Clonal Heterogeneity Reflected by PI3K-AKT-mTOR Signaling in Human Acute Myeloid Leukemia Cells and Its Association with Adverse Prognosis
Abstract
1. Introduction
2. Results
2.1. Clonal AML Cell Heterogeneity Reflected by PI3K-Akt-mTOR Signaling Is Seen for a Subset of Patients
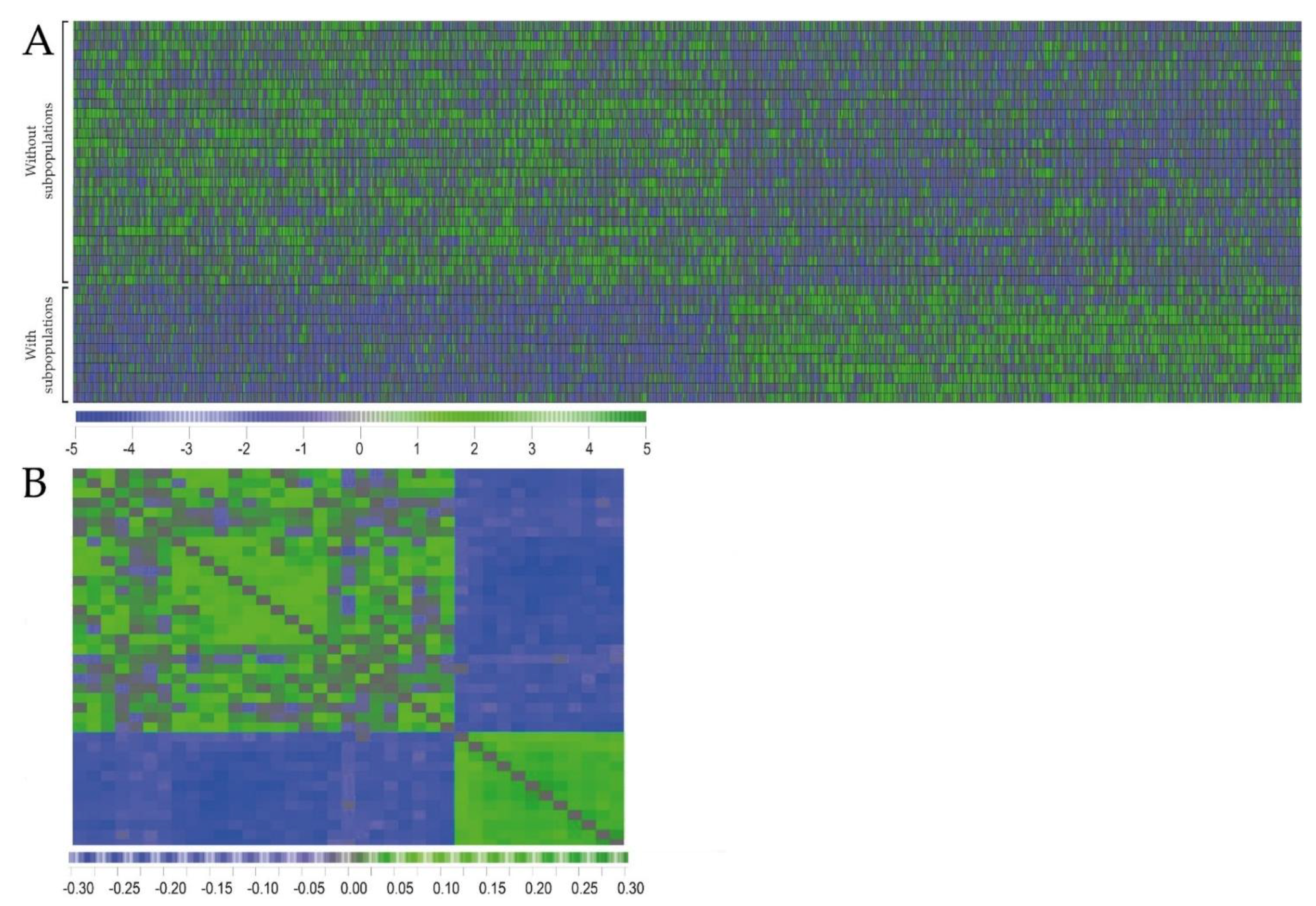
2.2. Primary AML Cells Derived from Patients with and without dual PI3K-Akt-mTOR Cell Populations Differ in Their Global Gene Expression Profiles
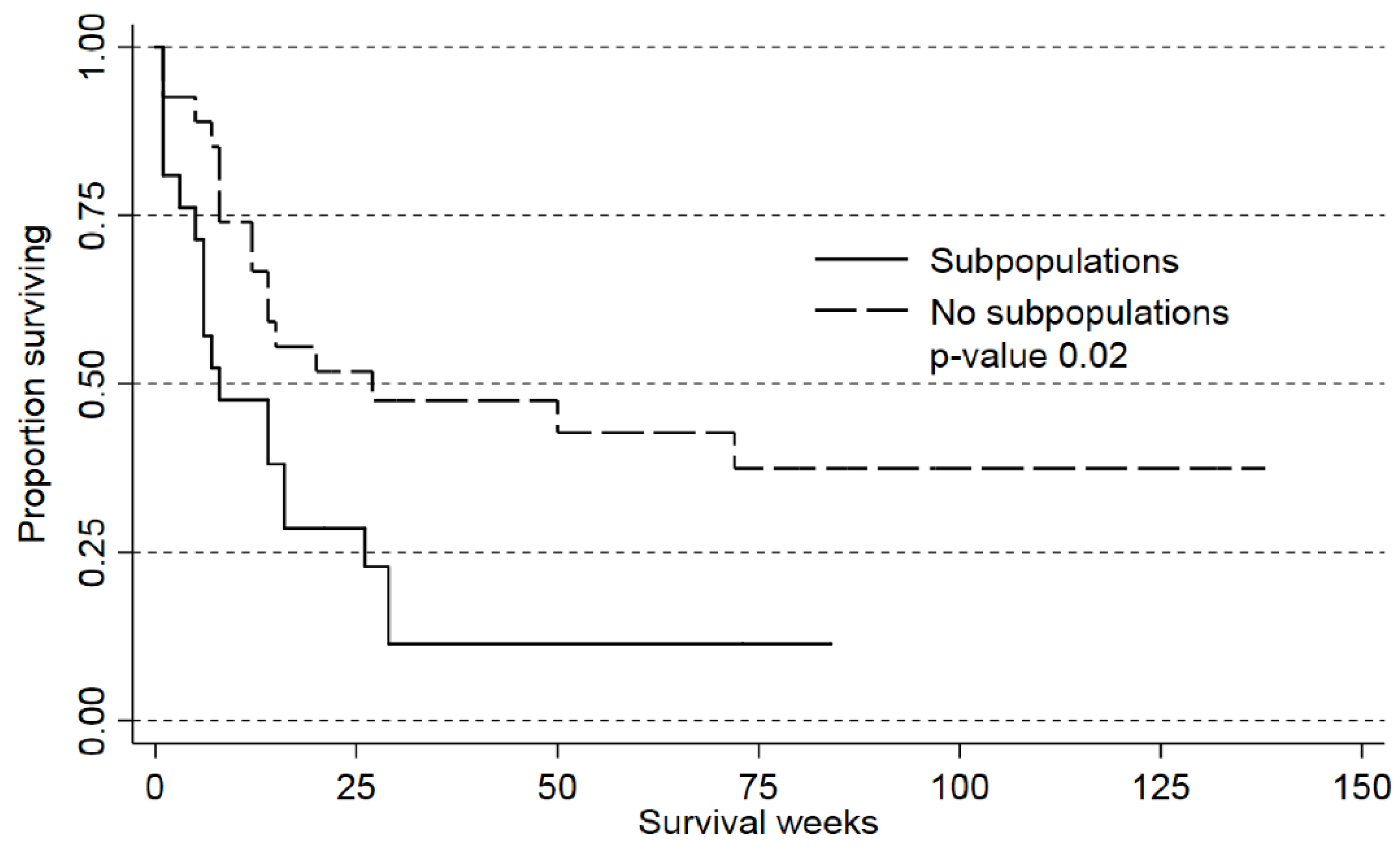
2.3. Detection of AML Subclones Based upon PI3K-Akt-mTOR Signaling Is Associated with Decreased Patient Survival
3. Discussion
4. Materials and Methods
4.1. AML Patients
4.2. Flow-Cytometric Analysis of PI3K-Akt-mTOR Activation
4.3. Analysis of Global Gene Expression Profiles and Mutation Analyses
4.4. Data Collection, Bioinformatical and Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Weisdorf, D.J.; Bloomfield, C.D. Acute Myeloid Leukemia. N. Engl. J. Med. 2015, 373, 1136–1152. [Google Scholar] [CrossRef] [PubMed]
- Martelli, A.M.; Evangelisti, C.; Chiarini, F.; McCubrey, J.A. The phosphatidylinositol 3-kinase/Akt/mTOR signaling network as a therapeutic target in acute myelogenous leukemia patients. Oncotarget 2010, 1, 89–103. [Google Scholar] [PubMed]
- Polak, R.; Buitenhuis, M. The PI3K/PKB signaling module as key regulator of hematopoiesis: Implications for therapeutic strategies in leukemia. Blood 2012, 119, 911–923. [Google Scholar] [CrossRef] [PubMed]
- Brenner, A.K.; Andersson Tvedt, T.H.; Bruserud, Ø. The Complexity of Targeting PI3K-Akt-mTOR Signalling in Human Acute Myeloid Leukaemia: The Importance of Leukemic Cell Heterogeneity, Neighbouring Mesenchymal Stem Cells and Immunocompetent Cells. Molecules 2016, 21, e1512. [Google Scholar] [CrossRef] [PubMed]
- Bruserud, Ø.; Aasebø, E.; Hernandez-Valladares, M.; Tsykunova, G.; Reikvam, H. Therapeutic targeting of leukemic stem cells in acute myeloid leukemia—The biological background for possible strategies. Expert Opin. Drug Discov. 2017, 12, 1053–1065. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Storer, B.; Wood, B.; Gyurkocza, B.; Sandmaier, B.M.; Appelbaum, F.R. Prognostic impact of discordant results from cytogenetics and flow cytometry in patients with acute myeloid leukemia undergoing hematopoietic cell transplantation. Cancer 2012, 118, 2411–2419. [Google Scholar] [CrossRef] [PubMed]
