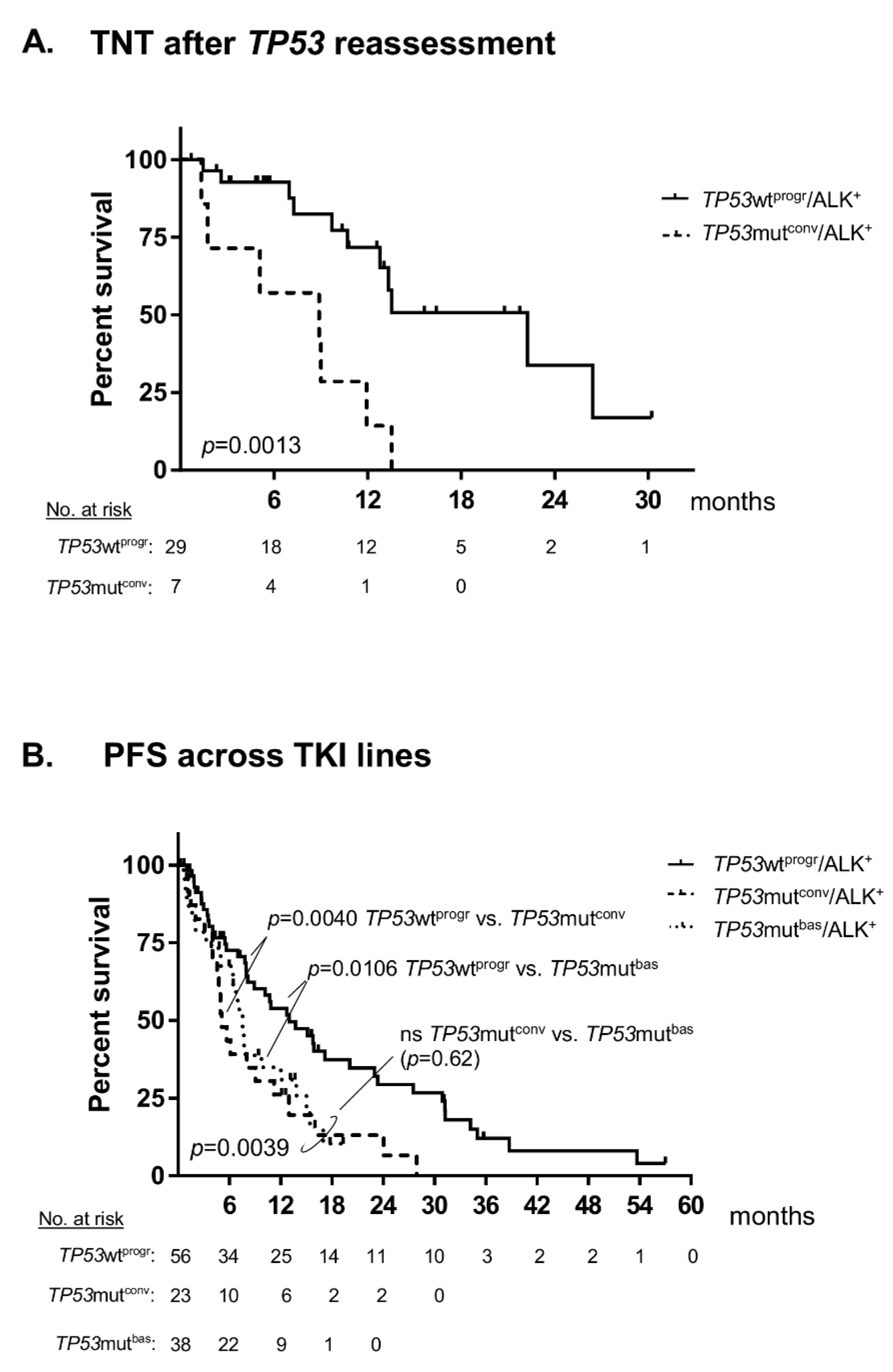
2. Results
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ALK | anaplastic lymphoma kinase |
| NSCLC | non-small cell lung cancer |
| TKI | tyrosine kinase inhibitor |
| TNT TP53 | time-to-next-treatment tumor protein p53 |
| PFS | progression-free survival |
| OS | overall survival |
References
- Soda, M.; Choi, Y.L.; Enomoto, M.; Takada, S.; Yamashita, Y.; Ishikawa, S.; Fujiwara, S.; Watanabe, H.; Kurashina, K.; Hatanaka, H.; et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007, 448, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Duruisseaux, M.; Besse, B.; Cadranel, J.; Perol, M.; Mennecier, B.; Bigay-Game, L.; Descourt, R.; Dansin, E.; Audigier-Valette, C.; Moreau, L.; et al. Overall survival with crizotinib and next-generation ALK inhibitors in ALK-positive non-small-cell lung cancer (IFCT-1302 CLINALK): A french nationwide cohort retrospective study. Oncotarget 2017, 8, 21903–21917. [Google Scholar] [CrossRef] [PubMed]
- Woo, C.G.; Seo, S.; Kim, S.W.; Jang, S.J.; Park, K.S.; Song, J.Y.; Lee, B.; Richards, M.W.; Bayliss, R.; Lee, D.H.; et al. Differential protein stability and clinical responses of EML4-ALK fusion variants to various alk inhibitors in advanced ALK-rearranged non-small cell lung cancer. Ann. Oncol. 2017, 28, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.J.; Zhu, V.W.; Yoda, S.; Yeap, B.Y.; Schrock, A.B.; Dagogo-Jack, I.; Jessop, N.A.; Jiang, G.Y.; Le, L.P.; Gowen, K.; et al. Impact of EML4-ALK variant on resistance mechanisms and clinical outcomes in ALK-positive lung cancer. J. Clin. Oncol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Christopoulos, P.; Endris, V.; Bozorgmehr, F.; Elsayed, M.; Kirchner, M.; Ristau, J.; Buchhalter, I.; Penzel, R.; Herth, F.J.; Heussel, C.P.; et al. EML4-ALK fusion variant V3 is a high-risk feature conferring accelerated metastatic spread, early treatment failure and worse overall survival in ALK+ NSCLC. Int. J. Cancer 2018, 142, 2589–2598. [Google Scholar] [CrossRef] [PubMed]
- Aisner, D.L.; Sholl, L.M.; Berry, L.D.; Rossi, M.R.; Chen, H.; Fujimoto, J.; Moreira, A.L.; Ramalingam, S.S.; Villaruz, L.C.; Otterson, G.A.; et al. The impact of smoking and TP53 mutations in lung adenocarcinoma patients with targetable mutations-the lung cancer mutation consortium (LCMC2). Clin. Cancer Res. 2018, 24, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Kron, A.; Alidousty, C.; Scheffler, M.; Merkelbach-Bruse, S.; Seidel, D.; Riedel, R.; Ihle, M.; Michels, S.; Nogova, L.; Fassunke, J.; et al. Impact of TP53 mutation status on systemic treatment outcome in ALK-rearranged non-small-cell lung cancer. Ann. Oncol. 2018. [Google Scholar] [CrossRef]
