Abstract
Metastatic dissemination of epithelial ovarian cancer (EOC) predominantly occurs through direct cell shedding from the primary tumor into the intra-abdominal cavity that is filled with malignant ascitic effusions. Facilitated by the fluid flow, cells distribute throughout the cavity, broadly seed and invade through peritoneal lining, and resume secondary tumor growth in abdominal and pelvic organs. At all steps of this unique metastatic process, cancer cells exist within a multidimensional tumor microenvironment consisting of intraperitoneally residing cancer-reprogramed fibroblasts, adipose, immune, mesenchymal stem, mesothelial, and vascular cells that exert miscellaneous bioactive molecules into malignant ascites and contribute to EOC progression and metastasis via distinct molecular mechanisms and epigenetic dysregulation. This review outlines basic epigenetic mechanisms, including DNA methylation, histone modifications, chromatin remodeling, and non-coding RNA regulators, and summarizes current knowledge on reciprocal interactions between each participant of the EOC cellular milieu and tumor cells in the context of aberrant epigenetic crosstalk. Promising research directions and potential therapeutic strategies that may encompass epigenetic tailoring as a component of complex EOC treatment are discussed.
1. Introduction
Epithelial ovarian cancer (EOC), a histopathologically, morphologically, and molecularly heterogeneous group of neoplasms [1], is the leading cause of gynecological malignancy-related deaths in women, with >14,000 deaths in the United States (US) and ~152,000 deaths worldwide yearly [2,3,4]. Most women have vastly disseminated intraperitoneal disease at the time of diagnosis contributing to a five-year survival rate of only 30% [5]. Development of multidrug resistant and essentially incurable tumor recurrence in the majority of patients after initial good response to standard platinum/taxane-based chemotherapy are also significant factors contributing to this deadly disease [6,7].
1.1. Tumor Microenvironment (TME) Associated with Ovarian Neoplasms
EOC initiation results from accumulation of genetic mutations and epigenetic changes resulting in malicious transformation of epithelial cells, stem cells, or transient metaplastic regions at the primary site, either ovary or the fallopian tube fimbriae [8,9,10,11,12,13,14,15,16,17,18]. While lymph node and hematogenous metastasis of ovarian cancer have been reported in human EOC cancer and/or model systems [19,20], the current consensus is that expansion of ovarian neoplastic masses occurs primarily via transcoelomic route, including the direct exfoliation of anoikis-resistant cancer cells and multi-cellular clusters from the original tumor, ascitic fluid-facilitated intraperitoneal dissemination, subsequent mesothelial adhesion and retraction, submesothelial extracellular matrix invasion, and ultimate establishment of secondary lesions in peritoneum-sheathed surfaces and organs [18,21,22,23]. During this metastasis process, ovarian cancer cells are confined to and nurtured by the complex host intraperitoneal cellular milieu, encompassing cells co-existing within the tumor bulk, freely available in ascitic effusions, and residing in peritoneal and adipose tissues—fibroblasts, mesothelial cells, adipocytes, infiltrating lymphocytes, macrophages, plasmacytoid dendritic cells, mesenchymal stem cells, and others (Figure 1) [24,25,26,27,28,29]. Both EOC and host non-cancerous cells secrete a plethora of bioactive soluble constituents—proteins, growth factors, phospholipids, hormones, cytokines—into the extracellular space and malignant ascites [23,27,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44], collectively generating a dynamic intraperitoneal TME that mediates ovarian cancer development, metastatic progression, and therapeutic response through receptor-ligand (autocrine, paracrine, endocrine) signaling, contact-dependent (juxtacrine) cell signaling, as well as epigenetic regulation (Figure 1B).
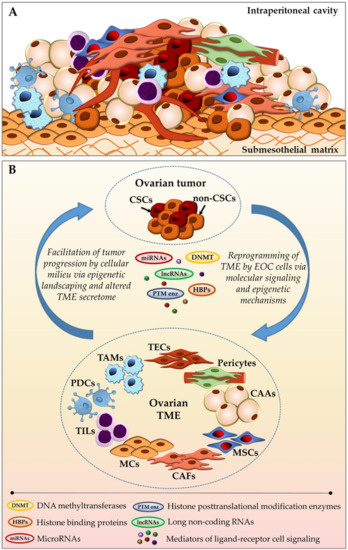
Figure 1.
Ovarian tumor-stroma bidirectional crosstalk. (A) Schematic representation of cellular diversity within the complex ovarian tumor bulk; and, (B) Reciprocal communication between ovarian cancer cells and intraperitoneally residing cancer-associated cellular milieu components via molecular signaling pathways and epigenetic regulation. CAAs—cancer-associated adipocytes; CAFs—cancer-associated fibroblasts; CSCs—cancer stem cells; EOC—epithelial ovarian cancer; MCs—mesothelial cells; MSCs—mesenchymal stem cells; PDCs—plasmacytoid dendritic cells; TAMs—tumor-associated macrophages; TECs—tumor-associated endothelial cells; TILs—tumor-infiltrating lymphocytes; TME—tumor microenvironment (see main text for details).
1.2. Basic Epigenetic Mechanisms at a Glance
Epigenetic modifications are heritable alterations in gene expression (activation or suppression) that occur as a result of perturbed chromatin organization and altered gene accessibility for transcriptional machinery in the absence of changes to the DNA itself [45]. Additionally, epigenetic mediation encompasses the modulation of gene expression at the posttranscriptional level via altered mRNA translation into protein (Figure 2). Fundamental epigenetic regulatory mechanisms include:
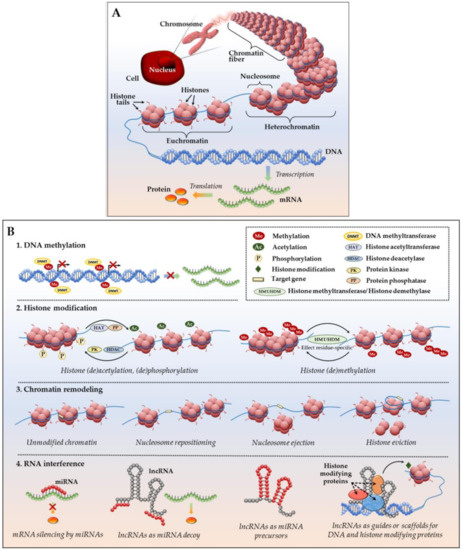
Figure 2.
Epigenetic regulation of gene expression. (A) DNA packing in a eukaryotic cell: a DNA molecule (chromosome) located inside the cell nucleus is composed of chromatin fibers which are made of nucleosomes—histone octamers wrapped by a DNA helix. Condensed chromatin (heterochromatin) is transcriptionally silent; loosely packed euchromatin allows access to DNA and active transcription into mRNA, followed by translation into protein; (B) Major epigenetic mechanisms of gene expression include: (1) DNA methylation; (2) histone modifications, such as histone acetylation, methylation, phosphorylation, etc.; (3) chromatin remodeling, including nucleosome sliding, nucleosome ejection and histone eviction; and, (4) mRNA interference with miRNAs and lncRNAs (see main text for details).
- DNA methylation—addition of methyl groups to DNA CpG sites without altering DNA nucleotide sequence. Methylation occurs by means of enzymes called DNA methyltransferases (DNMTs), which place methyl groups on symmetric cytosine residues in double-stranded CpG sites [46,47]. Hypermethylation of CpG islands (nucleotide sequences enriched for CpG sites) in the promoter regions of tumor suppressor genes (TSGs) and growth regulatory genes prompts gene silencing [46,47] as attached methyl groups physically block binding of transcription factors to the gene promoters. Alternatively, dense DNA methylation interferes with the proper nucleosome positioning [48]. Within the DNMT family (including three active enzymes, DNMT1, DNMT3a, and DNMT3b), DNMT1 exhibits high preference for hemimethylated DNA (in which one of two complimentary DNA strands already possess attached methyl groups), and is therefore responsible for so called “maintenance methylation” [49,50]. DNMT3a and DNMT3b are primarily responsible for the “de novo” methylation of previously unmethylated CpG regions [51,52], but both of these methyltransferases have been shown to carry out maintenance methylation as well [53]. Importantly, in human neoplastic cells, it has been shown that DNMT1 provides both de novo and maintenance DNA methylation of TSGs [54,55,56]. The demethylating agents (or hypomethylating agents (HMAs) that inhibit these enzymes (azacitidine or AZA; decitabine or DAC; SGI-110 or guadecitabine) are discussed below).
- Histone modifications—various posttranslational modifications (PTMs) at histone protein N-terminal tails, which impair proper interactions between adjacent nucleosomes to affect the compact packing of the chromatin and impede the binding ability of other factors/enzymes that are involved in gene transcription [57,58]. The most common and well-characterized PTM, histone acetylation, is a dynamic, reversible process in which positively charged histone lysine residues are neutralized via the addition of acetyl groups by histone acetyltransferases (HATs), resulting in the attenuation of bonds between negatively charged DNA string and a histone complex. In the reverse reaction, deacetylation, enzymes histone deacetylases (HDACs) remove acetyl groups, and reinforce positive charge of the lysines, securing compact wrapping of DNA around histones [59,60]. Similarly, histone (de)phosphorylation utilizes protein kinases and phosphatases to attach or remove negatively charged phosphate groups, respectively, influencing chemical attraction between DNA and histone tails (reviewed in [60,61]). Histone (de)methylation—addition/removal of methyl groups by histone-specific methyltransferases and demethylases—can either activate or silence gene transcription. Remarkably, the functional consequences of each histone (de)methylation event depend on the histone, amino acid and residue methylated, degree of modification (mono-, di- or tri-methylation), and attraction of additional function-specific protein cofactors to the site, as well as existence of other methyl or acetyl groups in close proximity (reviewed in [62]). Comprehensive analyses of currently known histone PTMs, including those less common (ubiquitylation, sumoylation, deamination, etc.), their functional outcomes and complex interplay between the DNA methylation and histone modifications have been recently published [63,64].
- Chromatin remodeling—rearrangement of chromatin organization through complete or partial nucleosome repositioning and altering gene access for transcription. Chromatin remodeling can occur via nucleosome sliding (movement of the core histone octamer nexus across DNA segment with no evident disintegration of the octamer itself), nucleosome ejection (nucleosome segregation from the chromatin chain), or histone eviction (removal of histone H2A/H2B dimers from the DNA-associated nucleosome, sometimes with an alternative histone replacement) [65,66,67]. These processes are mediated by a number of ATP-dependent chromatin remodelers with high binding affinity to modified core histone tails, as well as transcriptional enzymes, which are extensively described in [65,66,67]. In particular, ARID1A and SMARCA4 are prominent chromatin remodeler examples in ovarian cancer. ARID1A is frequently mutated in ovarian clear cell (~50%) and low grade ovarian endometrioid (30%) carcinomas [68,69]. Most interestingly, tumors with ARID1A mutations acquire sensitivity to pan-HDAC inhibitors, thus making ARID1A-bearing cancers attractive for HDAC-based therapy [70]. SMARCA4 is frequently (over 90%) mutated in ovarian small cell carcinomas of the hypercalcemic type [71,72], however, to our knowledge, the first case of a germline SMARCA4 mutation in a patient with HGSOC was recently reported [73]. Further investigation on the role and clinical applicability of SMARCA4 and ARID1A in HGSOC is warranted [74,75].Altogether, the three epigenetic mechanisms that are described above work closely to mediate DNA (un)coiling around the core histones and ensure dynamic chromatin reassembly between heterochromatin (condensed or closed, silent) and euchromatin (loose or open, transcription-permissive) states.
- Non-coding RNA interference—a group of epigenetic regulatory mechanisms that involves microRNAs (miRNAs; miR) and long non-coding RNAs (lncRNAs). MiRNAs are short (~22 nucleotides) non-messenger RNAs that act primarily at a posttranscriptional level by base pairing with their complimentary mRNA targets to alter mRNA translation into protein [76]. Remarkably, one miRNA may complement a variety of mRNAs, whereas the same mRNA transcript might be a target of multiple miRNAs. Additionally, miRNAs may act as mRNA destabilizers causing poly-A-tail shortening [77] or interfere at the gene transcriptional level by means of PTMs (e.g., initiation of histone H3 lysine9 methylation with RNA interference machinery, followed by DNA methylation and gene transcription repression) and heterochromatic silencing [78]. LncRNAs are long (>200 for up to a hundred thousand nucleotides) non-messenger RNA that execute epigenetic regulation via several mechanisms: engage in post-translational histone modifications through association with chromatin-modifying proteins as an obligatory active player in the complex or as a scaffold that brings different protein complexes in close vicinity for proper functioning; serve as endogenous competitors to mRNA by base pairing with miRNAs and uncovering mRNAs for effective protein translation; or, serve as precursor RNAs for miRNAs (all mechanisms are detailed in [79,80,81]).
The importance of epigenetic dysregulation for cancer progression cannot be understated, as various aberrant epigenetic modifications trigger activation of oncogenes, repression of tumor suppressor genes, and altered transcription of protein-encoding genes that are collectively responsible for proper expression of signaling proteins that are necessary for physiological cell life span and functions. In the light of significant advances in technical tools and the rapid emergence of large “omics” data, the current knowledge in the field of cancer epigenetics and understanding their translational potential have vastly expanded. Multiple elegant reviews illustrate a wealth of currently available data on cancer epigenomics [46,82,83,84,85,86,87]. In the context of EOC specifically, several recent works highlight major epigenetic players—DNA methylation [88,89,90], histone PTMs [88,89], miRNAs [88,89,91,92], lncRNAs [93,94], and discuss current and potential therapeutic implication strategies and challenges [85,95,96,97,98].
2. Epigenetic Crosstalk between EOC Cells and TME Cellular Components
The biological importance of TME as a reactive platform orchestrating diverse aspects of tumor initiation, evolvement, metastatic progression, altered immune response, development of therapeutic resilience, and cancer recurrence is unquestionable and continuously reaffirmed, as highlighted in a multitude of studies [99,100,101,102,103,104]. Given the unique ovarian cancer intraperitoneal TME and the emerging evidence of benefits from epigenetic-targeted therapeutics, we systematize epigenetic dysregulations linked to EOC progression from the perspective of complex reciprocal relationship between ovarian tumor cells and tumor-associated microenvironmental non-malignant cell setting. These interactions will allow for researchers to evaluate, comprehensively, the potential for exploiting specific epigenetic vulnerabilities in both cancerous and TME cells as molecular biomarkers, prognostic indicators, and complementation to conventional chemotherapy and TME-targeted interventions.
2.1. Cancer-Associated Fibroblasts (CAFs)
CAFs are traditionally abundantly present within the tumor stroma (Figure 1A). They serve a tumor-supporting role via remodeling of extracellular matrix, a scaffold for the tumor bulk. CAFs perform endocrine/paracrine communication with the surrounding tumor and other stromal components via the excretion of a variety of growth factors and chemokines, hence contributing to tumor growth, immune response, angiogenesis, chemoresistance, and cell stemness; furthermore, CAFs differentiate into other cell types, as comprehensively discussed elsewhere [105,106]. CAFs exhibit substantial heterogeneity, which is attributed to existence of multiple proposed CAF origins, including reprogramming of the normal resident fibroblasts, conversion from adipocytes, endothelial or epithelial cells, or differentiation of the bone marrow derived mesenchymal or hematopoietic stem cells (summarized in [107]). Gene expression analysis of CAFs purified from different types of cancers, including ovarian carcinomas, revealed normal genotype, and acquisition of somatic mutations is considered to be an extremely infrequent event [108,109,110]. Hence, epigenetic changes likely play an essential role in CAF regulation.
Studies have reported altered DNA methylation status of genes (either at the gene promoter regions or global) in stromal fibroblasts dissected from various cancer tissues that corresponded to methylation profiles that are found in adjacent malignant cells [111,112]. In turn, as was shown by Mathot et al. for breast cancer [113], malignant cells maintained in the presence of CAF soluble mediators exhibit a wide gene upregulation pattern (372 genes upregulated total in the study) epigenetically modulated via DNA hypermethylation that could be reversed by the DNMT inhibitor decitabine. Pistone and team [114] demonstrated through global transcriptome and DNA methylation analysis that CAF-secreted factors trigger combinatorial DNA hyper/hypomethylation changes, collectively responsible for epithelial-to-mesenchymal transition (EMT) and stemness phenotype in prostate cancer cells. Albrengues and colleagues [115] reported the transformation of fibroblasts into pro-invasive CAFs in response to a cytokine termed leukemia inhibitory factor (LIF), which triggers the continued activation of JAK1/STAT3 signaling pathway via the histone acetylation of STAT3, followed by the activation of DNMT3b, methylation and abrogation of the Src homology region 2 domain-containing phosphatase-1 (SHP-1), and ultimate sustained phosphorylation of JAK1. The process is chaperoned by DNMT1, whereas the inhibition of DNMTs and JAK signaling abolishes the CAF phenotype [115].