- Bochtler, T.; Stolzel, F.; Heilig, C.E.; Kunz, C.; Mohr, B.; Jauch, A.; Janssen, J.W.G.; Kramer, M.; Benner, A.; Bornhauser, M.; et al. Clonal heterogeneity as detected by metaphase karyotyping is an indicator of poor prognosis in acute myeloid leukemia. J. Clin. Oncol. 2013, 31, e3898. [Google Scholar] [CrossRef] [PubMed]
- Skavland, J.; Jorgensen, K.M.; Hadziavdic, K.; Hovland, R.; Jonassen, I.; Bruserud, Ø.; Gjertsen, B.T. Specific cellular signal-transduction responses to in vivo combination therapy with ATRA, valproic acid and theophylline in acute myeloid leukemia. Blood Cancer J. 2011, 1, e4. [Google Scholar] [CrossRef] [PubMed]
- Tabe, Y.; Tafuri, A.; Sekihara, K.; Yang, H.; Konopleva, M. Inhibition of mTOR kinase as a therapeutic target for acute myeloid leukemia. Expert Opin. Ther. Targets 2017, 21, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Buchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [PubMed]
- Herschbein, L.; Liesveld, J.L. Dueling for dual inhibition: Means to enhance effectiveness of PI3K/Akt/mTOR inhibitors in AML. Blood Rev. 2017, 32, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Reikvam, H.; Tamburini, J.; Skrede, S.; Holdhus, R.; Poulain, L.; Ersvaer, E.; Hatfield, K.J.; Bruserud, Ø. Antileukaemic effect of PI3K-mTOR inhibitors in acute myeloid leukaemia-gene expression profiles reveal CDC25B expression as determinate of pharmacological effect. Br. J. Haematol. 2014, 164, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Li, X.; Ma, J.; Zhao, J.; Liu, S.; Wang, G.; Edwards, H.; Taub, J.W.; Lin, H.; Ge, Y. Targeting PI3K, mTOR, ERK, and Bcl-2 signaling network shows superior antileukemic activity against AML ex vivo. Biochem. Pharmacol. 2018, 148, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Kanda, Y.; Hamaki, T.; Yamamoto, R.; Chizuka, A.; Suguro, M.; Matsuyama, T.; Takezako, N.; Miwa, A.; Kami, M.; Hirai, H.; et al. The clinical significance of CD34 expression in response to therapy of patients with acute myeloid leukemia: An overview of 2483 patients from 22 studies. Cancer 2000, 88, 2529–2533. [Google Scholar] [CrossRef]