- Christopoulos, P.; Kirchner, M.; Bozorgmehr, F.; Endris, V.; Elsayed, M.; Budczies, J.; Ristau, J.; Penzel, R.; Herth, F.J.; Heussel, C.P.; et al. Identification of a highly lethal V3+ TP53+ subset in ALK+ lung adenocarcinoma. Int. J. Cancer 2018. [Google Scholar] [CrossRef]
- Lin, J.J.; Riely, G.J.; Shaw, A.T. Targeting alk: Precision medicine takes on drug resistance. Cancer Discov. 2017, 7, 137–155. [Google Scholar] [CrossRef]
- McCoach, C.E.; Blakely, C.M.; Banks, K.C.; Levy, B.; Chue, B.M.; Raymond, V.M.; Le, A.T.; Lee, C.E.; Diaz, J.; Waqar, S.N.; et al. Clinical utility of cell-free DNA for the detection of alk fusions and genomic mechanisms of alk inhibitor resistance in non-small cell lung cancer. Clin. Cancer Res. 2018, 24, 2758–2770. [Google Scholar] [CrossRef]
- Amirouchene-Angelozzi, N.; Swanton, C.; Bardelli, A. Tumor evolution as a therapeutic target. Cancer Discov. 2017. [Google Scholar] [CrossRef] [PubMed]
- Gainor, J.F.; Dardaei, L.; Yoda, S.; Friboulet, L.; Leshchiner, I.; Katayama, R.; Dagogo-Jack, I.; Gadgeel, S.; Schultz, K.; Singh, M.; et al. Molecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK-rearranged lung cancer. Cancer Discov. 2016, 6, 1118–1133. [Google Scholar] [CrossRef] [PubMed]
- Malcikova, J.; Stano-Kozubik, K.; Tichy, B.; Kantorova, B.; Pavlova, S.; Tom, N.; Radova, L.; Smardova, J.; Pardy, F.; Doubek, M.; et al. Detailed analysis of therapy-driven clonal evolution of TP53 mutations in chronic lymphocytic leukemia. Leukemia 2015, 29, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Chin, M.; Sive, J.I.; Allen, C.; Roddie, C.; Chavda, S.J.; Smith, D.; Blombery, P.; Jones, K.; Ryland, G.L.; Popat, R.; et al. Prevalence and timing of TP53 mutations in del(17p) myeloma and effect on survival. Blood Cancer J. 2017, 7, e610. [Google Scholar] [CrossRef] [PubMed]
- Bykov, V.J.N.; Eriksson, S.E.; Bianchi, J.; Wiman, K.G. Targeting mutant p53 for efficient cancer therapy. Nat. Rev. Cancer 2018, 18, 89–102. [Google Scholar] [CrossRef]
- Zhang, L.L.; Kan, M.; Zhang, M.M.; Yu, S.S.; Xie, H.J.; Gu, Z.H.; Wang, H.N.; Zhao, S.X.; Zhou, G.B.; Song, H.D.; et al. Multiregion sequencing reveals the intratumor heterogeneity of driver mutations in TP53-driven non-small cell lung cancer. Int. J. Cancer 2017, 140, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Jamal-Hanjani, M.; Wilson, G.A.; McGranahan, N.; Birkbak, N.J.; Watkins, T.B.K.; Veeriah, S.; Shafi, S.; Johnson, D.H.; Mitter, R.; Rosenthal, R.; et al. Tracking the evolution of non-small-cell lung cancer. N. Engl. J. Med. 2017, 376, 2109–2121. [Google Scholar] [CrossRef] [PubMed]