Noteworthy, even brief transitory communication between cancer cells and normal fibroblasts leads to an elevated release of transforming growth factor beta 1 (TGF-β1) by activated fibroblasts in response to cancer cell presence [116]. This may be of particular relevance in the context of ovarian cancer epigenetic regulation, as TGF-β is known to catalyze global DNA hypermethylation alterations in EOC cells, promoting EMT and metastasis [117]. Moreover, Cardenas and co-workers found that TGF-β enhances both expression and enzymatic activity of DNMT-1, -3a, and -3b in EOC cells [117]. Importantly, the hypermethylation effect and EMT may be abrogated by treatment with DNMT inhibitors [117]. On the other hand, TGF-β assists in the reprogramming of CAFs from other cell types [118] and it favors further aggressive invasion of HGSOC through TGF-β-induced secretion of prometastatic mediators by CAFs [118,119]. Collectively, these data suggest formation of a constitutively active positive feedback loop in EOC, where initial fibroblast-malignant cell interaction triggers secretion of TGF-β by stromal cells, followed by EMT-associated global epigenetic alterations in EOC cells, as well as promotion of CAF phenotype and secretory activity, which closes the cycle and further aggravates EOC. In this scenario, the combined usage of demethylating agents and drugs targeting TGF-β signaling requires further investigation.
To our knowledge, posttranscriptional histone modifications in EOC-associated CAFs have not been directly addressed, whereas similar studies in the context of other cancer types are very limited. The histone mark H3K27me3 in breast CAFs was shown to be reduced, along with decreased expression of enhancer of zeste homolog 2 (EZH2), a methyltransferase that is responsible for the H3K27me3 histone mark [120], resulting in promotion of cancer cell invasion by CAFs via upregulated thrombospondin type 1 motif 1 [121]. In EOC, however, EZH2 is commonly upregulated [122], and thereby, such a mechanism is not likely. Undoubtedly, studies are needed in elucidating histone PTMs possibly taking place during malignant cell-CAF interplay.
Malignant TME-driven implication of miRNAs in CAF phenotype and functioning has been dissected for various cancer types (breast [123], cervical [124], gastric [125], prostate [126], colorectal [127,128], and bladder [129]), including EOC. In an elegant gain/loss-of-function study employing the co-transfection of multiple miRNA mimics and inhibitors, Mitra et al. [130] discovered a combination of three dysregulated miRNAs (upregulation of miR-155 plus repression of miR-31 and miR-214) that are capable of converting quiescent fibroblasts into ovarian CAFs with extensive expression of chemokines, in particular, direct target of miR-214 CCL-5 (C-C motif ligand 5), CXCL-10 (C-X-C motif ligand 10), CCL-7 and CCL-8, among others, accompanied by the substantial augmentation of ovarian tumor growth. The respective reverse experiments have shown the restoration of wild-type fibroblast phenotype and alleviation of ovarian tumor growth in co-culture [130], suggesting these miRNAs as novel therapeutic targets for halting EOC progression by means of manipulating stromal signals.
Zhao and coworkers [131] have recently discerned prognostically unfavorable upregulation of lncRNA LINC00092 in EOC cells which is induced by high CXCL-14 (C-X-C motif ligand 14) chemokine expression in ovarian CAFs. Expressed LINC00092 binds to a glycolytic enzyme and it facilitates glycolysis in EOC cells, boosts metastatic activity, and reciprocally supports pro-active CAF phenotype [131]. A differential expression analysis of CAFs purified from 67 high grade serous ovarian carcinomas (HGSOC) and 10 normal ovarian fibroblast samples identified 39 divergently expressed lncRNAs (out of 1970 lncRNAs total analyzed) in HGSOC CAFs in comparison with normal fibroblasts [132]. Subsequent context-specific regulatory network construction and pathway analyses have linked the upregulation of seven lncRNAs (FLJ39739, GAS5, H19, LOC100499466, MALAT1, NEAT1, and TUG1) and downregulation of four lncRNAs (CASC2, DLEU2, HCG18, and LOC100133669) to the furtherance of metastasis-associated pathways in HGSOC [132]. Mechanistic insight on the interaction of these differentially expressed lncRNAs in CAFs with proteins/pathways is essential for the further translation of these findings into diagnostic and/or therapeutic strategies.
2.2. Cancer-Associated Adipocytes (CAAs)
One of the distinguishing characteristics of the ovarian cancer TME is tumor cells that are in close vicinity to adipose tissue at all stages of development and metastasis. Adipose tissue exists at the site of the primary tumor (ovarian fat pad, mesosalpingeal/mesoovarian adipose layers) and around the intra-abdominal cavity in the form of substantial omental adipose nodes, large adipose bundles in the mesentery (both serve as prevalent EOC metastasis locations), as well as smaller fat depots in the parietal and diaphragmatic peritoneum (Figure 1A). It is now commonly accepted that adipose tissue is a highly communicative metabolic and secretory organ. Aside from functioning as energy (lipid) storage, it produces adipokines, metabolic substrates, growth factors, hormones, and immune mediators, and it contains other stromal components, such as fibroblasts, stromal vascular fraction, macrophages and other immune cells, nerve tissue, and extracellular matrix [133]. In presence of malignant setting, normal adipocytes rapidly acquire a highly active phenotype (CAAs) and respond by metabolic and secretory profile changes, causing pro-inflammatory, pro-invasive, proliferative, and radio- and chemoresistance effect on cancer cells [28,134,135,136,137,138,139].
There is accumulating evidence for tumor-adipocyte epigenetic interactions. During maturation, murine preadipocytes demonstrate substantial upregulation of miR-17-92 cluster, which further stimulates adipocyte differentiation via negative targeting of tumor-suppressor Rb2/p130 and is known to boost cell proliferation in various cancers [140]. In breast cancer (another type of adipose tissue-rich neoplasm), the transition of adipocytes into inflammatory CAAs in the vicinity of malignant milieu is dependent on miRNA regulatory mechanism. In particular, in the presence of breast cancer cells, reprogramming CAA increase expression of miR-5112, which suppresses the translation of Cpeb1, a negative regulator of interleukin (IL)-6. As a result, the pro-inflammatory CAAs exhibit an increased expression of IL-6 and a proliferation-promoting effect on breast cancer cells [141]. During ovarian cancer metastatic progression, omental CAAs and CAFs deliver miR-21-containing exosomes to cancer cells. Mir-21 targets apoptotic protease activating factor 1, inhibits ovarian cancer apoptosis, and confers paclitaxel chemoresistance [142], suggesting stromal-derived miR-21 blockade as a potential therapeutic strategy for metastatic and refractory ovarian cancer. Profiling of miRNAs in tumor interstitial fluid of human breast tumor tissues identified a list of 23 miRNAs that were associated with the presence of adipocytes and immune cells in tissues, suggesting epigenetic tumor-stroma crosstalk [143].
A genome-wide expression profiling of peri-prostate adipose tissue samples taken from prostate cancer patients revealed shifted expression of genes collectively accounting for increased proliferative and anti-apoptotic activity and mitigated immunosurveillance, suggesting that the peri-prostate adipose tissue cultivates prostate cancer development [144]. A related epigenome-wide DNA methylation pilot study of peri-prostate adipose tissue in normal weight and obese prostate cancer patients, performed by the same research group [145], revealed abundant DNA hypermethylation in cancer patients with excessive adiposity, with the epigenetically altered genes contributing to altered fatty acid metabolism, immune perturbations (including those providing tumor immune evasion), pluripotency of stem cells, and other pathways that are advantageous for cancer support. Concordantly, analysis of DNA methylation in omental tissue of obese women revealed significant DNA hypermethylation, whereas subsequent weight loss had a substantial hypomethylating effect [146]. Interestingly, obesity-induced proinflammatory cytokines triggered expression of DNMT1 and methylation of adipokine gene, whereas treatment with the DNMT inhibitor reverted the process in adipocytes [147]. Recent work by Tang et al. [148] demonstrated that treatment of adipocytes alone with the DNMT inhibitor resulted in the re-expression of tumor suppressor genes (e.g., SUSD2, TFP12, GREM1, TRIM29), altered expression of EMT mediators (e.g., CDH1, CDH2, FN1, and SLUG), and diminished migrative and invasive properties of co-cultured EOC cells. These data provide clear evidence that therapeutic tailoring of epigenetic aberrations in CAAs may, in turn, have anti-metastatic effect on ovarian malignant cell behavior [148].
Taking into consideration that discovery of lncRNAs as a class occurred very recently, there is an ample gap in evidence of their regulatory role in tumor-stroma interplay. However, recent studies report that lncRNAs are capable of mediating the expression of genes that are associated with lipid adipogenesis and metabolism via RNA, DNA or miRNA complementation, or recruitment of proteins involved in modulation of histone markers [149,150,151]. A recent study, including transcriptome profiling of primary brown and white adipocytes, preadipocytes, and cultured adipocytes reported a list of 175 lncRNAs that are specifically regulated during adipogenesis [152]. Given a constantly increasing number of lncRNAs with documented significance in the development of a variety of cancers, including ovarian [94,153,154], investigation of lncRNA-associated epigenetic modifications in the context of interrelationship between EOC and CAAs will undoubtedly be a very fruitful and promising direction for further basic/translational research.
2.3. Mesenchymal Stem Cells (MSCs)
MSCs encompass a diverse multipotent cell subgroup that was recruited to the tumor stroma from such sources as bone marrow, adipose tissue, umbilical cord, endometrium (menstrual blood), pericytes, as well as other organ-specific locations. The existence of MSCs has been reported for the majority of organs and tissues, including ovary (Figure 1A) [155]. Due to their capacity to differentiate into other active pro-cancerous stromal components (such as CAFs and CAAs) as well as sustain a cancer stem cell (CSC) population, MSCs are strongly associated with cancer progression yet retain a normal (non-malignant) genotype. In particular, ovarian carcinoma-associated MSCs (CA-MSCs) exhibit multipotent potential and strong EOC growth-permissive and stemness-promoting properties, increasing resistance to platinum-based chemotherapy, providing tumor stromal support and neovascularization [156,157,158,159,160,161]. Contradictory to those reports are studies demonstrating tumor-restricting effect of human MSCs on EOC cells via stimulation of EOC cell cycle arrest, enhanced apoptosis, altered mitochondria membrane potential and suppressed neoangiogenesis [162], and the inhibition of cisplatin-resistant ovarian carcinoma xenograft growth [163]. Given a wealth of controversial data on the assisting vs restricting role of MSCs on EOC development and the extensive interest in therapeutic applications of MSCs as vehicles for EOC-targeted drug delivery due to their high tumor site tropism [164,165,166,167,168], further investigations to determine the underlying mechanisms of MSC-EOC interactions are clearly necessary.
While the overwhelming majority of research work is focused on the elucidation of cell signaling pathway implications, the epigenetic interplay between MSCs and EOC remains largely unexplored. While considering the well accepted role of epigenetic aberrations in CSC reprogramming (comprehensively outlined in [169]), emerging evidence on efficient epigenetic transformation of MSCs into CSCs via tailoring chromatin remodeling (imbalanced DNA methylation of cancer-implicated genes, application/loss of histone marks, etc.) [170,171], and epigenetic modulation of MSC differentiation into diverse stromal cell lineages [172,173,174], the involvement of EOC-mediated epigenetic factors on MSC phenotype and functioning, and the subsequent reciprocal effect of reprogrammed MSCs on ovarian carcinoma, is highly plausible. Toward these possibilities, Reza et al. [175] reported the anti-proliferative and pro-apoptotic effect of adipose-derived MSC exosomes on ovarian cancer A2780 and SKOV3 cells; subsequent exosomal RNA sequencing identified a list of enriched miRNAs targeting EOC cell survival pathway genes [175]. MiRNA expression profiling in aging MSCs revealed altered expression of two miRNAs, miR-638 and miR-572, both of which have been reported to be upregulated in ovarian carcinoma; however, distinct studies display controversial findings on the impact of these miRNAs on EOC [176,177,178,179]. Among other non-coding RNAs, abundant in MSC-derived extracellular vesicles and associated with ovarian cancer development are miR-21, miR-92a and miR-221 [180]. Alternatively, Ho and colleagues [181] demonstrated, that ovarian cancer stromal progenitor cells isolated from tumor tissues and ascitic effusions of EOC patients displayed 40 hypermethylated tumor suppressor genes (TSGs) (with DLC1, RASS382, CDH13, BRCA1, TIMP3, HIN-1, ESR1, CDKN2A, CCND2, CDKN2B, as most frequently hypermethylated and correlating with validated mRNA expression decrease in DLC1, RASSF1A, CCND2, and CDKN2B) in comparison with matching patient ovarian cancer cells and were capable of promoting tumor growth in vivo when co-injected with SKOV3 cells. Most importantly, treatment with the hypomethylating agent (HMA) 5-aza-2-deoxycytidine (decitabine or DAC, a desoxyribonucloside that exclusively incorporates into DNA and inhibits DNMTs by irreversible binding of the latter [182]) resulted in efficient demethylation of TSGs in stromal progenitor cells, repressed their self-renewal and growth and mitigated proliferation of ovarian tumor cells [181]. Collectively, these studies suggest a direct or indirect epigenetic relationship between MSCs and ovarian tumor cells and warrant additional research that can potentially lead to the identification of novel therapeutic targets for ovarian carcinomas.
2.4. Tumor-Associated Endothelial Cells (TECs)
TECs refer to endothelial cells that line the inner walls of newly formed blood vessels in tumors. TECs support blood flow, tumor tissue trophics, and accelerate tumor progression. TECs are genetically non-malignant cells, however, they differ from normal endothelial cells by exhibiting cytogenetic abnormalities [183], distorted morphology [184], and altered molecular profiles [185], as well as improper functional characteristics. TECs form a disorganized, excessively sprouted, branched, fragile, and gap-prone endothelial network, which allows for chaotic non-laminar blood flow, increased vascular permeability and escape of primary tumor cells into blood circulation. TECs help circulating cancer cells overcome anoikis and escort their movement to metastatic niches [186]. Finally, TECs are able to develop taxol resistance in the presence of surrounding malignant milieu [187].
TECs exhibit considerable heterogeneity (reviewed in [188]), and diverse features of TECs in various tumors are attributed to the specific malignant settings [189]. In the context of epigenetic regulation, Maishi et al. [190] implanted TECs that were isolated from the high metastatic (HM) melanomas into low metastatic (LM) melanomas and achieved metastatic enhancement as a result of elevated levels of proteoglycan biglycan, attracting tumor cells to intravasate. Strikingly, upregulation of biglycan was due to DNA demethylation of its promoter region in HM-TECs as opposed to normal endothelial cells, LM-TECs, and tumor cells [190]. Furthermore, exposure of LM-TECs to conditioned medium from HM-tumors and treatment with the HMA decitabine proved the upregulation of biglycan in LM-TECs via DNA demethylation triggered in the presence of HM-tumor [190]. The study elegantly reaffirmed epigenetically-mediated bidirectional communication between tumor cells and TECs, and further suggested epigenetic perturbations as attractive targets to manipulate neoangiogenesis and tumor metastasis.
Epigenetic regulation of TECs has been reported. DNMT and HDAC inhibitors efficiently mitigated TEC growth in vitro and in vivo [191,192]. Moreover, epigenetic modifications were shown to regulate TEC-mediated immune response [193]. Downregulation of intercellular adhesion molecule-1 (ICAM-1) in TECs due to promoter histone H3 deacetylation and the loss of histone H3 Lysine4 methylation was reported, and re-expression of ICAM-1 in TECs by DNMT and HDAC inhibitors successfully restored leukocyte-TEC adhesion and leukocyte infiltration of vessel walls in vitro and in vivo, respectively [193]. In a separate study, the same group identified additional anti-angiogenesis genes (including clusterin, fibrillin 1, and quiescin Q6) downregulated in normal endothelial cells subjected to a tumor-mimicking setting (conditioned medium) via epigenetic silencing by promoter histone H3 deacetylation and loss of histone H3 Lysine4 methylation [194]. Furthermore, subsequent treatment with DNMT and HDAC inhibitors reversed the gene silencing effects [194].