- Ryningen, A.; Ersvaer, E.; Oyan, A.M.; Kalland, K.H.; Vintermyr, O.K.; Gjertsen, B.T.; Bruserud, O. Stress-induced in vitro apoptosis of native human acute myelogenous leukemia (AML) cells shows a wide variation between patients and is associated with low BCL-2:Bax ratio and low levels of heat shock protein 70 and 90. Leuk. Res. 2006, 30, 1531–1540. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, I.; Garcia-Esparcia, P.; Carmona, M.; Carro, E.; Aronica, E.; Kovacs, G.G.; Grison, A.; Gustincich, S. Olfactory Receptors in Non-Chemosensory Organs: The Nervous System in Health and Disease. Front Aging Neurosci. 2016, 8, e163. [Google Scholar] [CrossRef] [PubMed]
- Lachen-Montes, M.; Fernandez-Irigoyen, J.; Santamaria, E. Deconstructing the molecular architecture of olfactory areas using proteomics. Proteom. Clin. Appl. 2016, 10, 1178–1190. [Google Scholar] [CrossRef] [PubMed]
- Antunes, G.; de Souzax, F.M.S. Olfactory receptor signaling. Method Cell. Biol. 2016, 132, 127–145. [Google Scholar]
- Malki, A.; Fiedler, J.; Fricke, K.; Ballweg, I.; Pfaffl, M.W.; Krautwurst, D. Class I odorant receptors, TAS1R and TAS2R taste receptors, are markers for subpopulations of circulating leukocytes. J. Leukocyte Biol. 2015, 97, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Maßberg, D.; Hatt, H. Human Olfactory Receptors: Novel Cellular Functions Outside of the Nose. Physiol. Rev. 2018, 98, 1739–1763. [Google Scholar] [CrossRef] [PubMed]
- Ranzani, M.; Iyer, V.; Ibarra-Soria, X.; Del Castillo Velasco-Herrera, M.; Garnett, M.; Logan, D.; Adams, D.J. Revisiting olfactory receptors as putative drivers of cancer. Wellcome Open Res. 2017, 2, e9. [Google Scholar] [CrossRef] [PubMed]
- Gelis, L.; Jovancevic, N.; Bechara, F.G.; Neuhaus, E.M.; Hatt, H. Functional expression of olfactory receptors in human primary melanoma and melanoma metastasis. Exp. Dermatol. 2017, 26, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Morita, R.; Hirohashi, Y.; Torigoe, T.; Ito-Inoda, S.; Takahashi, A.; Mariya, T.; Asanuma, H.; Tamura, Y.; Tsukahara, T.; Kanaseki, T.; et al. Olfactory Receptor Family 7 Subfamily C Member 1 Is a Novel Marker of Colon Cancer-Initiating Cells and Is a Potent Target of Immunotherapy. Clin. Cancer Res. 2016, 22, 3298–3309. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, E.M.; Zhang, W.Y.; Gelis, L.; Deng, Y.; Noldus, J.; Hatt, H. Activation of an Olfactory Receptor Inhibits Proliferation of Prostate Cancer Cells. J. Biol. Chem. 2009, 284, 16218–16225. [Google Scholar] [CrossRef] [PubMed]