- Passiglia, F.; Rizzo, S.; Di Maio, M.; Galvano, A.; Badalamenti, G.; Listi, A.; Gulotta, L.; Castiglia, M.; Fulfaro, F.; Bazan, V.; et al. The diagnostic accuracy of circulating tumor DNA for the detection of EGFR-T790M mutation in nsclc: A systematic review and meta-analysis. Sci. Rep. 2018, 8, 13379. [Google Scholar] [CrossRef]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New Response Evaluation Criteria In Solid Tumours: Revised recist guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Pfarr, N.; Stenzinger, A.; Penzel, R.; Warth, A.; Dienemann, H.; Schirmacher, P.; Weichert, W.; Endris, V. High-throughput diagnostic profiling of clinically actionable gene fusions in lung cancer. Genes Chromosomes Cancer 2016, 55, 30–44. [Google Scholar] [CrossRef]
- Volckmar, A.-L.; Leichsenring, J.; Kirchner, M.; Christopoulos, P.; Neumann, O.; Budczies, J.; de Oliveira, C.M.M.; Rempel, E.; Buchhalter, I.; Brandt, R.; et al. Combined targeted DNA and RNA sequencing of advanced NSCLC in routine molecular diagnostics: Analysis of the first 3,000 Heidelberg cases. Int. J. Cancer 2019. [Google Scholar] [CrossRef] [PubMed]
- Endris, V.; Penzel, R.; Warth, A.; Muckenhuber, A.; Schirmacher, P.; Stenzinger, A.; Weichert, W. Molecular diagnostic profiling of lung cancer specimens with a semiconductor-based massive parallel sequencing approach: Feasibility, costs, and performance compared with conventional sequencing. J. Mol. Diagn. 2013, 15, 765–775. [Google Scholar] [CrossRef] [PubMed]
| All Study Patients (N = 62) | TP53wtbas (n = 39) | TP53mutbas (n = 23) | ||
|---|---|---|---|---|
| TP53wtprogr (n = 30) | TP53mutconv (n = 9) | |||
| Age (median; IQR) | 51; 17 | 63; 20 | 65; 19 | |
| Sex (male/female) | 15/16 | 5/4 | 12/11 | |
| ECOG PS at diagnosis of stage IV (median; IQR) | 0; 0 | 0; 0 | 1; 0 | |
| Histology | adenocarcinoma 1 | 29/30 | 9/9 | 23/23 |
| ALK status | positive | all cases by inclusion criteria | ||
| EML4-ALK V3 2 | 8/24 | 5/9 | 8/20 | |
| Stage IV NSCLC | at initial diagnosis | 20/30 * | 9/9 | 21/23 |
| M1a | 7/20 | 1/9 | 5/21 | |
| by relapse of M0 NSCLC | 10/30 | 0/9 | 2/23 | |
| TP53 assessment at baseline + at progression 3 | ||||
| method | FFPE at BL +FFPE at PD 4 | 8/30 (neg + neg) | 2/9 (neg + pos) | See Table S1 |
| FFPE at BL +ctDNA at PD | 11/30 (neg + neg) | 7/9 (neg + pos) | ||
| FFPE at PD 4 | 6/30 (neg) | |||
| only ctDNA at PD | 5/30 (neg) | |||
| TKI line (start) at 2nd assessment (median; IQR) | 2; 1 | 2; 1 | ||
| treatment line at 2nd assessment (median; IQR) | 2; 3 | 4; 1 | ||
| - days after diagnosis of stage IV (median; IQR) | 702; 1056 | 752; 600 | ||
| ALK TKI treatment, next-line | ||||
| crizotinib | 14 | 2 | ||