An influential study assessing the role of EOC on TECs was conducted by Sood group [195], who performed a comparative genome-wide gene expression analysis of endothelial cells from five different normal ovarian tissues and TECs isolated from 10 invasive ovarian carcinomas and revealed a list of 400 differentially expressed genes, among which EZH2 was significantly upregulated. EZH2, a histone-lysine N-methyltransferase, is frequently overexpressed in ovarian tumor tissues and is a well-established epigenetic mediator (stimulator) of EOC tumor growth, invasion, metastasis, neoangiogenesis, platinum-resistance and ovarian cancer stem cell renewal through transcriptional repression of signaling pathways, as well as direct/indirect interaction with multiple miRNAs and lncRNAs (reviewed in [196]). Overexpression of EZH2 in TECs by the surrounding EOC setting occurred in a VEGF-mediated paracrine manner, which in turn promotes EZH2 direct binding, methylation, and transcriptional repression of vasohibin-1 (a selective negative regulator of TEC migration, proliferation, and vessel tube formation), stimulating further angiogenesis [197]. Remarkably, the silencing of the EZH2 gene in vitro and in vivo (via systemic siRNA nanoparticle delivery in both EOC cells and TECs in mouse EOC xenografts) substantially increased vasohibin-1 expression and reduced neoangiogenesis and tumor growth [197], underlining the additional therapeutic potential of concomitant EZH2 targeting in TECs and EOCs. Noteworthy, the expression of EZH2 in ovarian cancers has been shown to be regulated by different non-coding RNAs, for example miR-298 [198] and lncRNA HOTAIR (HOX transcript antisense RNA) [199], providing additional druggable epigenetic targets in EOC cells and TECs in order to suppress neoangiogenesis and ovarian tumor progression.
2.5. Pericytes
Another type of vessel-associated cells, pericytes (Figure 1), play a key role in normal vasculature development, including neoangiogenesis, and in cancer contribute to tumor development and metastasis. These perivascular cells reside in the microvessel walls, immediately beyond the basement membrane, and provide physical support to endothelial cells, control integrity of the vessel endothelial layer, vascular permeability, and blood flow [200], serve as an origin of mesenchymal stem cells (MSCs) for some human tissues [201] and as multipotent precursors for other types of cells [202], and exert immune mediators [203]. During cancer progression, pericytes essentially contribute to rapid neoangiogenesis and tumor growth, expansion of the CSC population, on one hand, but also strengthen immune defense against tumor invading and restrict metastatic seeding, on the other hand, as comprehensively outlined in [204,205]. In EOC patients, high pericyte score (carrying a pericyte gene signature) was shown to be an unequivocal predictor of earlier relapse and poor prognosis [206]. Dual targeting of pericytes in combination with endothelial cells [207], as well as the disruption of connections between pericytes and endothelial cells via insertion of N-cadherin blocking peptides [208], are potential anti-angiogenesis therapeutic strategies in EOC.
Intriguingly, in a HGSOC xenograft model, injection of pericytes concomitantly with EOC cells amplified tumor growth and metastasis rate without altering tumor vasculature, highlighting a direct impact of perycites on EOC cells, independent of neoangiogenesis [206]. No studies have adequately described epigenetic tumor-perycite crosstalk in ovarian carcinomas. However, proximity of the malignant (glioblastoma) setting [209], hypoxic conditions [210] and inflammatory stimulation [211] trigger an epigenetically mediated (through altered non-coding RNA) pericyte response. Though these findings are quite limited, they raise the possibility of dynamic epigenetic cooperation between pericytes and EOC cells (and other cancer-associated stromal cells) within a highly reactive, proinfammatory, and hypoxia-prone ascitic TME. In addition, a very interesting preclinical study by Kratzsch et al. [212] assessed the effect of 5-azacitidine (AZA; a ribonucleoside that is capable to incorporate into cellular RNA and DNA and acts as a HMA interfering with the DNMT activity [182,213]), valproic acid (HDAC inhibitor), temozolomide (a standard DNA alkylating chemotherapeutic positive control), and a bevacizumab (an angiogenesis inhibitor targeted therapy positive control) on the tumor growth and neovasculature status in the murine glioblastoma multiforme models. Strikingly, besides the suppressive effect on glioma tumor growth, the HMA also resulted in a notable antiangiogenic effect via the substantial diminishing of endothelial cells and the decreasing number of pericytes [212]. Contrarily, HDAC inhibitor valproic acid had no mitigating effect either on tumor growth or on the vasculature [212]. These results suggest concordant epigenetic expression changes taking place within the malignant tissue and the adjacent TME cells as a result of their mutual communication (as both tumorous and angiogenesis cells selectively responded to the same epigenetic-tailoring drug, but not to the other). In dissonance with that is work by Karén et al. [214], who reported successful inhibition of pericyte differentiation, migration, and proliferation in response to HDAC inhibitors in vitro, and HDAC suppression stimulated expression of genes regulating vessel stabilization and maturation in pericytes. Collectively, these data highlight translational potential of cancer-prone epigenetic signatures not only in the neoplastic component per se, but in the surrounding stroma as well.
2.6. Tumor-Associated Macrophages (TAMs)
TAMs within malignant stroma (Figure 1A) are considered to be a fundamental immune cell subpopulation responsible for cancer-associated inflammation, matrix remodeling, tumor immune escape, growth, invasion, angiogenesis, metastasis, cancer cell stemness, and drug resistance. Macrophage diversity and ability to transfer between M1 (classic, host immune defense activating, tumoricidal) and M2 (alternative, immunosuppressive, pro-tumorigenic) phenotypes are defined by the distinct tumor microenvironments [215]. Several recent reviews [216,217] elegantly describe bidirectional TAM interactions with cancer cells, other immune cell subpopulations and non-malignant stromal components, such as CAFs, TECs, B cells, eosinophils, basophils, dendritic cells, natural killer cells, and others.
In addition to ample evidence on receptor-ligand signaling interrelationship between EOC and TAMs [218,219,220,221,222,223], a better understanding of their epigenetic cooperation has begun to accrue. Ying et al. [224] reported initiation of M2 polarization and tumor-promoting capabilities in ovarian TAMs by EOC-released exosomal miR-222-3p through SOCS3/STAT3 pathway. Similarly, macrophage M2 phenotype shift was induced by exosomal miR-940 released from hypoxic epithelial ovarian tumors [225]. Importantly, TAM-secreted exosomes suppress endothelial cell migration by targeting miR-146b-5p/TRAF6/NF-κB/MMP2 pathway, whereas the EOC-released exosomal delivery of lncRNAs to the site efficiently restores the endothelial cell movement [226]. Alternatively, Hu et al. [227] observed altered expression of 19 miRNAs in TAMs exposed to tumor necrosis factor-related inducer of apoptosis (TWEAK; commonly expressed by immune cells, such as dendritic cells and natural killer cells) and demonstrated that the exosomal transportation of overexpressed miRNAs, and in particular, miR-7, from TAMs to EOC cells significantly repressed their metastatic activity in vitro and in vivo via repression of EGFR/AKT/ERK1/2 pathway. Furthermore, the insertion of antagomiR-7 into TAMs repressed levels of miR-7 in TAMs, in released exosome vesicles and in the recipient ovarian malignant cells, and stimulated EOC metastasis [227].
Chromatin remodeling-related epigenetic modifications resulting from TAM-EOC interaction remain largely unknown. However, as reported for gastric cancer, TAMs are capable of silencing TSG gelsolin in cancer cells by increased DNMT1 expression and DNA methylation of gelsolin promoter via activation of CCL5/CCR5/STAT3 signaling [228]. Most importantly, either suppressing DNMT enzyme activity, or treatment with demethylation agent, or interfering with CCL5/CCR5/STAT3 pathway led to decreased in vivo tumor growth, suggesting several possible routes of epigenetic inhibition of gastric cancer development [228]. In oral cancer, interferon-γ mRNA expression in TAMs was substantially decreased in comparison with normal or benign oral tumor specimens as a result of promoter region methylation, with the level of methylation strongly correlating with the clinical stage, histopathology grade, and primary tumor scale [229]. Promoter hypermethylation and downregulation of the follistatin like-1 (FSTL-1) glycoprotein was associated with metastatic activity of nasopharyngeal carcinomas and dysfunction of macrophages, whereas treatment with recombinant human FSTL-1 protein elevated IL-1β and tumor necrosis factor-α in TAMs and repressed cancer cell immune evasion [230]. Using a panel of distinct cancer type cells, including cervical, hepatocellular, epidermoid carcinomas, glioblastoma, rabdomyosarcoma, and murine melanoma, Osawa and co-authors [231] demonstrated that cancer cell hypoxia and nutrient starvation lead to activation of histone demethylase JMJD1A (Jumonji domain-containing 1A), followed by increased AKT phosphorylation, cancer cell metastatic properties, increased angiogenesis, and infiltration of macrophages into cervical cancer and muscle sarcoma tissues in vivo. Remarkably, JMJD1A repression mitigated tumor progression through decreased neovascularization and alleviated TAM infiltration, and importantly, enhanced the anti-tumor effect of anti-angiogenesis agents bevacizumab and sunitinib [231]. Ishii et al. [232] described M2 macrophage polarization via reciprocal epigenetic changes in histone H3 lysine4 and histone H3 lysine27 methylation through STAT6 mediation, which collectively lead to transcriptional activation of specific M2 maker genes. Altogether, these findings underline the immediate importance of continued research accessing TAM-modulated epigenetic changes in EOC, as they may reveal novel diagnostic and therapeutic approaches to mitigate TAM-associated pro-tumoral inflammation and cancer progression, and boost host immune defense mechanisms.
2.7. Tumor-Infiltrating Lymphocytes (TILs)
TILs are collectively represented by varying amounts of T and B lymphocytes recruited from the circulatory system to the tumor site to fulfill the host immune response function. In EOC, TILs may be present in primary tumors and advanced metastatic lesions, both within malignant (intratumoral) bulk and stromal compartment, and differential representation of certain TIL subsets (helpers, killers, regulatory/suppressors, effectors, memory cells, and such) depends on the disease stage, therapeutic management, chemotherapy response status, and possesses prognostic significance (Figure 1A) [29,233,234,235,236,237,238].
The extensive evidence on signaling molecules and pathways implicated in TIL functioning and ovarian cancer prognosis, as well as current attempts to differentially manipulate TIL subsets and signaling networks towards boosting adequate anti-tumor immunity, have been comprehensively summarized by Santoiemma and Powell in a recent review [239]. The epigenetic regulation of TIL functioning in ovarian cancer and epigenetic-based immunotherapeutic strategies are emerging areas of interest. Sehouli et al. [240] proposed the concept of “epigenetic immunophenotyping” (identification of certain epigenetic marks) of both overall and specialized TIL populations. By using matching healthy and cancer tissues, including ovarian, they established a strong correlation between epigenetics and cancer prognosis [240]. In support of this concept, treatment with DNMT inhibitor 5-azacitidine led to the substantial enrichment for immunomodulatory pathways (interferon signaling, antigen processing and presentation, and cytokines/chemokines) in ovarian and other cancers [241]. Similarly, with the use of global gene expression profiling of EOC treated with DNMT inhibitor decitabine, Wang et al. [242] discovered prominent upregulation of immunoregulatory genes cohort in decitabine-exposed malignant cells. The group further showed that decitabine treatment stimulated TIL infiltration and anti-tumor function in both subcutaneous and intraperitoneal syngeneic murine ovarian cancer models [242]. Moreover, combining a DNMT inhibitor with the standard immune checkpoint blockade antibody stimulated conversion of naïve T cells into T effectors and contributed to better mouse survival [242]. Based on this work, Stone et al. [243] further demonstrated that the activation of type I interferon signaling in response to DNMT inhibitor 5-azacytidine was a key requirement for efficient stimulation of CD45+ (leukocyte common antigen) immune cells, CD8+ cytotoxic T cells, natural killer cells, restriction of macrophages, and myeloid-derived suppressor cells in the ovarian TME in vivo, prompting the inhibition of tumor growth and increased survival. In support, Adair and Hogan [244] demonstrated the enhanced expression of cancer-testis antigens and class I major histocompatibility complex (MHC)-encoded molecules in EOC cells that were treated with DNMT inhibitors and subsequent infiltration of antigen-reactive CD8+ cytotoxic T cells to EOC. Consistent with that, seminal work by Chiappinelli and co-authors [245] established that DNMT inhibitors activate interferon signaling, TIL infiltration, and EOC cell death via the upregulation of viral defense pathway, as hypomethylation of endogenous retrovirus (ERV) genes leads to upregulated viral defense gene expression and boosts immune response.
In the context of non-coding RNA interference, two prognostic miRNA signatures—malignancy signature and immunological signature—have been recently identified in advanced EOC [246]. Briefly, using integrative mRNA/miRNA co-expression analysis of primary tumor tissues from advanced EOC patients, two modules were established: a malignancy module (let-7f, let-7g, miR-106a, miR-17, miR200c, miR-26a, miR-26b, and miR-328), which was associated with the more aggressive EOC growth and an immunological module (miR-197, miR-22, miR-22#, miR-28, miR-339–5p, miR-340#, miR-628–5p, miR-629, miR-661, and miR-98), which strongly correlated with intratumoral infiltration by T cells, natural killer cells, cytotoxic lymphocytes, and macrophages [246]. These microRNA signatures may serve as prognostic and treatment efficacy biomarkers as well as potential targets for epigenetic-based immunotherapy of advanced ovarian cancer [246].
2.8. Plasmacytoid Dendritic Cells (PDCs)
PDCs constitute a rare, yet critically important and highly specialized immune cell subpopulation whose main role in immune surveillance is rapid recognition of foreign pathogens via selectively expressed toll-like receptors and the immediate activation of both innate and acquired immune systems (Figure 1A) [247]. Incessant stimulation of PDCs by self-DNA (a situation when PDCs, which do not normally react to inert DNA of organism’s cells, become continually activated by the altered DNA of transformed cells) is a characteristic of a variety of neoplasms, including ovarian carcinomas [248,249,250]. Accumulation of PDCs within the epithelial ovarian tumor bulk promotes vasculogenesis [251,252] and immune tolerance [253], and it is associated with unfavorable disease prognosis [254]. Importantly, the elimination of immature PDCs in mice bearing ovarian tumors via the targeting of specific markers led to vascular ablation, substantially reduced tumor growth, and triggered anti-tumor immune response and tumor re-sensitization to chemotherapeutic agents [255].
Epigenetic regulation of hematopoietic stem cell lineage commitment and subsequent differentiation of dendritic cell precursors into specific dendritic subtypes (including PDCs) and interferon response in the context of chromatin remodeling, miRNA and lncRNAs interference are well documented [256,257,258,259,260] and not focused on herein. Meanwhile to our knowledge, the epigenetic impact on and by PDCs in relation to EOC remains largely undefined. A single study demonstrated that exogenous supplementation of miR-155 (which is downregulated in tumor-associated PDCs) via targeted nanoparticle delivery to EOC-associated PDCs results in the genome-wide silencing of multiple immunosuppressive mediators, following stimulation of immune defense mechanisms and abolishment of ovarian carcinoma progression in vivo [261]. Importantly, a comparison of methylome profiles of dendritic cells and myeloid-derived suppressor cells (MDSCs, an immune cell subtype bearing same progenitor with PDCs) grown under tumor-associated conditions (exposure to prostaglandin E2 or malignant conditioned medium) revealed MDSC-specific DNMT3A enzyme activation, DNA methylation signature, and silencing of immunogenic-associated genes analogous to changes that were observed in primary MDSCs dissected from ovarian cancer patients [262]. Moreover, the suppression of DNMT3A activity abrogates MDSC-specific hypermethylation and MDSC immunosuppressive properties [262]. These findings support the epigenetic tuning of immune surveillance by ovarian TME and underline the necessity for further investigations in this direction.
2.9. Mesothelial Cells (MCs)
MCs are simple squamous epithelial cells that line the intra-abdominal cavity as an upper single layer (mesothelium) of the peritoneum, immediately supported by the basal membrane, underneath which is the collagen-rich extracellular matrix (Figure 1A). MCs function as an active physical barrier against the invasion of ovarian neoplastic cells into submesothelial matrix and metastasis formation, and mesothelial disruption (clearance) imposes a higher level of EOC peritoneal adhesion and dispersal [263,264]. Besides, MCs serve a secretory role by releasing bioactive soluble factors into malignant ascites, such as phospholipid lysophosphatidic acid [265] and VEGF [266], which are known to enhance EOC tumor growth, metastasis, apoptotic resistance, and angiogenesis [265,267,268,269]. MCs also produce large amounts of tumor-associated immunostimulatory protein K90 [270], associated with the poor prognosis in ovarian and breast cancers [271,272] and implicated in drug resistance [273].
The bidirectional interplay between EOC cells and MCs has been recently assessed in a study by Matte et al. [274], who reported global gene expression changes (649 altered genes primarily related to cell growth and proliferation, apoptosis, cell cycle, cell assembly, and organization) in MCs exposed to EOC ascitic effusions [274]; reciprocally, EOC cells exposed to EOC-associated MC setting exhibited an enhanced resistance to induced apoptosis [274]. However, epigenetic regulation underlying these gene expression alterations are unknown. Clearly, DNA methylation, histone modification profiling, and non-coding RNA profiling are further key steps necessary to distinguish specific epigenetic mechanisms responsible for gene expression switches occurring in EOC cells and cancer-affected mesothelium because of their interdependence. Gaining better understanding of these events may provide valuable insight on cellular adjustment during tumor-mesothelial adhesion step and suggest novel approaches to block metastatic seeding of the peritoneum.