- Pavan, B.; Capuzzo, A.; Dalpiaz, A. Potential therapeutic effects of odorants through their ectopic receptor in pigmented cells. Drug Discov. Today 2017, 22, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhao, H.; Fu, N.; Chen, L. The diversified function and potential therapy of ectopic olfactory receptors in non-olfactory tissues. J. Cell Physiol. 2018, 233, 2104–2115. [Google Scholar] [CrossRef] [PubMed]
- Manteniotis, S.; Wojcik, S.; Brauhoff, P.; Mollmann, M.; Petersen, L.; Gothert, J.R.; Schmiegel, W.; Duhrsen, U.; Gisselmann, G.; Hatt, H. Functional characterization of the ectopically expressed olfactory receptor 2AT4 in human myelogenous leukemia. Cell. Death Discov. 2016, 2, e15070. [Google Scholar] [CrossRef] [PubMed]
- Manteniotis, S.; Wojcik, S.; Gothert, J.R.; Durig, J.; Duhrsen, U.; Gisselmann, G.; Hatt, H. Deorphanization and characterization of the ectopically expressed olfactory receptor OR51B5 in myelogenous leukemia cells. Cell. Death Discov. 2016, 2, e16010. [Google Scholar] [CrossRef] [PubMed]
- Sanz, G.; Leray, I.; Muscat, A.; Acquistapace, A.; Cui, T.; Rivière, J.; Vincent-Naulleau, S.; Giandomenico, V.; Mir, L.M. Gallein, a Gβγ subunit signalling inhibitor, inhibits metastatic spread of tumour cells expressing OR51E2 and exposed to its odorant ligand. BMC Res. Notes 2017, 10, e541. [Google Scholar] [CrossRef] [PubMed]
- Maßberg, D.; Simon, A.; Haussinger, D.; Keitel, V.; Gisselmann, G.; Conrad, H.; Hatt, H. Monoterpene (−)-citronellal affects hepatocarcinoma cell signaling via an olfactory receptor. Arch. Biochem. Biophys. 2015, 566, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Sanz, G.; Leray, I.; Grebert, D.; Antoine, S.; Acquistapace, A.; Muscat, A.; Boukadiri, A.; Mir, L.M. Structurally related odorant ligands of the olfactory receptor OR51E2 differentially promote metastasis emergence and tumor growth. Oncotarget 2017, 8, 4330–4341. [Google Scholar] [CrossRef] [PubMed]
- Sanz, G.; Leray, I.; Dewaele, A.; Sobilo, J.; Lerondel, S.; Bouet, S.; Grebert, D.; Monnerie, R.; Pajot-Augy, E.; Mir, L.M. Promotion of Cancer Cell Invasiveness and Metastasis Emergence Caused by Olfactory Receptor Stimulation. PloS ONE 2014, 9, e85110. [Google Scholar] [CrossRef] [PubMed]
- Irish, J.M.; Hovland, R.; Krutzik, P.O.; Perez, O.D.; Bruserud, Ø.; Gjertsen, B.T.; Nolan, G.P. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell 2004, 118, 217–228. [Google Scholar] [CrossRef] [PubMed]