| ceritinib | 7 | 4 | ||
| alectinib | 6 | 1 | ||
| brigatinib | 2 | - | ||
| - no. of patients 5 | 29/30 (97%) | 7/9 (78%) | ||
| - no. of patients with CBDP | 15/30 | 4/7 | ||
| ALK TKI treatment, all lines (1–8) | ||||
| crizotinib | 23 | 9 | 19 | |
| ceritinib | 12 | 9 | 5 | |
| alectinib | 14 | 4 | 10 | |
| brigatinib | 4 | 0 | 3 | |
| lorlatinib | 3 | 1 | 1 | |
| - no. of patients 5,6 | 29/30 (97%) | 9/9 (100%) | 22/23 (96%) | |
| Chemotherapy, all lines (1–8) | ||||
| platin-doublets | 15 | 8 | 7 | |
| monotherapy | 6 | 4 | 6 | |
| - no. of patients | 14/30 (47%) | 8/9 (89%) | 8/23 (35%) | |
| Summary of the complete treatment | ||||
| no. of treatment lines (mean; SD) | 3.0; 1.5 | 4.0; 1.7 | 2.4; 1.6 | |
| no. of TKI treatment lines (mean; SD) | 1.9; 1.2 | 2.6; 1.0 | 1.7; 1.1 | |
| patients with additional radiotherapy | 18/30 (60%) | 6/9 (67%) | 12/21 (57%) | |
| patients with additional surgical treatment 7 | 5/30 (17%) | 1/9 (11%) | 5/21 (24%) | |
| Follow-up in months (median (25th–75th percentile)) | 36 (28–94) | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Christopoulos, P.; Dietz, S.; Kirchner, M.; Volckmar, A.-L.; Endris, V.; Neumann, O.; Ogrodnik, S.; Heussel, C.-P.; Herth, F.J.; Eichhorn, M.; et al. Detection of TP53 Mutations in Tissue or Liquid Rebiopsies at Progression Identifies ALK+ Lung Cancer Patients with Poor Survival. Cancers 2019, 11, 124. https://doi.org/10.3390/cancers11010124
Christopoulos P, Dietz S, Kirchner M, Volckmar A-L, Endris V, Neumann O, Ogrodnik S, Heussel C-P, Herth FJ, Eichhorn M, et al. Detection of TP53 Mutations in Tissue or Liquid Rebiopsies at Progression Identifies ALK+ Lung Cancer Patients with Poor Survival. Cancers. 2019; 11(1):124. https://doi.org/10.3390/cancers11010124
Chicago/Turabian StyleChristopoulos, Petros, Steffen Dietz, Martina Kirchner, Anna-Lena Volckmar, Volker Endris, Olaf Neumann, Simon Ogrodnik, Claus-Peter Heussel, Felix J. Herth, Martin Eichhorn, and et al. 2019. "Detection of TP53 Mutations in Tissue or Liquid Rebiopsies at Progression Identifies ALK+ Lung Cancer Patients with Poor Survival" Cancers 11, no. 1: 124. https://doi.org/10.3390/cancers11010124
APA StyleChristopoulos, P., Dietz, S., Kirchner, M., Volckmar, A.-L., Endris, V., Neumann, O., Ogrodnik, S., Heussel, C.-P., Herth, F. J., Eichhorn, M., Meister, M., Budczies, J., Allgäuer, M., Leichsenring, J., Zemojtel, T., Bischoff, H., Schirmacher, P., Thomas, M., Sültmann, H., & Stenzinger, A. (2019). Detection of TP53 Mutations in Tissue or Liquid Rebiopsies at Progression Identifies ALK+ Lung Cancer Patients with Poor Survival. Cancers, 11(1), 124. https://doi.org/10.3390/cancers11010124