3. Ovarian TME: Potential Epigenomic-Based Therapeutic Strategies
The growing body of information on epigenetic control of ovarian cancer metastatic advancement and acquisition of resistance to standard-of-care chemotherapy make epigenomic alterations attractive prognostic markers and druggable targets to improve therapeutic outcomes in EOC patients especially with platinum-resistant and recurrent disease. Our group and others are actively exploiting DNA hypermethylation, modifications of histone marks, and aberrant expression of non-coding RNAs in malignant EOC cells in an attempt to treat EOC, re-sensitize ovarian tumors to conventional chemotherapy, and prevent/delay disease recurrence (Figure 3A). We and others aim to discern global DNA methylation patterns and methylation states of specific candidate genes to prognosticate and improve patient response to chemotherapies with hypomethylating agents (HMAs) [15,88,275,276,277]. We recently demonstrated successful in vitro and in vivo re-sensitization of multiple cisplatin-resistant ovarian cancer cell lines using a novel small-molecule DNMT inhibitor guadecitabine (SGI-110) [275]. Efficient epigenetic targeting of ovarian CSC population by the HMA with induction of differentiation, restriction of tumor-initiating capacity, and re-sensitization to platinum was also reported [278]. In a recently completed phase I clinical trial, guadecitabine “priming” in combination with carboplatin induced clinical response in patients with platinum-resistant, recurrent HGSOC [279]. Similarly, HDAC inhibitors are being examined in advanced, refractory, and recurrent ovarian cancer [280,281,282]. However, despite some promising results, cell adaptation to HDAC inhibitor-mediated epigenomic disruption has been frequently observed [283,284,285]. Finally, the recent identification of large miRNA and lncRNA classes as epigenetic regulators of gene expression granted new opportunities for miRNA- and lncRNA-employed prognostic evaluation and targeting of EOC [92,93,94,153,286].
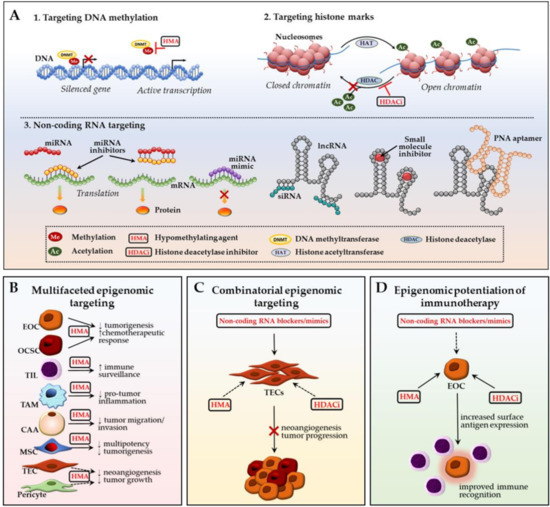
Figure 3.
Ovarian TME: potential epigenomic-based therapeutic strategies. (A) Main epigenetic therapy mechanisms include: (1) DNA demethylation with hypomethylating agents (HMAs) to re-activate transcription of silenced genes; (2) restoration of open, transcription-permissive euchromatin state with histone deacetylase (HDAC) inhibitors preventing removal of acetyl groups from the histone tails; (3) targeting non-coding RNA-mediated epigenetic perturbations via delivery of exogenous miRNA inhibitors (antagomirs/blockmirs/sponges) or mimics, lncRNA siRNAs, small molecules or peptide nucleic acid aptamers; (B) Multifaceted epigenomic targeting approach suggests simultaneous targeting of several malignant and TME cell types using one epigenetic drug; (C) Combinatorial epigenomic targeting involves employment of multiple epigenetic mechanisms to revert cancer-associated phenotype in TME cells and inhibit their tumor-promoting effect on EOC cells; (D) Epigenomic potentiation of immunotherapy involves epigenetic stimulation of cancer cell immune gene/pathway representation which allows for increased immune surveillance efficacy. CAA—cancer-associated adipocyte; HMA—hypomethylating agent; HDACi—histone deacetylase inhibitor; EOC—epithelial ovarian cancer cell; MSC—mesenchymal stem cell; OCSC—ovarian cancer stem cell; PNA—peptide nucleic acid; TAM—tumor-associated macrophage; TEC—tumor-associated endothelial cell; TIL—tumor-infiltrating lymphocyte. Black arrows represent interactions reported in EOC; dashed arrows designate patterns that were observed in other cancer types and may potentially be applicable towards ovarian cancer (see main text for details).
Continuous accumulation of knowledge on epigenomic perturbations in ovarian TME cellular compartments in the context of their communication with EOC cells holds great translational promise. The fact that non-malignant stromal cells lack genetic mutations and acquire potentially reversible tumor-specific molecular traits and cancer-indulgent functions via epigenetic reprogramming by EOC cells suggests the epigenetic tailoring of ovarian TME as a promising strategy in ovarian cancer management. Given the varying stroma representation in ovarian malignancies (stromal compartment may range from 7 to 83% in different ovarian tumors [287]) and high level of TME cell heterogeneity, it is pivotal to correctly evaluate TME composition within the tumor bulk in each disease case. Prevalence of one or another type of TME cells may dictate activation of certain mechanistic pathways, help to dissect “first choice” epigenetic therapy candidates, and possibly lead to the identification of epigenetic alterations in the TME that are specific for each patient.
Treatment of different stromal components with HMAs demonstrated tumor-restrictive potential. In particular, the exposure of adipocytes to guadecitabine attenuated HGSOC cells metastasis-associated behaviors [148]. HMA decitabine effectively inhibited the multipotent and tumor-promoting capabilities of ovarian cancer-associated MSCs and led to decrease in cancer cell proliferation [181]. While not specified for EOC, treatment with the DNMT inhibitor azacitidine showed tumor-inhibiting and anti-angiogenesis effect with a decreased amount of TECs and pericytes in other cancer types [212]. HMA therapy also showed change in immunoregulatory cell response within the tumor bulk with increased infiltration of tumor-restrictive TIL subsets and suppression of TAM-mediated tumor progression in EOC and other cancers [228,242,243,244]. Such multifaceted epigenomic targeting of multiple stromal cell types in parallel with primary EOC cell targeting through assessing the same epigenetic mechanism (Figure 3B) may improve efficacy, while also diminishing the toxic effects that are associated with multi-drug treatment regimens.
Alternatively, similarly to an improved anticancer effect of a DNMT/HDAC inhibitor combination on ovarian tumors [288] and other malignancies [284,289], the combinatorial epigenomic targeting of ovarian TME components using several epigenomic approaches is plausible (Figure 3C). For example, anti-angiogenesis and anti-proliferative effect of simultaneous utilization of an HDAC inhibitor to target pro-tumorigenic histone marks and an HMA to demethylate DNA promoter CpG islands in colon tumor-conditioned TECs was reported [194]. Analogous drug combination may have therapeutic prospective to block ovarian cancer neovascularization. Moreover, systemic nanoparticle delivery of siRNAs or antagomirs targeting tumor-assisting non-coding RNAs (such as EZH2, HOTAIR, and miR-298) into ovarian TECs may provide an additional option for combinatorial blocking of EOC tumor growth and angiogenesis [197,198,199]. Current data provide strong rationale for combining HDAC and DNMT inhibitors to affect EOC-associated pericytes [212,214].
Given that CAF reprogramming is shown to repress ovarian tumor advancement [290], prevention or reversion of CAF phenotype may potentially be accomplished through combined treatment with a HMA [113,115] and a CAF-delivered mixture of miRNA miR-31/miR-214 mimics plus miR-155 antagonist to interfere with the EOC-associated CAF miRNA signature [130]. The recent discovery of a number of altered lncRNAs in HGSOC CAFs [132] may suggest novel options for restricting CAF-promoted EOC growth via targeted siRNA injections. Similarly, non-coding RNA signatures recently identified in EOC-related immune cell populations (discussed earlier in this review) warrant further siRNA/miRNAs targeted conveyance studies in this direction. Importantly, in contrast to CAFs, miR-155 was downregulated in EOC-associated PDCs and its exogenous delivery to these cells, in turn, boosted anti-tumor immune defense response and EOC suppression [261]. Taking into consideration such drastically opposite effects of the same epigenomic regulators on different ovarian TME cell compartments, the development of methodologies for highly specific nanoparticle transportation and precise cell type targeting is pivotal. Several studies investigating the potential of exosome-facilitated epigenomic (miRNA) cargo delivery for EOC treatment are also in progress [291].
Exploiting host immune system in managing ovarian cancer growth and metastatic spread has emerged as another key research and therapeutic direction [292,293,294,295,296]. A number of comprehensive analyses of epigenomic strategies employed to advance cancer immunotherapies [297,298,299] provide detailed illustration on how currently available epigenetic drugs may potentially contribute to modulation of cancer immunosurveillance and anti-tumor responses (Figure 3D). In ovarian cancer in particular, Stone et al. [243] demonstrated while using preclinical in vivo models that combining DNMT inhibitor and HDAC inhibitor with immune checkpoint blockade resulted in the most notable anti-cancer effect and survival due to activation of type I interferon signaling, more efficient recruitment of anti-tumor TIL subsets and the restriction of tumor-indulgent TAMs and myeloid-derived suppressor cells to the ovarian TME. Alternatively, Song and coworkers [300] reported HDAC inhibitor-stimulated increase of NKG2D ligand expression on the surface of ovarian cancer cells, which allowed for better recognition and killing of the latter by engineered NGK2DL-specific chimeric antigen receptor T cells. Finally, Wargo and colleagues [301] reported improved recognition of tumor cells by engineered peripheral blood lymphocytes in response to increased cancer-testis antigen NY-ESO-1 expression (typically expressed in the majority of epithelial tumors, including ovarian [302]), catalyzed by DNMT inhibitor (alone or combined with HDAC inhibitor). Furthermore, the results of a recent phase I clinical trial by Odunsi et al. [303] showed improved efficacy of NY-ESO-1 vaccine therapy when combined with escalated doses of HMA decitabine in patients with EOC relapse. Current clinical trials combining immunotherapeutic approach with epigenetic drug(s) are summarized in Table 1. In addition to epigenomic potentiation of immunotherapy by chromatin remodelers, boosting regulatory T cell-mediated immune response may be achieved via targeting non-coding RNAs [304] and it requires further testing for applicability in EOC (Figure 3D).

Table 1.
Clinical trials evaluating safety and efficacy of immunotherapeutic agents in combination with epigenetic drugs in patients with ovarian cancer (https://clinicaltrials.gov).
4. Conclusions
To summarize, a deeper understanding of epigenomic involvement in reciprocal EOC tumor-stroma interrelationship is essential and it will help to determine pharmacological routes to alter the TME, which in turn could inhibit EOC metastatic progression and the development of chemoresistance and tumor recurrence. Furthermore, we believe epigenetically-mediated pharmacological engagement of certain TME players, such as immune cells, to boost immune responses towards complete malignant cell elimination has tremendous potential.
Author Contributions
Y.K. contributed to the conceptualization and writing (original draft and figure preparation, review and editing). K.P.N. contributed to conceptualization and writing (review and editing).
Funding
This work was supported in part by the Ovarian Cancer Research Fund Alliance [Epigenetic Vulnerabilities of Ovarian Cancer Stem Cells, Grant Number 458788] (K.P.N.); the Leo and Anne Albert Charitable Trust (K.P.N.); and the Indiana Clinical and Translational Sciences Institute Grant Award (Y.K.).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Vaughan, S.; Coward, J.I.; Bast, R.C.; Berchuck, A.; Berek, J.S.; Brenton, J.D.; Coukos, G.; Crum, C.C.; Drapkin, R.; Etemadmoghadam, D. Rethinking Ovarian Cancer: Recommendations for Improving Outcomes. Nat. Rev. Cancer 2011, 11, 719–725. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Cancer Facts & Figures 2018; American Cancer Society: Atlanta, GA, USA, 2018. [Google Scholar]
- Siegel, R.L.; Miller, K.D.; Ahmedin, J. Cancer Statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Jacques, F.; Isabelle, S.; Rajesh, D.; Sultan, E.; Colin, M.; Marise, R.; Parkin, D.M.; David, F.; Freddie, B. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar]
- Howlader, N.; Noone, A.M.; Krapcho, M.; Garshell, J.; Miller, D.; Altekruse, S.F.; Kosary, C.L.; Yu, M.; Ruhl, J.; Tatalovich, Z.; et al. (Eds.) SEER Cancer Statistics Review, 1975–2011; National Cancer Institute: Bethesda, MD, USA, 2013. Available online: http://Seer.Cancer.Gov/Csr/1975_2011/ (accessed on 20 April 2014).