- van Giesen, L.; Garrity, P.A. More than meets the IR: The expanding roles of variant Ionotropic Glutamate Receptors in sensing odor, taste, temperature and moisture. F1000 Res. 2017, 6, e1753. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Zhuang, H.; Chi, Q.; Vosshall, L.B.; Matsunami, H. Genetic variation in a human odorant receptor alters odour perception. Nature 2007, 449, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.L.; Wang, J.H.; Zhao, A.H.; Xu, X.; Wang, Y.H.; Chen, T.L.; Li, J.M.; Mi, J.Q.; Zhu, Y.M.; Liu, Y.F.; et al. A distinct glucose metabolism signature of acute myeloid leukemia with prognostic value. Blood 2014, 124, 1645–1654. [Google Scholar] [CrossRef] [PubMed]
- Schwob, J.E.; Jang, W.; Holbrook, E.H.; Lin, B.; Herrick, D.B.; Peterson, J.N.; Hewitt Coleman, J. Stem and progenitor cells of the mammalian olfactory epithelium: Taking poietic license. J. Comp. Neurol. 2017, 525, 1034–1054. [Google Scholar] [CrossRef] [PubMed]
- Feingold, E.A.; Penny, L.A.; Nienhuis, A.W.; Forget, B.G. An olfactory receptor gene is located in the extended human beta-globin gene cluster and is expressed in erythroid cells. Genomics 1999, 61, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Gjertsen, B.T.; Oyan, A.M.; Marzolf, B.; Hovland, R.; Gausdal, G.; Doskeland, S.O.; Dimitrov, K.; Golden, A.; Kalland, K.H.; Hood, L.; et al. Analysis of acute myelogenous leukemia: preparation of samples for genomic and proteomic analyses. J. Hematother. Stem Cell Res. 2002, 11, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Reikvam, H.; Hovland, R.; Forthun, R.B.; Erdal, S.; Gjertsen, B.T.; Fredly, H.; Bruserud, Ø. Disease-stabilizing treatment based on all-trans retinoic acid and valproic acid in acute myeloid leukemia–identification of responders by gene expression profiling of pretreatment leukemic cells. BMC Cancer 2017, 17, e630. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]

| Covariate | Crude Analysis | Adjusted Analysis | ||||
|---|---|---|---|---|---|---|
| p-Value | HR | 95%-CI | p-Value | HR | 95%-CI | |
| Age (per decade) | <0.01 | 1.64 | 1.24–2.17 | <0.01 | 1.69 | 1.22–2.36 |
| Subpopulation versus no subpopulation | 0.03 | 2.15 | 1,01–4,26 | 0.04 | 2.28 | 1.03–5.04 |
| Adverse cytogenetics | 0.519 | 0.759 | 0.33–1.75 | 0.11 | 0.33 | 0.11–1.04 |
| NPM1-wt and Flt3-wt | NA | 1 (reference) | NA | 1 (reference) | ||