- Marcus, C.S.; Maxwell, G.L.; Darcy, K.M.; Hamilton, C.A.; McGuire, W.P. Current Approaches and Challenges in Managing and Monitoring Treatment Response in Ovarian Cancer. J. Cancer 2014, 5, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Bowtell, D.D.; Böhm, S.; Ahmed, A.A.; Aspuria, P.; Bast, R.C., Jr.; Beral, V.; Berek, J.S.; Birrer, M.J.; Blagden, S.; Bookman, M.A. Rethinking Ovarian Cancer II: Reducing Mortality from High-Grade Serous Ovarian Cancer. Nat. Rev. Cancer 2015, 15, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Auersperg, N.; Wong, A.S.T.; Choi, K.; Kang, S.K.; Leung, P.C.K. Ovarian Surface Epithelium: Biology, Endocrinology, and Pathology. Endocr. Rev. 2001, 22, 255–288. [Google Scholar] [CrossRef] [PubMed]
- Auersperg, N. The Origin of Ovarian Cancers—Hypotheses and Controversies. Front. Biosci. 2013, 5, 709–719. [Google Scholar] [CrossRef]
- Bowen, N.J.; Walker, L.D.; Matyunina, L.V.; Logani, S.; Totten, K.A.; Benigno, B.B.; McDonald, J.F. Gene Expression Profiling Supports the Hypothesis that Human Ovarian Surface Epithelia are Multipotent and Capable of Serving as Ovarian Cancer Initiating Cells. BMC Med. Genom. 2009, 2, 71. [Google Scholar] [CrossRef] [PubMed]
- Chene, G.; Dauplat, J.; Radosevic-Robin, N.; Cayre, A.; Penault-Llorca, F. Tu-be Or Not Tu-be: That is the Question… About Serous Ovarian Carcinogenesis. Crit. Rev. Oncol. 2013, 88, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Flesken-Nikitin, A.; Hwang, C.; Cheng, C.; Michurina, T.V.; Enikolopov, G.; Nikitin, A.Y. Ovarian Surface Epithelium at the Junction Area Contains a Cancer-Prone Stem Cell Niche. Nature 2013, 495, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Seidman, J.D.; Yemelyanova, A.; Zaino, R.J.; Kurman, R.J. The Fallopian Tube-Peritoneal Junction: A Potential Site of Carcinogenesis. Int. J. Gynecol. Pathol. 2011, 30, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Earp, M.A.; Cunningham, J.M. DNA Methylation Changes in Epithelial Ovarian Cancer Histotypes. Genomics 2015, 106, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Barton, C.A.; Hacker, N.F.; Clark, S.J.; O’Brien, P.M. DNA Methylation Changes in Ovarian Cancer: Implications for Early Diagnosis, Prognosis and Treatment. Gynecol. Oncol. 2008, 109, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Montavon, C.; Gloss, B.S.; Warton, K.; Barton, C.A.; Statham, A.L.; Scurry, J.P.; Tabor, B.; Nguyen, T.V.; Qu, W.; Samimi, G. Prognostic and Diagnostic Significance of DNA Methylation Patterns in High Grade Serous Ovarian Cancer. Gynecol. Oncol. 2012, 124, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Robles, A.I.; Jen, J.; Harris, C.C. Clinical Outcomes of TP53 Mutations in Cancers. Cold Spring Harb. Perspect. Med. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Klymenko, Y.; Kim, O.; Stack, M.S. Complex Determinants of Epithelial: Mesenchymal Phenotypic Plasticity in Ovarian Cancer. Cancers 2017, 9, 104. [Google Scholar] [CrossRef] [PubMed]
- Pu, T.; Xiong, L.; Liu, Q.; Zhang, M.; Cai, Q.; Liu, H.; Sood, A.K.; Li, G.; Kang, Y.; Xu, C. Delineation of Retroperitoneal Metastatic Lymph Nodes in Ovarian Cancer with Near-infrared Fluorescence Imaging. Oncol. Lett. 2017, 14, 2869–2877. [Google Scholar] [CrossRef] [PubMed]
- Pradeep, S.; Kim, S.W.; Wu, S.Y.; Nishimura, M.; Chaluvally-Raghavan, P.; Miyake, T.; Pecot, C.V.; Kim, S.; Choi, H.J.; Bischoff, F.Z. Hematogenous Metastasis of Ovarian Cancer: Rethinking Mode of Spread. Cancer Cell 2014, 26, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Lengyel, E. Ovarian Cancer Development and Metastasis. Am. J. Pathol. 2010, 177, 1053–1064. [Google Scholar] [CrossRef] [PubMed]
- Shield, K.; Ackland, M.L.; Ahmed, N.; Rice, G.E. Multicellular Spheroids in Ovarian Cancer Metastases: Biology and Pathology. Gynecol. Oncol. 2009, 113, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Hudson, L.G.; Zeineldin, R.; Stack, M.S. Phenotypic Plasticity of Neoplastic Ovarian Epithelium: Unique Cadherin Profiles in Tumor Progression. Clin. Exp. Metastasis 2008, 25, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Davidson, B. Ovarian Carcinoma and Serous Effusions. Changing Views Regarding Tumor Progression and Review of Current Literature1. Anal. Cell. Pathol. 2001, 23, 107–128. [Google Scholar] [CrossRef] [PubMed]
- Burleson, K.M.; Casey, R.C.; Skubitz, K.M.; Pambuccian, S.E.; Oegema, T.R., Jr.; Skubitz, A.P.N. Ovarian Carcinoma Ascites Spheroids Adhere to Extracellular Matrix Components and Mesothelial Cell Monolayers. Gynecol. Oncol. 2004, 93, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.; Chang, S.; Hsiao, C.; Chien, T.; Shih, D.T. Isolation and Characterization of Stromal Progenitor Cells from Ascites of Patients with Epithelial Ovarian Adenocarcinoma. J. Biomed. Sci. 2012, 19, 23. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Stenvers, K. Getting to Know Ovarian Cancer Ascites: Opportunities for Targeted Therapy-Based Translational Research. Front. Oncol. 2013, 3, 256. [Google Scholar] [CrossRef] [PubMed]
- Nieman, K.M.; Romero, I.L.; Van Houten, B.; Lengyel, E. Adipose Tissue and Adipocytes Support Tumorigenesis and Metastasis. Biochim. Biophys. Acta 2013, 1831, 1533–1541. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Conejo-Garcia, J.R.; Katsaros, D.; Gimotty, P.A.; Massobrio, M.; Regnani, G.; Makrigiannakis, A.; Gray, H.; Schlienger, K.; Liebman, M.N. Intratumoral T Cells, Recurrence, and Survival in Epithelial Ovarian Cancer. N. Engl. J. Med. 2003, 348, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Becker, G.; Galandi, D.; Blum, H.E. Malignant Ascites: Systematic Review and Guideline for Treatment. Eur. J. Cancer 2006, 42, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Byrne, A.T.; Ross, L.; Holash, J.; Nakanishi, M.; Hu, L.; Hofmann, J.I.; Yancopoulos, G.D.; Jaffe, R.B. Vascular Endothelial Growth Factor-Trap Decreases Tumor Burden, Inhibits Ascites, and Causes Dramatic Vascular Remodeling in an Ovarian Cancer Model. Clin. Cancer Res. 2003, 9, 5721–5728. [Google Scholar] [PubMed]
- Fang, X.; Schummer, M.; Mao, M.; Yu, S.; Tabassam, F.H.; Swaby, R.; Hasegawa, Y.; Tanyi, J.L.; LaPushin, R.; Eder, A. Lysophosphatidic Acid is a Bioactive Mediator in Ovarian Cancer. Biochim. Biophys. Acta 2002, 1582, 257–264. [Google Scholar] [CrossRef]
- Garrison, R.N.; Galloway, R.H.; Heuser, L.S. Mechanisms of Malignant Ascites Production. J. Surg. Res. 1987, 42, 126–132. [Google Scholar] [CrossRef]
- Gotlieb, W.H.; Abrams, J.S.; Watson, J.M.; Velu, T.J.; Berek, J.S.; Martínez-Maza, O. Presence of Interleukin 10 (IL-10) in the Ascites of Patients with Ovarian and Other Intra-Abdominal Cancers. Cytokine 1992, 4, 385–390. [Google Scholar] [CrossRef]
- Hu, L.; Hofmann, J.; Zaloudek, C.; Ferrara, N.; Hamilton, T.; Jaffe, R.B. Vascular Endothelial Growth Factor Immunoneutralization Plus Paclitaxel Markedly Reduces Tumor Burden and Ascites in Athymic Mouse Model of Ovarian Cancer. Am. J. Pathol. 2002, 161, 1917–1924. [Google Scholar] [CrossRef]
- Mesiano, S.; Ferrara, N.; Jaffe, R.B. Role of Vascular Endothelial Growth Factor in Ovarian Cancer: Inhibition of Ascites Formation by Immunoneutralization. Am. J. Pathol. 1998, 153, 1249–1256. [Google Scholar] [CrossRef]
- Moradi, M.M.; Carson, L.F.; Twiggs, L.B.; Weinberg, J.B.; Haney, A.; Ramakrishnan, S. Serum and Ascitic Fluid Levels of Interleukin-1, Interleukin-6, and Tumor Necrosis Factor-alpha in Patients with Ovarian Epithelial Cancer. Cancer 1993, 72, 2433–2440. [Google Scholar] [CrossRef]
- Pedersen, N.; Schmitt, M.; Ronne, E.; Nicoletti, M.I.; Hoyer-Hansen, G.; Conese, M.; Giavazzi, R.; Dano, K.; Kuhn, W.; Janicke, F. A Ligand-Free, Soluble Urokinase Receptor is Present in the Ascitic Fluid from Patients with Ovarian Cancer. J. Clin. Investig. 1993, 92, 2160–2167. [Google Scholar] [CrossRef] [PubMed]
- Santin, A.D.; Hermonat, P.L.; Ravaggi, A.; Cannon, M.J.; Pecorelli, S.; Parham, G.P. Secretion of Vascular Endothelial Growth Factor in Ovarian Cancer. Eur. J. Gynaecol. Oncol. 1999, 20, 177–181. [Google Scholar] [PubMed]
- Topalak, O.; Saygili, U.; Soyturk, M.; Karaca, N.; Batur, Y.; Uslu, T.; Erten, O. Serum, Pleural Effusion, and Ascites CA-125 Levels in Ovarian Cancer and Nonovarian Benign and Malignant Diseases: A Comparative Study. Gynecol. Oncol. 2002, 85, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, O.; Hafter, R.; Coppenrath, E.; Pflanz, M.A.; Schmitt, M.; Babic, R.; Linke, R.; Gossner, W.; Graeff, H. Fibrin-Fibronectin Compounds in Human Ovarian Tumor Ascites and their Possible Relation to the Tumor Stroma. Cancer Res. 1988, 48, 3507–3514. [Google Scholar] [PubMed]
- Xu, Y.; Shen, Z.; Wiper, D.W.; Wu, M.; Morton, R.E.; Elson, P.; Kennedy, A.W.; Belinson, J.; Markman, M.; Casey, G. Lysophosphatidic Acid as a Potential Biomarker for Ovarian and Other Gynecologic Cancers. JAMA 1998, 280, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Yabushita, H.; Shimazu, M.; Noguchi, M.; Kishida, T.; Narumiya, H.; Sawaguchi, K.; Noguchi, M. Vascular Endothelial Growth Factor Activating Matrix Metalloproteinase in Ascitic Fluid during Peritoneal Dissemination of Ovarian Cancer. Oncol. Rep. 2003, 10, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Carduner, L.; Leroy-Dudal, J.; Picot, C.R.; Gallet, O.; Carreiras, F.; Kellouche, S. Ascites-induced shift along epithelial-mesenchymal spectrum in ovarian cancer cells: Enhancement of their invasive behavior partly dependant on αv integrins. Clin. Exp. Metastasis 2014, 31, 675–688. [Google Scholar] [CrossRef] [PubMed]
- Waddington, C. Embryology, Epigenetics and Biogenetics. Nature 1956, 177, 1241. [Google Scholar] [CrossRef]
- Esteller, M. Cancer Epigenomics: DNA Methylomes and Histone-Modification Maps. Nat. Rev. Genet. 2007, 8, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Lyko, F. The DNA Methyltransferase Family: A Versatile Toolkit for Epigenetic Regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Huff, J.; Zilberman, D. Dnmt1-Independent CG Methylation Contributes to Nucleosome Positioning in Diverse Eukaryotes. Cell 2014, 156, 1286–1297. [Google Scholar] [CrossRef] [PubMed]
- Bestor, T.; Laudano, A.; Mattaliano, R.; Ingram, V. Cloning and Sequencing of a cDNA Encoding DNA Methyltransferase of Mouse Cells: The Carboxyl-Terminal Domain of the Mammalian Enzymes is Related to Bacterial Restriction Methyltransferases. J. Mol. Biol. 1988, 203, 971–983. [Google Scholar] [CrossRef]
- Hermann, A.; Goyal, R.; Jeltsch, A. The Dnmt1 DNA-(Cytosine-C5)-Methyltransferase Methylates DNA Processively with High Preference for Hemimethylated Target Sites. J. Biol. Chem. 2004, 279, 48350–48359. [Google Scholar] [CrossRef] [PubMed]
- Lyko, F.; Ramsahoye, B.H.; Kashevsky, H.; Tudor, M.; Mastrangelo, M.; Orr-Weaver, T.L.; Jaenisch, R. Mammalian (Cytosine-5) Methyltransferases Cause Genomic DNA Methylation and Lethality in Drosophila. Nat. Genet. 1999, 23, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Bird, A. DNA Methylation De Novo. Science 1999, 286, 2287–2288. [Google Scholar] [CrossRef] [PubMed]
- Rhee, I.; Jair, K.; Yen, R.C.; Lengauer, C.; Herman, J.G.; Kinzler, K.W.; Vogelstein, B.; Baylin, S.B.; Schuebel, K.E. CpG Methylation is Maintained in Human Cancer Cells Lacking DNMT1. Nature 2000, 404, 1003–1007. [Google Scholar] [CrossRef] [PubMed]
- Jair, K.W.; Bachman, K.E.; Suzuki, H.; Ting, A.H.; Rhee, I.; Yen, R.W.; Baylin, S.B.; Schuebel, K.E. De Novo CpG Island Methylation in Human Cancer Cells. Cancer Res. 2006, 66, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Ting, A.H.; Jair, K.W.; Schuebel, K.E.; Baylin, S.B. Differential Requirement for DNA Methyltransferase 1 in Maintaining Human Cancer Cell Gene Promoter Hypermethylation. Cancer Res. 2006, 66, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Robert, M.; Morin, S.; Beaulieu, N.; Gauthier, F.; Chute, I.C.; Barsalou, A.; MacLeod, A.R. DNMT1 is Required to Maintain CpG Methylation and Aberrant Gene Silencing in Human Cancer Cells. Nat. Genet. 2003, 33, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal Structure of the Nucleosome Core Particle at 2.8 Å. Resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Ellis, L.; Atadja, P.W.; Johnstone, R.W. Epigenetics in Cancer: Targeting Chromatin Modifications. Mol. Cancer. Ther. 2009, 8, 1409–1420. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Seto, E. HATs and HDACs: From Structure, Function and Regulation to Novel Strategies for Therapy and Prevention. Oncogene 2007, 26, 5310–5318. [Google Scholar] [CrossRef] [PubMed]
- Zentner, G.E.; Henikoff, S. Regulation of Nucleosome Dynamics by Histone Modifications. Nat. Struct. Mol. Biol. 2013, 20, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Oki, M.; Aihara, H.; Ito, T. Role of histone phosphorylation in chromatin dynamics and its implications in diseases. In Chromatin and Disease; Springer: Berlin, Germany, 2007; pp. 323–340. [Google Scholar]
- Greer, E.L.; Shi, Y. Histone Methylation: A Dynamic Mark in Health, Disease and Inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of Chromatin by Histone Modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Cedar, H.; Bergman, Y. Linking DNA Methylation and Histone Modification: Patterns and Paradigms. Nat. Rev. Genet. 2009, 10, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Hamiche, A.; Sandaltzopoulos, R.; Gdula, D.A.; Wu, C. ATP-Dependent Histone Octamer Sliding Mediated by the Chromatin Remodeling Complex NURF. Cell 1999, 97, 833–842. [Google Scholar] [CrossRef]
- Clapier, C.R.; Cairns, B.R. Chromatin remodeling complexes. In Fundamentals of Chromatin; Springer: Berlin, Germany, 2014; pp. 69–146. [Google Scholar]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of Action and Regulation of ATP-Dependent Chromatin-Remodelling Complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, K.C.; Shah, S.P.; Al-Agha, O.M.; Zhao, Y.; Tse, K.; Zeng, T.; Senz, J.; McConechy, M.K.; Anglesio, M.S.; Kalloger, S.E. ARID1A Mutations in Endometriosis-Associated Ovarian Carcinomas. N. Engl. J. Med. 2010, 363, 1532–1543. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Wang, T.L.; Shih, I.; Mao, T.L.; Nakayama, K.; Roden, R.; Glas, R.; Slamon, D.; Diaz, L.A., Jr.; Vogelstein, B.; et al. Frequent Mutations of Chromatin Remodeling Gene ARID1A in Ovarian Clear Cell Carcinoma. Science 2010, 330, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, T.; Park, P.H.; Wu, S.; Fatkhutdinov, N.; Karakashev, S.; Nacarelli, T.; Kossenkov, A.V.; Speicher, D.W.; Jean, S.; Zhang, L. Repurposing Pan-HDAC Inhibitors for ARID1A-Mutated Ovarian Cancer. Cell Rep. 2018, 22, 3393–3400. [Google Scholar] [CrossRef] [PubMed]