| NPM1-mutated and Flt3-wt | 0.92 | 1.62 | 0.58–4.56 | 0.693 | 1.24 | 0.42–3.75 |
| NPM1-wt and FLT3-mutated | 0.21 | 1.86 | 0.71–4.92 | 0.03 | 3.88 | 1.18–12.71 |
| NPM1-mutated and FLT3-mutated | 0.05 | 2.33 | 1.00–5.34 | 0.48 | 1.44 | 0.57–3.61 |
| Patient characteristics | |||
|---|---|---|---|
| Age | Secondary AML | ||
| Median (years) | 67 | chemo | 5 |
| Range (years) | 18–87 | CML | 2 |
| CML-RELAPSE | 1 | ||
| Gender | CMML | 4 | |
| Females | 49 | de novo | 81 |
| Males | 65 | LiFraum, chemo | 1 |
| MDS | 8 | ||
| MDS, AML relapse | 1 | ||
| MDS, CHEMO | 1 | ||
| Myelofibrosis | 3 | ||
| Polycytemia vera | 1 | ||
| Relapse | 5 | ||
| Total | 114 | Relapse, chemo | 1 |
| Cell Morphology | |||
| FAB Classification | CD34 Receptor | ||
| M0 | 8 | Negative (<20%) | 30 |
| M1 | 28 | Positive (>20%) | 78 |
| M2 | 22 | n.d. | 6 |
| M4 | 27 | ||
| M5 | 21 | ||
| M7 | 1 | ||
| n.d. | 7 | ||
| Cell Genetics | |||
| Cytogenetics | Mutations | ||
| Adverse | 20 | NPM1 mutationsMutated | 35 |
| Favorable | 11 | Wild-type | 62 |
| Intermediate | 11 | n.d. | 17 |
| Normal | 60 | ||
| n.d. | 12 | Flt3 mutations | |
| ITD | 41 | ||
| Wild-type | 55 | ||
| n.d. | 18 | ||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nepstad, I.; Hatfield, K.J.; Tvedt, T.H.A.; Reikvam, H.; Bruserud, Ø. Clonal Heterogeneity Reflected by PI3K-AKT-mTOR Signaling in Human Acute Myeloid Leukemia Cells and Its Association with Adverse Prognosis. Cancers 2018, 10, 332. https://doi.org/10.3390/cancers10090332
Nepstad I, Hatfield KJ, Tvedt THA, Reikvam H, Bruserud Ø. Clonal Heterogeneity Reflected by PI3K-AKT-mTOR Signaling in Human Acute Myeloid Leukemia Cells and Its Association with Adverse Prognosis. Cancers. 2018; 10(9):332. https://doi.org/10.3390/cancers10090332
Chicago/Turabian StyleNepstad, Ina, Kimberley Joanne Hatfield, Tor Henrik Anderson Tvedt, Håkon Reikvam, and Øystein Bruserud. 2018. "Clonal Heterogeneity Reflected by PI3K-AKT-mTOR Signaling in Human Acute Myeloid Leukemia Cells and Its Association with Adverse Prognosis" Cancers 10, no. 9: 332. https://doi.org/10.3390/cancers10090332
APA StyleNepstad, I., Hatfield, K. J., Tvedt, T. H. A., Reikvam, H., & Bruserud, Ø. (2018). Clonal Heterogeneity Reflected by PI3K-AKT-mTOR Signaling in Human Acute Myeloid Leukemia Cells and Its Association with Adverse Prognosis. Cancers, 10(9), 332. https://doi.org/10.3390/cancers10090332