- Jelinic, P.; Mueller, J.J.; Olvera, N.; Dao, F.; Scott, S.N.; Shah, R.; Gao, J.; Schultz, N.; Gonen, M.; Soslow, R.A.; et al. Recurrent SMARCA4 Mutations in Small Cell Carcinoma of the Ovary. Nat. Genet. 2014, 46, 424–426. [Google Scholar] [CrossRef] [PubMed]
- Witkowski, L.; Carrot-Zhang, J.; Albrecht, S.; Fahiminiya, S.; Hamel, N.; Tomiak, E.; Grynspan, D.; Saloustros, E.; Nadaf, J.; Rivera, B. Germline and Somatic SMARCA4 Mutations Characterize Small Cell Carcinoma of the Ovary, Hypercalcemic Type. Nat. Genet. 2014, 46, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Muppala, R.; Donenberg, T.; Huang, M.S.; Schlumbrecht, M.P. SMARCA4 Germline Gene Mutation in a Patient with Epithelial Ovarian: A Case Report. Gynecol. Oncol. Rep. 2017, 22, 45–47. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Fukumoto, T.; Magno, E. SWI/SNF Complexes in Ovarian Cancer: Mechanistic Insights and Therapeutic Implications. Mol. Cancer Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Bitler, B.G.; Aird, K.M.; Garipov, A.; Li, H.; Amatangelo, M.; Kossenkov, A.V.; Schultz, D.C.; Liu, Q.; Shih, I.; Conejo-Garcia, J.R. Synthetic Lethality by Targeting EZH2 Methyltransferase Activity in ARID1A-Mutated Cancers. Nat. Med. 2015, 21, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Eichhorn, S.W.; Guo, H.; McGeary, S.E.; Rodriguez-Mias, R.A.; Shin, C.; Baek, D.; Hsu, S.; Ghoshal, K.; Villén, J.; Bartel, D.P. mRNA Destabilization is the Dominant Effect of Mammalian microRNAs by the Time Substantial Repression Ensues. Mol. Cell 2014, 56, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Volpe, T.A.; Kidner, C.; Hall, I.M.; Teng, G.; Grewal, S.I.; Martienssen, R.A. Regulation of Heterochromatic Silencing and Histone H3 Lysine-9 Methylation by RNAi. Science 2002, 297, 1833–1837. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Engreitz, J.M.; Ollikainen, N.; Guttman, M. Long Non-Coding RNAs: Spatial Amplifiers that Control Nuclear Structure and Gene Expression. Nat. Rev. Mol. Cell Biol. 2016, 17, 756–770. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, S.; Imai-Sumida, M.; Tanaka, Y.; Dahiya, R. Interaction and Cross-Talk between Non-Coding RNAs. Cell. Mol. Life Sci. 2018, 75, 467–484. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Baylin, S.B. The Epigenomics of Cancer. Cell 2007, 128, 683–692. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Jones, P. Cancer Genetics and Epigenetics: Two Sides of the Same Coin? Cancer Cell 2012, 22, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.P. The Epigenetics of Cancer Etiology. Semin. Cancer Biol. 2004, 14, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Waldmann, T.; Schneider, R. Targeting Histone Modifications—Epigenetics in Cancer. Curr. Opin. Cell Biol. 2013, 25, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Sawan, C.; Herceg, Z. Histone modifications and cancer. In Advances in Genetics; Elsevier: Amsterdam, The Netherlands, 2010; pp. 57–85. [Google Scholar]
- Audia, J.E.; Campbell, R.M. Histone Modifications and Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019521. [Google Scholar] [CrossRef] [PubMed]
- Balch, C.; Fang, F.; Matei, D.E.; Huang, T.H.; Nephew, K.P. Minireview: Epigenetic Changes in Ovarian Cancer. Endocrinology 2009, 150, 4003–4011. [Google Scholar] [CrossRef] [PubMed]
- Balch, C.; Matei, D.E.; Huang, T.H.; Nephew, K.P. Role of Epigenomics in Ovarian and Endometrial Cancers. Epigenomics 2010, 2, 419–447. [Google Scholar] [CrossRef] [PubMed]
- Natanzon, Y.; Goode, E.L.; Cunningham, J.M. Epigenetics in Ovarian Cancer. Semin. Cancer Biol. 2018, 51, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Volinia, S.; Bonome, T.; Calin, G.A.; Greshock, J.; Yang, N.; Liu, C.; Giannakakis, A.; Alexiou, P.; Hasegawa, K. Genomic and Epigenetic Alterations Deregulate microRNA Expression in Human Epithelial Ovarian Cancer. Proc. Natl. Acad. Sci. USA 2008, 105, 7004–7009. [Google Scholar] [CrossRef] [PubMed]
- Kinose, Y.; Sawada, K.; Nakamura, K.; Kimura, T. The Role of microRNAs in Ovarian Cancer. Biomed. Res. Int. 2014, 2014, 249393. [Google Scholar] [CrossRef] [PubMed]
- Meryet-Figuiere, M.; Lambert, B.; Gauduchon, P.; Vigneron, N.; Brotin, E.; Poulain, L.; Denoyelle, C. An Overview of Long Non-Coding RNAs in Ovarian Cancers. Oncotarget 2016, 7, 44719–44734. [Google Scholar] [CrossRef] [PubMed]
- Worku, T.; Bhattarai, D.; Ayers, D.; Wang, K.; Wang, C.; Rehman, Z.U.; Talpur, H.S.; Yang, L. Long Non-Coding RNAs: The New Horizon of Gene Regulation in Ovarian Cancer. Cell. Physiol. Biochem. 2017, 44, 948–966. [Google Scholar] [CrossRef] [PubMed]
- Asadollahi, R.; Hyde, C.A.; Zhong, X.Y. Epigenetics of Ovarian Cancer: From the Lab to the Clinic. Gynecol. Oncol. 2010, 118, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Balch, C.; Huang, T.H.; Brown, R.; Nephew, K.P. The Epigenetics of Ovarian Cancer Drug Resistance and Resensitization. Am. J. Obstet. Gynecol. 2004, 191, 1552–1572. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.L.; Licht, J.D. Targeting Epigenetics in Cancer. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 187–207. [Google Scholar] [CrossRef] [PubMed]
- Pfister, S.X.; Ashworth, A. Marked for Death: Targeting Epigenetic Changes in Cancer. Nat. Rev. Drug Discov. 2017, 16, 241–263. [Google Scholar] [CrossRef] [PubMed]
- Balkwill, F.R.; Capasso, M.; Hagemann, T. The Tumor Microenvironment at a Glance. J. Cell. Sci. 2012, 125, 5591–5596. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental Regulation of Tumor Progression and Metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Zhuang, X.; Lin, L.; Yu, P.; Wang, Y.; Shi, Y.; Hu, G.; Sun, Y. New Horizons in Tumor Microenvironment Biology: Challenges and Opportunities. BMC Med. 2015, 13, 45. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, J.; Zhang, L.; Wei, F.; Lian, Y.; Wu, Y.; Gong, Z.; Zhang, S.; Zhou, J.; Cao, K.; et al. Role of Tumor Microenvironment in Tumorigenesis. J. Cancer 2017, 8, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Lyssiotis, C.A.; Kimmelman, A.C. Metabolic Interactions in the Tumor Microenvironment. Trends Cell Biol. 2017, 27, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Fearon, D.T. Immune-Suppressing Cellular Elements of the Tumor Microenvironment. Annu. Rev. Cancer Biol. 2017, 1, 241–255. [Google Scholar] [CrossRef]
- Kalluri, R. The Biology and Function of Fibroblasts in Cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Lin, J. Seed-in-Soil: Pancreatic Cancer Influenced by Tumor Microenvironment. Cancers 2017, 9, 93. [Google Scholar] [CrossRef] [PubMed]
- Shiga, K.; Hara, M.; Nagasaki, T.; Sato, T.; Takahashi, H.; Takeyama, H. Cancer-Associated Fibroblasts: Their Characteristics and Their Roles in Tumor Growth. Cancers 2015, 7, 2443–2458. [Google Scholar] [CrossRef] [PubMed]
- Allinen, M.; Beroukhim, R.; Cai, L.; Brennan, C.; Lahti-Domenici, J.; Huang, H.; Porter, D.; Hu, M.; Chin, L.; Richardson, A. Molecular Characterization of the Tumor Microenvironment in Breast Cancer. Cancer Cell 2004, 6, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Walter, K.; Omura, N.; Hong, S.; Griffith, M.; Goggins, M. Pancreatic Cancer Associated Fibroblasts Display Normal Allelotypes. Cancer Biol. Ther. 2008, 7, 882–888. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Hu, M.; Sridhar, A.; Opeskin, K.; Fox, S.; Shipitsin, M.; Trivett, M.; Thompson, E.R.; Ramakrishna, M.; Gorringe, K.L. No Evidence of Clonal Somatic Genetic Alterations in Cancer-Associated Fibroblasts from Human Breast and Ovarian Carcinomas. Nat. Genet. 2008, 40, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Hanson, J.A.; Gillespie, J.W.; Grover, A.; Tangrea, M.A.; Chuaqui, R.F.; Emmert-Buck, M.R.; Tangrea, J.A.; Libutti, S.K.; Linehan, W.M.; Woodson, K.G. Gene Promoter Methylation in Prostate Tumor–Associated Stromal Cells. J. Natl. Cancer Inst. 2006, 98, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Yao, J.; Cai, L.; Bachman, K.E.; van den Brûle, F.; Velculescu, V.; Polyak, K. Distinct Epigenetic Changes in the Stromal Cells of Breast Cancers. Nat. Genet. 2005, 37, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Mathot, P.; Grandin, M.; Devailly, G.; Souaze, F.; Cahais, V.; Moran, S.; Campone, M.; Herceg, Z.; Esteller, M.; Juin, P. DNA Methylation Signal Has a Major Role in the Response of Human Breast Cancer Cells to the Microenvironment. Oncogenesis 2017, 6, e390. [Google Scholar] [CrossRef] [PubMed]
- Pistore, C.; Giannoni, E.; Colangelo, T.; Rizzo, F.; Magnani, E.; Muccillo, L.; Giurato, G.; Mancini, M.; Rizzo, S.; Riccardi, M. DNA Methylation Variations are Required for Epithelial-to-Mesenchymal Transition Induced by Cancer-Associated Fibroblasts in Prostate Cancer Cells. Oncogene 2017, 36, 5551–5566. [Google Scholar] [CrossRef] [PubMed]
- Albrengues, J.; Bertero, T.; Grasset, E.; Bonan, S.; Maiel, M.; Bourget, I.; Philippe, C.; Serrano, C.H.; Benamar, S.; Croce, O. Epigenetic Switch Drives the Conversion of Fibroblasts into Proinvasive Cancer-Associated Fibroblasts. Nat. Commun. 2015, 6, 10204. [Google Scholar] [CrossRef] [PubMed]
- Stuelten, C.H.; Busch, J.I.; Tang, B.; Flanders, K.C.; Oshima, A.; Sutton, E.; Karpova, T.S.; Roberts, A.B.; Wakefield, L.M.; Niederhuber, J.E. Transient Tumor-Fibroblast Interactions Increase Tumor Cell Malignancy by a TGF-B Mediated Mechanism in a Mouse Xenograft Model of Breast Cancer. PLoS ONE 2010, 5, e9832. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, H.; Vieth, E.; Lee, J.; Segar, M.; Liu, Y.; Nephew, K.P.; Matei, D. TGF-B Induces Global Changes in DNA Methylation during the Epithelial-to-Mesenchymal Transition in Ovarian Cancer Cells. Epigenetics 2014, 9, 1461–1472. [Google Scholar] [CrossRef] [PubMed]
- Calon, A.; Tauriello, D.V.F.; Batlle, E. TGF-Beta in CAF-Mediated Tumor Growth and Metastasis. Semin. Cancer Biol. 2014, 25, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Yeung, T.L.; Leung, C.S.; Wong, K.K.; Samimi, G.; Thompson, M.S.; Liu, J.; Zaid, T.M.; Ghosh, S.; Birrer, M.J.; Mok, S.C. TGF-Beta Modulates Ovarian Cancer Invasion by Upregulating CAF-Derived Versican in the Tumor Microenvironment. Cancer Res. 2013, 73, 5016–5028. [Google Scholar] [CrossRef] [PubMed]
- Bracken, A.P.; Dietrich, N.; Pasini, D.; Hansen, K.H.; Helin, K. Genome-Wide Mapping of Polycomb Target Genes Unravels their Roles in Cell Fate Transitions. Genes Dev. 2006, 20, 1123–1136. [Google Scholar] [CrossRef] [PubMed]
- Tyan, S.; Hsu, C.; Peng, K.; Chen, C.; Kuo, W.; Eva, Y.L.; Shew, J.; Chang, K.; Juan, L.; Lee, W. Breast Cancer Cells Induce Stromal Fibroblasts to Secrete ADAMTS1 for Cancer Invasion through an Epigenetic Change. PLoS ONE 2012, 7, e35128. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, R. Role of EZH2 in Epithelial Ovarian Cancer: From Biological Insights to Therapeutic Target. Front. Oncol. 2013, 3, 47. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Sun, Y.; Hou, Y.; Peng, Q.; Wang, L.; Luo, H.; Tang, X.; Zeng, Z.; Liu, M. MiRNA Expression Analysis of Cancer-Associated Fibroblasts and Normal Fibroblasts in Breast Cancer. Int. J. Biochem. Cell Biol. 2012, 44, 2051–2059. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tang, S.; Le, S.; Lu, R.; Rader, J.S.; Meyers, C.; Zheng, Z. Aberrant Expression of Oncogenic and Tumor-Suppressive microRNAs in Cervical Cancer is Required for Cancer Cell Growth. PLoS ONE 2008, 3, e2557. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Sakamoto, N.; Oue, N.; Yashiro, M.; Sentani, K.; Yanagihara, K.; Hirakawa, K.; Yasui, W. MicroRNA-143 Regulates Collagen Type III Expression in Stromal Fibroblasts of Scirrhous Type Gastric Cancer. Cancer Sci. 2014, 105, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, M.; Coppola, V.; Addario, A.; Patrizii, M.; Maugeri-Sacca, M.; Memeo, L.; Colarossi, C.; Francescangeli, F.; Biffoni, M.; Collura, D. Control of Tumor and Microenvironment Cross-Talk by miR-15a and miR-16 in Prostate Cancer. Oncogene 2011, 30, 4231–4242. [Google Scholar] [CrossRef] [PubMed]
- Yamamichi, N.; Shimomura, R.; Inada, K.; Sakurai, K.; Haraguchi, T.; Ozaki, Y.; Fujita, S.; Mizutani, T.; Furukawa, C.; Fujishiro, M.; et al. Locked Nucleic Acid in Situ Hybridization Analysis of miR-21 Expression during Colorectal Cancer Development. Clin. Cancer Res. 2009, 15, 4009–4016. [Google Scholar]
- Schepeler, T.; Reinert, J.T.; Ostenfeld, M.S.; Christensen, L.L.; Silahtaroglu, A.N.; Dyrskjot, L.; Wiuf, C.; Sorensen, F.J.; Kruhoffer, M.; Laurberg, S.; et al. Diagnostic and Prognostic microRNAs in Stage II Colon Cancer. Cancer Res. 2008, 68, 6416–6424. [Google Scholar] [CrossRef] [PubMed]
- Enkelmann, A.; Heinzelmann, J.; von Eggeling, F.; Walter, M.; Berndt, A.; Wunderlich, H.; Junker, K. Specific Protein and miRNA Patterns Characterise Tumour-Associated Fibroblasts in Bladder Cancer. J. Cancer Res. Clin. Oncol. 2011, 137, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.K.; Zillhardt, M.; Hua, Y.; Tiwari, P.; Murmann, A.E.; Peter, M.E.; Lengyel, E. MicroRNAs Reprogram Normal Fibroblasts into Cancer-Associated Fibroblasts in Ovarian Cancer. Cancer Discov. 2012, 2, 1100–1108. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Ji, G.; Le, X.; Wang, C.; Xu, L.; Feng, M.; Zhang, Y.; Yang, H.; Xuan, Y.; Yang, Y.; et al. Long Noncoding RNA LINC00092 Acts in Cancer-Associated Fibroblasts to Drive Glycolysis and Progression of Ovarian Cancer. Cancer Res. 2017, 77, 1369–1382. [Google Scholar] [CrossRef] [PubMed]
- Vafaee, F.; Colvin, E.K.; Mok, S.C.; Howell, V.M.; Samimi, G. Functional Prediction of Long Non-Coding RNAs in Ovarian Cancer-Associated Fibroblasts Indicate a Potential Role in Metastasis. Sci. Rep. 2017, 7, 10374. [Google Scholar] [CrossRef] [PubMed]
- Kershaw, E.E.; Flier, J.S. Adipose Tissue as an Endocrine Organ. J. Clin. Endocrinol. Metab. 2004, 89, 2548–2556. [Google Scholar] [CrossRef] [PubMed]
- Rio, M. The role of cancer-associated adipocytes (CAA) in the dynamic interaction between the tumor and the host. In Tumor-Associated Fibroblasts and Their Matrix; Springer: Berlin, Germany, 2011; pp. 111–123. [Google Scholar]
- Hoy, A.J.; Balaban, S.; Saunders, D.N. Adipocyte–Tumor Cell Metabolic Crosstalk in Breast Cancer. Trends Mol. Med. 2017, 23, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Toren, P.; Venkateswaran, V. Periprostatic Adipose Tissue and Prostate Cancer Progression: New Insights into the Tumor Microenvironment. Clin. Genitourin. Cancer 2014, 12, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Dirat, B.; Bochet, L.; Dabek, M.; Daviaud, D.; Dauvillier, S.; Majed, B.; Wang, Y.Y.; Meulle, A.; Salles, B.; Le Gonidec, S.; et al. Cancer-Associated Adipocytes Exhibit an Activated Phenotype and Contribute to Breast Cancer Invasion. Cancer Res. 2011, 71, 2455–2465. [Google Scholar] [CrossRef] [PubMed]
- Bochet, L.; Meulle, A.; Imbert, S.; Salles, B.; Valet, P.; Muller, C. Cancer-Associated Adipocytes Promotes Breast Tumor Radioresistance. Biochem. Biophys. Res. Commun. 2011, 411, 102–106. [Google Scholar]
- Tabuso, M.; Homer-Vanniasinkam, S.; Adya, R.; Arasaradnam, R.P. Role of Tissue Microenvironment Resident Adipocytes in Colon Cancer. World J. Gastroenterol. 2017, 23, 5829–5835. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, Y.C.; Wang, J.; Kong, J.; Qi, Y.; Quigg, R.J.; Li, X. miR-17-92 Cluster Accelerates Adipocyte Differentiation by Negatively Regulating Tumor-Suppressor Rb2/p130. Proc. Natl. Acad. Sci. USA 2008, 105, 2889–2894. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Hong, B.S.; Ryu, H.S.; Lee, H.; Lee, M.; Park, I.A.; Kim, J.; Han, W.; Noh, D.; Moon, H. Transition into Inflammatory Cancer-Associated Adipocytes in Breast Cancer Microenvironment Requires microRNA Regulatory Mechanism. PLoS ONE 2017, 12, e0174126. [Google Scholar] [CrossRef] [PubMed]
- Au Yeung, C.L.; Co, N.N.; Tsuruga, T.; Yeung, T.L.; Kwan, S.Y.; Leung, C.S.; Li, Y.; Lu, E.S.; Kwan, K.; Wong, K.K.; et al. Exosomal Transfer of Stroma-Derived miR21 Confers Paclitaxel Resistance in Ovarian Cancer Cells through Targeting APAF1. Nat. Commun. 2016, 7, 11150. [Google Scholar] [CrossRef] [PubMed]
- Halvorsen, A.R.; Helland, Å.; Gromov, P.; Wielenga, V.T.; Talman, M.M.; Brunner, N.; Sandhu, V.; Børresen-Dale, A.; Gromova, I.; Haakensen, V.D. Profiling of microRNAs in Tumor Interstitial Fluid of Breast Tumors—A Novel Resource to Identify Biomarkers for Prognostic Classification and Detection of Cancer. Mol. Oncol. 2017, 11, 220–234. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, R.; Monteiro, C.; Catalán, V.; Hu, P.; Cunha, V.; Rodríguez, A.; Gómez-Ambrosi, J.; Fraga, A.; Príncipe, P.; Lobato, C. Obesity and Prostate Cancer: Gene Expression Signature of Human Periprostatic Adipose Tissue. BMC Med. 2012, 10, 108. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Monteiro, C.; Matos, A.; You, J.; Fraga, A.; Pereira, C.; Catalán, V.; Rodríguez, A.; Gómez-Ambrosi, J.; Frühbeck, G. Epigenome-Wide DNA Methylation Profiling of Periprostatic Adipose Tissue in Prostate Cancer Patients with Excess Adiposity—A Pilot Study. Clin. Epigenet. 2018, 10, 54. [Google Scholar] [CrossRef] [PubMed]
- Benton, M.C.; Johnstone, A.; Eccles, D.; Harmon, B.; Hayes, M.T.; Lea, R.A.; Griffiths, L.; Hoffman, E.P.; Stubbs, R.S.; Macartney-Coxson, D. An Analysis of DNA Methylation in Human Adipose Tissue Reveals Differential Modification of Obesity Genes before and After Gastric Bypass and Weight Loss. Genome Biol. 2015, 16, 8. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.Y.; Park, Y.J.; Pan, X.; Shin, K.C.; Kwak, S.; Bassas, A.F.; Sallam, R.M.; Park, K.S.; Alfadda, A.A.; Xu, A. Obesity-Induced DNA Hypermethylation of the Adiponectin Gene Mediates Insulin Resistance. Nat. Commun. 2015, 6, 7585. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Pulliam, N.; Ozes, A.; Buechlein, A.; Ding, N.; Rusch, D.; Keer, H.; O’Hagan, H.; Stack, M.S.; Nephew, K.P. Epigenetic Targeting of Adipocytes Inhibits High-Grade Serous Ovarian Cancer Cell Migration and Invasion. Mol. Cancer. Res. 2018, 16, 1226–1240. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z. Progress and Prospects of Long Noncoding RNAs in Lipid Homeostasis. Mol. Metab. 2016, 5, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.; Liu, L.; Li, H.; Sun, Y.; Luo, H.; Li, T.; Wang, S.; Dalton, S.; Zhao, R.C.; Chen, R. Long Noncoding RNA ADINR Regulates Adipogenesis by Transcriptionally Activating C/EBPα. Stem Cell Rep. 2015, 5, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Jin, C.; Zheng, Y.; Li, X.; Zhang, S.; Zhang, Y.; Jia, L.; Li, W. Knockdown of lncRNA MIR31HG Inhibits Adipocyte Differentiation of Human Adipose-Derived Stem Cells Via Histone Modification of FABP4. Sci. Rep. 2017, 7, 8080. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Goff, L.A.; Trapnell, C.; Alexander, R.; Lo, K.A.; Hacisuleyman, E.; Sauvageau, M.; Tazon-Vega, B.; Kelley, D.R.; Hendrickson, D.G.; et al. Long Noncoding RNAs Regulate Adipogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 3387–3392. [Google Scholar] [CrossRef] [PubMed]
- Nikpayam, E.; Tasharrofi, B.; Sarrafzadeh, S.; Ghafouri-Fard, S. The Role of Long Non-Coding RNAs in Ovarian Cancer. Iran. Biomed. J. 2017, 21, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, E.S.; Meryet-Figuiere, M.; Sabzalipoor, H.; Kashani, H.H.; Nikzad, H.; Asemi, Z. Dysregulated Expression of Long Noncoding RNAs in Gynecologic Cancers. Mol. Cancer 2017, 16, 107. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Meirelles, L.; Chagastelles, P.C.; Nardi, N.B. Mesenchymal Stem Cells Reside in Virtually all Post-Natal Organs and Tissues. J. Cell Sci. 2006, 119, 2204–2213. [Google Scholar] [CrossRef] [PubMed]
- McLean, K.; Gong, Y.; Choi, Y.; Deng, N.; Yang, K.; Bai, S.; Cabrera, L.; Keller, E.; McCauley, L.; Cho, K.R.; et al. Human Ovarian Carcinoma-Associated Mesenchymal Stem Cells Regulate Cancer Stem Cells and Tumorigenesis Via Altered BMP Production. J. Clin. Investig. 2011, 121, 3206–3219. [Google Scholar] [CrossRef] [PubMed]
- Coffman, L.G.; Choi, Y.J.; McLean, K.; Allen, B.L.; di Magliano, M.P.; Buckanovich, R.J. Human Carcinoma-Associated Mesenchymal Stem Cells Promote Ovarian Cancer Chemotherapy Resistance Via a BMP4/HH Signaling Loop. Oncotarget 2016, 7, 6916–6932. [Google Scholar] [CrossRef] [PubMed]
- Touboul, C.; Lis, R.; Al Farsi, H.; Raynaud, C.M.; Warfa, M.; Althawadi, H.; Mery, E.; Mirshahi, M.; Rafii, A. Mesenchymal Stem Cells Enhance Ovarian Cancer Cell Infiltration through IL6 Secretion in an Amniochorionic Membrane Based 3D Model. J. Transl. Med. 2013, 11, 28. [Google Scholar] [CrossRef] [PubMed]
- Lis, R.; Touboul, C.; Mirshahi, P.; Ali, F.; Mathew, S.; Nolan, D.J.; Maleki, M.; Abdalla, S.A.; Raynaud, C.M.; Querleu, D. Tumor Associated Mesenchymal Stem Cells Protects Ovarian Cancer Cells from Hyperthermia through CXCL12. Int. J. Cancer 2011, 128, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Spaeth, E.L.; Dembinski, J.L.; Sasser, A.K.; Watson, K.; Klopp, A.; Hall, B.; Andreeff, M.; Marini, F. Mesenchymal Stem Cell Transition to Tumor-Associated Fibroblasts Contributes to Fibrovascular Network Expansion and Tumor Progression. PLoS ONE 2009, 4, e4992. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.A.; Park, H.; Lim, E.H.; Kim, K.H.; Choi, J.S.; Lee, J.H.; Shin, J.W.; Lee, K.W. Exosomes from Ovarian Cancer Cells Induce Adipose Tissue-Derived Mesenchymal Stem Cells to Acquire the Physical and Functional Characteristics of Tumor-Supporting Myofibroblasts. Gynecol. Oncol. 2011, 123, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Bu, S.; Wang, Q.; Zhang, Q.; Sun, J.; He, B.; Xiang, C.; Liu, Z.; Lai, D. Human Endometrial Mesenchymal Stem Cells Exhibit Intrinsic Anti-Tumor Properties on Human Epithelial Ovarian Cancer Cells. Sci. Rep. 2016, 6, 37019. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Chen, M.; Wang, L.; Ning, Y.; Liang, J.; Zhang, H.; Xu, C.; Chen, S.; Yao, L. Systemic Mesenchymal Stem Cells Reduce Growth Rate of Cisplatin-Resistant Ovarian Cancer. Int. J. Clin. Exp. Pathol. 2013, 6, 2506–2514. [Google Scholar] [PubMed]
- Zhang, Y.; Wang, J.; Ren, M.; Li, M.; Chen, D.; Chen, J.; Shi, F.; Wang, X.; Dou, J. Gene Therapy of Ovarian Cancer Using IL-21-Secreting Human Umbilical Cord Mesenchymal Stem Cells in Nude Mice. J. Ovar. Res. 2014, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Chen, W.; Zhuang, R.; Song, T.; Li, P. The Effect of Endostatin Mediated by Human Mesenchymal Stem Cells on Ovarian Cancer Cells in Vitro. J. Cancer Res. Clin. Oncol. 2010, 136, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Dembinski, J.L.; Wilson, S.M.; Spaeth, E.L.; Studeny, M.; Zompetta, C.; Samudio, I.; Roby, K.; Andreeff, M.; Marini, F.C. Tumor Stroma Engraftment of Gene-Modified Mesenchymal Stem Cells as Anti-Tumor Therapy Against Ovarian Cancer. Cytotherapy 2013, 15, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Wei, D.; Sun, L.; Wang, Y.; Wu, X.; Li, Y.; Fang, Z.; Shang, H.; Wei, Z. A Preliminary Study on the Construction of Double Suicide Gene Delivery Vectors by Mesenchymal Stem Cells and the in Vitro Inhibitory Effects on SKOV3 Cells. Oncol. Rep. 2014, 31, 781–787. [Google Scholar] [CrossRef] [PubMed]
- Amara, I.; Touati, W.; Beaune, P.; de Waziers, I. Mesenchymal Stem Cells as Cellular Vehicles for Prodrug Gene Therapy Against Tumors. Biochimie 2014, 105, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, P.; Iliou, M.S.; Esteller, M. Epigenetic Alterations Involved in Cancer Stem Cell Reprogramming. Mol. Oncol. 2012, 6, 620–636. [Google Scholar] [CrossRef] [PubMed]
- Leu, Y.; Huang, T.H.; Hsiao, S. Epigenetic reprogramming of mesenchymal stem cells. In Epigenetic Alterations in Oncogenesis; Springer: Berlin, Germany, 2013; pp. 195–211. [Google Scholar]
- Teven, C.M.; Liu, X.; Hu, N.; Tang, N.; Kim, S.H.; Huang, E.; Yang, K.; Li, M.; Gao, J.L.; Liu, H.; et al. Epigenetic Regulation of Mesenchymal Stem Cells: A Focus on Osteogenic and Adipogenic Differentiation. Stem Cells Int. 2011, 2011, 201371. [Google Scholar] [CrossRef] [PubMed]
- Mortada, I.; Mortada, R. Epigenetic Changes in Mesenchymal Stem Cells Differentiation. Eur. J. Med. Genet. 2017, 61, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.B.; Benkusky, N.A.; Sen, B.; Rubin, J.; Pike, J.W. Epigenetic Plasticity Drives Adipogenic and Osteogenic Differentiation of Marrow-Derived Mesenchymal Stem Cells. J. Biol. Chem. 2016, 291, 17829–17847. [Google Scholar] [CrossRef] [PubMed]
- Collino, F.; Bruno, S.; Deregibus, M.C.; Tetta, C.; Camussi, G. MicroRNAs and mesenchymal stem cells. In Vitamins & Hormones; Elsevier: Amsterdam, The Netherlands, 2011; pp. 291–320. [Google Scholar]
- Reza, A.M.M.T.; Choi, Y.-J.; Yasuda, H.; Kim, J.-H. Human Adipose Mesenchymal Stem Cell-Derived Exosomal-miRNAs are Critical Factors for Inducing Anti-Proliferation Signalling to A2780 and SKOV-3 Ovarian Cancer Cells. Sci. Rep. 2016, 6, 38498. [Google Scholar] [CrossRef] [PubMed]
- Bellayr, I.H.; Kumar, A.; Puri, R.K. MicroRNA Expression in Bone Marrow-Derived Human Multipotent Stromal Cells. BMC Genom. 2017, 18, 605. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Chen, L.; Chen, Y.; Shao, Q.; Qin, W. Bafilomycin A1 Inhibits the Growth and Metastatic Potential of the BEL-7402 Liver Cancer and HO-8910 Ovarian Cancer Cell Lines and Induces Alterations in their microRNA Expression. Exp. Ther. Med. 2015, 10, 1829–1834. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, J.; Zang, D.; Wu, S.; Liu, A.; Zhu, J.; Wu, G.; Li, J.; Jiang, L. Upregulation of miR-572 Transcriptionally Suppresses SOCS1 and p21 and Contributes to Human Ovarian Cancer Progression. Oncotarget 2015, 6, 15180–15193. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Huang, Y.; Zhang, L.; Tian, G.; Liao, Q.; Chen, S. MiR-572 Prompted Cell Proliferation of Human Ovarian Cancer Cells by Suppressing PPP2R2C Expression. Biomed. Pharmacother. 2016, 77, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Lopatina, T.; Gai, C.; Deregibus, M.C.; Kholia, S.; Camussi, G. Cross Talk between Cancer and Mesenchymal Stem Cells through Extracellular Vesicles Carrying Nucleic Acids. Front. Oncol. 2016, 6, 125. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.; Shih, D.T.; Hsiao, C.; Huang, S.; Chang, S.; Cheng, W. Gene Methylation of Human Ovarian Carcinoma Stromal Progenitor Cells Promotes Tumorigenesis. J. Transl. Med. 2015, 13, 367. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Taylor, S.M. Cellular Differentiation, Cytidine Analogs and DNA Methylation. Cell 1980, 20, 85–93. [Google Scholar] [CrossRef]
- Akino, T.; Hida, K.; Hida, Y.; Tsuchiya, K.; Freedman, D.; Muraki, C.; Ohga, N.; Matsuda, K.; Akiyama, K.; Harabayashi, T.; et al. Cytogenetic Abnormalities of Tumor-Associated Endothelial Cells in Human Malignant Tumors. Am. J. Pathol. 2009, 175, 2657–2667. [Google Scholar] [CrossRef] [PubMed]
- Konerding, M.; Malkusch, W.; Klapthor, B.; Van Ackern, C.; Fait, E.; Hill, S.; Parkins, C.; Chaplin, D.; Presta, M.; Denekamp, J. Evidence for Characteristic Vascular Patterns in Solid Tumours: Quantitative Studies using Corrosion Casts. Br. J. Cancer 1999, 80, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Seaman, S.; Stevens, J.; Yang, M.Y.; Logsdon, D.; Graff-Cherry, C.; Croix, B.S. Genes that Distinguish Physiological and Pathological Angiogenesis. Cancer Cell 2007, 11, 539–554. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.; Kumar, B.; Yu, J.; Old, M.; Teknos, T.N.; Kumar, P. Tumor-Associated Endothelial Cells Promote Tumor Metastasis by Chaperoning Circulating Tumor Cells and Protecting them from Anoikis. PLoS ONE 2015, 10, e0141602. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, K.; Ohga, N.; Hida, Y.; Kawamoto, T.; Sadamoto, Y.; Ishikawa, S.; Maishi, N.; Akino, T.; Kondoh, M.; Matsuda, A. Tumor Endothelial Cells Acquire Drug Resistance by MDR1 Up-Regulation Via VEGF Signaling in Tumor Microenvironment. Am. J. Pathol. 2012, 180, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Hida, K.; Ohga, N.; Akiyama, K.; Maishi, N.; Hida, Y. Heterogeneity of Tumor Endothelial Cells. Cancer Sci. 2013, 104, 1391–1395. [Google Scholar] [CrossRef] [PubMed]
- Ohga, N.; Ishikawa, S.; Maishi, N.; Akiyama, K.; Hida, Y.; Kawamoto, T.; Sadamoto, Y.; Osawa, T.; Yamamoto, K.; Kondoh, M. Heterogeneity of Tumor Endothelial Cells: Comparison between Tumor Endothelial Cells Isolated from High-and Low-Metastatic Tumors. Am. J. Pathol. 2012, 180, 1294–1307. [Google Scholar] [CrossRef] [PubMed]
- Maishi, N.; Ohba, Y.; Akiyama, K.; Ohga, N.; Hamada, J.; Nagao-Kitamoto, H.; Alam, M.T.; Yamamoto, K.; Kawamoto, T.; Inoue, N. Tumour Endothelial Cells in High Metastatic Tumours Promote Metastasis Via Epigenetic Dysregulation of Biglycan. Sci. Rep. 2016, 6, 28039. [Google Scholar] [CrossRef] [PubMed]
- Hellebrekers, D.M.; Jair, K.W.; Vire, E.; Eguchi, S.; Hoebers, N.T.; Fraga, M.F.; Esteller, M.; Fuks, F.; Baylin, S.B.; van Engeland, M.; et al. Angiostatic Activity of DNA Methyltransferase Inhibitors. Mol. Cancer Ther. 2006, 5, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Kwon, H.J.; Lee, Y.M.; Baek, J.H.; Jang, J.; Lee, S.; Moon, E.; Kim, H.; Lee, S.; Chung, H.Y. Histone Deacetylases Induce Angiogenesis by Negative Regulation of Tumor Suppressor Genes. Nat. Med. 2001, 7, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Hellebrekers, D.M.; Castermans, K.; Vire, E.; Dings, R.P.; Hoebers, N.T.; Mayo, K.H.; Oude Egbrink, M.G.; Molema, G.; Fuks, F.; van Engeland, M.; et al. Epigenetic Regulation of Tumor Endothelial Cell Anergy: Silencing of Intercellular Adhesion Molecule-1 by Histone Modifications. Cancer Res. 2006, 66, 10770–10777. [Google Scholar] [CrossRef] [PubMed]
- Hellebrekers, D.M.; Melotte, V.; Vire, E.; Langenkamp, E.; Molema, G.; Fuks, F.; Herman, J.G.; Van Criekinge, W.; Griffioen, A.W.; van Engeland, M. Identification of Epigenetically Silenced Genes in Tumor Endothelial Cells. Cancer Res. 2007, 67, 4138–4148. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Bonome, T.; Li, Y.; Kamat, A.A.; Han, L.Y.; Schmandt, R.; Coleman, R.L.; Gershenson, D.M.; Jaffe, R.B.; Birrer, M.J.; et al. Gene Alterations Identified by Expression Profiling in Tumor-Associated Endothelial Cells from Invasive Ovarian Carcinoma. Cancer Res. 2007, 67, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.A.; Varambally, S.; Arend, R.C. Histone Methyltransferase EZH2: A Therapeutic Target for Ovarian Cancer. Mol. Cancer Ther. 2018, 17, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Han, H.D.; Mangala, L.S.; Ali-Fehmi, R.; Newton, C.S.; Ozbun, L.; Armaiz-Pena, G.N.; Hu, W.; Stone, R.L.; Munkarah, A. Regulation of Tumor Angiogenesis by EZH2. Cancer Cell 2010, 18, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Chen, J.; Wang, H. MicroRNA-298 Inhibits Malignant Phenotypes of Epithelial Ovarian Cancer by Regulating the Expression of EZH2. Oncol. Lett. 2016, 12, 3926–3932. [Google Scholar] [CrossRef] [PubMed]
- Özeş, A.R.; Wang, Y.; Zong, X.; Fang, F.; Pilrose, J.; Nephew, K.P. Therapeutic Targeting using Tumor Specific Peptides Inhibits Long Non-Coding RNA HOTAIR Activity in Ovarian and Breast Cancer. Sci. Rep. 2017, 7, 894. [Google Scholar] [CrossRef] [PubMed]
- Bergers, G.; Song, S. The Role of Pericytes in Blood-Vessel Formation and Maintenance. Neuro-Oncology 2005, 7, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Crisan, M.; Yap, S.; Casteilla, L.; Chen, C.; Corselli, M.; Park, T.S.; Andriolo, G.; Sun, B.; Zheng, B.; Zhang, L. A Perivascular Origin for Mesenchymal Stem Cells in Multiple Human Organs. Cell Stem Cell 2008, 3, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Farrington-Rock, C.; Crofts, N.J.; Doherty, M.J.; Ashton, B.A.; Griffin-Jones, C.; Canfield, A.E. Chondrogenic and Adipogenic Potential of Microvascular Pericytes. Circulation 2004, 110, 2226–2232. [Google Scholar] [CrossRef] [PubMed]
- Balabanov, R.; Washington, R.; Wagnerova, J.; Dore-Duffy, P. CNS Microvascular Pericytes Express Macrophage-Like Function, Cell Surface Integrin αM, and Macrophage Marker ED-2. Microvasc. Res. 1996, 52, 127–142. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.L.; Okamoto, O.K. Combined Effects of Pericytes in the Tumor Microenvironment. Stem Cells Int. 2015, 2015, 868475. [Google Scholar] [CrossRef] [PubMed]
- Ferland-McCollough, D.; Slater, S.; Richard, J.; Reni, C.; Mangialardi, G. Pericytes, an Overlooked Player in Vascular Pathobiology. Pharmacol. Ther. 2017, 171, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Sinha, D.; Chong, L.; George, J.; Schluter, H.; Monchgesang, S.; Mills, S.; Li, J.; Parish, C.; Bowtell, D.; Kaur, P.; et al. Pericytes Promote Malignant Ovarian Cancer Progression in Mice and Predict Poor Prognosis in Serous Ovarian Cancer Patients. Clin. Cancer Res. 2016, 22, 1813–1824. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Shahzad, M.M.; Moreno-Smith, M.; Lin, Y.; Jennings, N.B.; Allen, J.K.; Landen, C.N.; Mangala, L.S.; Armaiz-Pena, G.N.; Schmandt, R. Targeting Pericytes with a PDGF-B Aptamer in Human Ovarian Carcinoma Models. Cancer Biol. Ther. 2010, 9, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Perotti, A.; Sessa, C.; Mancuso, A.; Noberasco, C.; Cresta, S.; Locatelli, A.; Carcangiu, M.L.; Passera, K.; Braghetti, A.; Scaramuzza, D.; et al. Clinical and Pharmacological Phase I Evaluation of Exherin (ADH-1), a Selective Anti-N-Cadherin Peptide in Patients with N-Cadherin-Expressing Solid Tumours. Ann. Oncol. 2009, 20, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Yi, D.; Xiang, W.; Zhang, Q.; Cen, Y.; Su, Q.; Zhang, F.; Lu, Y.; Zhao, H.; Fu, P. Human Glioblastoma-Derived Mesenchymal Stem Cell to Pericytes Transition and Angiogenic Capacity in Glioblastoma Microenvironment. Cell Physiol. Biochem. 2018, 46, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Truettner, J.S.; Katyshev, V.; Esen-Bilgin, N.; Dietrich, W.D.; Dore-Duffy, P. Hypoxia Alters MicroRNA Expression in Rat Cortical Pericytes. Microrna 2013, 2, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Niida, S.; Azuma, E.; Yanagibashi, T.; Muramatsu, M.; Huang, T.T.; Sagara, H.; Higaki, S.; Ikutani, M.; Nagai, Y. Inflammation-Induced Endothelial Cell-Derived Extracellular Vesicles Modulate the Cellular Status of Pericytes. Sci. Rep. 2015, 5, 8505. [Google Scholar] [CrossRef] [PubMed]
- Kratzsch, T.; Kuhn, S.A.; Joedicke, A.; Hanisch, U.K.; Vajkoczy, P.; Hoffmann, J.; Fichtner, I. Treatment with 5-Azacitidine Delay Growth of Glioblastoma Xenografts: A Potential New Treatment Approach for Glioblastomas. J. Cancer Res. Clin. Oncol. 2018, 144, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Olin, E.; Buskirk, H.; Reineke, L. Cytotoxicity and Mode of Action of 5-Azacytidine on L1210 Leukemia. Cancer Res. 1970, 30, 2760–2769. [Google Scholar] [PubMed]
- Karén, J.; Rodriguez, A.; Friman, T.; Dencker, L.; Sundberg, C.; Scholz, B. Effects of the Histone Deacetylase Inhibitor Valproic Acid on Human Pericytes in Vitro. PLoS ONE 2011, 6, e24954. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.E.; Pollard, J.W. Distinct Role of Macrophages in Different Tumor Microenvironments. Cancer Res. 2006, 66, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Turkowski, K.; Mora, J.; Brune, B.; Seeger, W.; Weigert, A.; Savai, R. Redirecting Tumor-Associated Macrophages to Become Tumoricidal Effectors as a Novel Strategy for Cancer Therapy. Oncotarget 2017, 8, 48436–48452. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Marchesi, F.; Malesci, A.; Laghi, L.; Allavena, P. Tumour-Associated Macrophages as Treatment Targets in Oncology. Nat. Rev. Clin. Oncol. 2017, 14, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Hagemann, T.; Wilson, J.; Burke, F.; Kulbe, H.; Li, N.F.; Pluddemann, A.; Charles, K.; Gordon, S.; Balkwill, F.R. Ovarian Cancer Cells Polarize Macrophages Toward a Tumor-Associated Phenotype. J. Immunol. 2006, 176, 5023–5032. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, K.; Komohara, Y.; Takaishi, K.; Katabuchi, H.; Takeya, M. Detection of M2 Macrophages and Colony-stimulating Factor 1 Expression in Serous and Mucinous Ovarian Epithelial Tumors. Pathol. Int. 2009, 59, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Colvin, E.K. Tumor-Associated Macrophages Contribute to Tumor Progression in Ovarian Cancer. Front. Oncol. 2014, 4, 137. [Google Scholar] [CrossRef] [PubMed]
- Reinartz, S.; Finkernagel, F.; Adhikary, T.; Rohnalter, V.; Schumann, T.; Schober, Y.; Nockher, W.A.; Nist, A.; Stiewe, T.; Jansen, J.M. A Transcriptome-Based Global Map of Signaling Pathways in the Ovarian Cancer Microenvironment Associated with Clinical Outcome. Genome Biol. 2016, 17, 108. [Google Scholar] [CrossRef] [PubMed]
- Worzfeld, T.; Finkernagel, F.; Reinartz, S.; Konzer, A.; Adhikary, T.; Nist, A.; Stiewe, T.; Wagner, U.; Looso, M.; Graumann, J.; et al. Proteotranscriptomics Reveal Signaling Networks in the Ovarian Cancer Microenvironment. Mol. Cell Proteom. 2018, 17, 270–289. [Google Scholar] [CrossRef] [PubMed]
- Adhikary, A.; Chakraborty, S.; Mazumdar, M.; Ghosh, S.; Mukherjee, S.; Manna, A.; Mohanty, S.; Nakka, K.K.; Joshi, S.; De, A.; et al. Inhibition of Epithelial to Mesenchymal Transition by E-Cadherin Up-Regulation Via Repression of Slug Transcription and Inhibition of E-Cadherin Degradation: Dual Role of Scaffold/Matrix Attachment Region-Binding Protein 1 (SMAR1) in Breast Cancer Cells. J. Biol. Chem. 2014, 289, 25431–25444. [Google Scholar] [CrossRef] [PubMed]
- Ying, X.; Wu, Q.; Wu, X.; Zhu, Q.; Wang, X.; Jiang, L.; Chen, X.; Wang, X. Epithelial Ovarian Cancer-Secreted Exosomal miR-222-3p Induces Polarization of Tumor-Associated Macrophages. Oncotarget 2016, 7, 43076–43087. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ying, X.; Wang, X.; Wu, X.; Zhu, Q.; Wang, X. Exosomes Derived from Hypoxic Epithelial Ovarian Cancer Deliver microRNA-940 to Induce Macrophage M2 Polarization. Oncol. Rep. 2017, 38, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Wu, X.; Ying, X.; Zhu, Q.; Wang, X.; Jiang, L.; Chen, X.; Wu, Y.; Wang, X. Suppression of Endothelial Cell Migration by Tumor Associated Macrophage-Derived Exosomes is Reversed by Epithelial Ovarian Cancer Exosomal lncRNA. Cancer Cell Int. 2017, 17, 62. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Li, D.; Wu, A.; Qiu, X.; Di, W.; Huang, L.; Qiu, L. TWEAK-Stimulated Macrophages Inhibit Metastasis of Epithelial Ovarian Cancer Via Exosomal Shuttling of microRNA. Cancer Lett. 2017, 393, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, C.; Yang, C.; Tsai, I.; Hou, Y.; Chen, C.; Shan, Y. Tumor-Associated Macrophages Promote Epigenetic Silencing of Gelsolin through DNA Methyltransferase 1 in Gastric Cancer Cells. Cancer Immunol. Res. 2017, 5, 885–897. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Jiang, C.; Liu, X.; Xu, S.; Zhang, Z.; Chen, H.; Zhang, Y.; Liu, Y.; Ma, D. Hypermethylation of IFN-Γ in Oral Cancer Tissues. Clin. Oral Investig. 2017, 21, 2535–2542. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Xiao, X.; Huang, T.; Du, C.; Wang, S.; Mo, Y.; Ma, N.; Murata, M.; Li, B.; Wen, W.; et al. Epigenetic Inactivation of Follistatin-Like 1 Mediates Tumor Immune Evasion in Nasopharyngeal Carcinoma. Oncotarget 2016, 7, 16433–16444. [Google Scholar] [CrossRef] [PubMed]
- Osawa, T.; Tsuchida, R.; Muramatsu, M.; Shimamura, T.; Wang, F.; Suehiro, J.; Kanki, Y.; Wada, Y.; Yuasa, Y.; Aburatani, H.; et al. Inhibition of Histone Demethylase JMJD1A Improves Anti-Angiogenic Therapy and Reduces Tumor-Associated Macrophages. Cancer Res. 2013, 73, 3019–3028. [Google Scholar] [CrossRef] [PubMed]
- Ishii, M.; Wen, H.; Corsa, C.A.; Liu, T.; Coelho, A.L.; Allen, R.M.; Carson, W.F., 4th; Cavassani, K.A.; Li, X.; Lukacs, N.W.; et al. Epigenetic Regulation of the Alternatively Activated Macrophage Phenotype. Blood 2009, 114, 3244–3254. [Google Scholar] [CrossRef] [PubMed]
- Leffers, N.; Gooden, M.J.; de Jong, R.A.; Hoogeboom, B.; Klaske, A.; Hollema, H.; Boezen, H.M.; van der Zee, A.G.J.; Daemen, T.; Nijman, H.W. Prognostic Significance of Tumor-Infiltrating T-Lymphocytes in Primary and Metastatic Lesions of Advanced Stage Ovarian Cancer. Cancer Immunol. Immunother. 2009, 58, 449–459. [Google Scholar] [CrossRef] [PubMed]
- James, F.R.; Jiminez-Linan, M.; Alsop, J.; Mack, M.; Song, H.; Brenton, J.D.; Pharoah, P.D.; Ali, H.R. Association between Tumour Infiltrating Lymphocytes, Histotype and Clinical Outcome in Epithelial Ovarian Cancer. BMC Cancer 2017, 17, 657. [Google Scholar] [CrossRef] [PubMed]
- Barnett, J.C.; Bean, S.M.; Whitaker, R.S.; Kondoh, E.; Baba, T.; Fujii, S.; Marks, J.R.; Dressman, H.K.; Murphy, S.K.; Berchuck, A. Ovarian Cancer Tumor Infiltrating T-Regulatory (T(Reg)) Cells are Associated with a Metastatic Phenotype. Gynecol. Oncol. 2010, 116, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Tsiatas, M.L.; Gyftaki, R.; Liacos, C.; Politi, E.; Rodolakis, A.; Dimopoulos, M.A.; Bamias, A. Study of T Lymphocytes Infiltrating Peritoneal Metastases in Advanced Ovarian Cancer: Associations with Vascular Endothelial Growth Factor Levels and Prognosis in Patients Receiving Platinum-Based Chemotherapy. Int. J. Gynecol. Cancer 2009, 19, 1329–1334. [Google Scholar] [CrossRef] [PubMed]
- Napoletano, C.; Bellati, F.; Landi, R.; Pauselli, S.; Marchetti, C.; Visconti, V.; Sale, P.; Liberati, M.; Rughetti, A.; Frati, L. Ovarian Cancer Cytoreduction Induces Changes in T Cell Population Subsets Reducing Immunosuppression. J. Cell. Mol. Med. 2010, 14, 2748–2759. [Google Scholar] [CrossRef] [PubMed]
- Curiel, T.J.; Coukos, G.; Zou, L.; Alvarez, X.; Cheng, P.; Mottram, P.; Evdemon-Hogan, M.; Conejo-Garcia, J.R.; Zhang, L.; Burow, M. Specific Recruitment of Regulatory T Cells in Ovarian Carcinoma Fosters Immune Privilege and Predicts Reduced Survival. Nat. Med. 2004, 10, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Santoiemma, P.P.; Powell, D.J., Jr. Tumor Infiltrating Lymphocytes in Ovarian Cancer. Cancer Biol. Ther. 2015, 16, 807–820. [Google Scholar] [CrossRef] [PubMed]
- Sehouli, J.; Loddenkemper, C.; Cornu, T.; Schwachula, T.; Hoffmüller, U.; Grützkau, A.; Lohneis, P.; Dickhaus, T.; Gröne, J.; Kruschewski, M. Epigenetic Quantification of Tumor-Infiltrating T-Lymphocytes. Epigenetics 2011, 6, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chiappinelli, K.B.; Guzzetta, A.A.; Easwaran, H.; Yen, R.W.; Vatapalli, R.; Topper, M.J.; Luo, J.; Connolly, R.M.; Azad, N.S.; et al. Immune Regulation by Low Doses of the DNA Methyltransferase Inhibitor 5-Azacitidine in Common Human Epithelial Cancers. Oncotarget 2014, 5, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Amoozgar, Z.; Huang, J.; Saleh, M.H.; Xing, D.; Orsulic, S.; Goldberg, M.S. Decitabine Enhances Lymphocyte Migration and Function and Synergizes with CTLA-4 Blockade in a Murine Ovarian Cancer Model. Cancer. Immunol. Res. 2015, 3, 1030–1041. [Google Scholar] [CrossRef] [PubMed]
- Stone, M.L.; Chiappinelli, K.B.; Li, H.; Murphy, L.M.; Travers, M.E.; Topper, M.J.; Mathios, D.; Lim, M.; Shih, I.M.; Wang, T.L.; et al. Epigenetic Therapy Activates Type I Interferon Signaling in Murine Ovarian Cancer to Reduce Immunosuppression and Tumor Burden. Proc. Natl. Acad. Sci. USA 2017, 114, E10981–E10990. [Google Scholar] [CrossRef] [PubMed]
- Adair, S.J.; Hogan, K.T. Treatment of Ovarian Cancer Cell Lines with 5-Aza-2′-Deoxycytidine Upregulates the Expression of Cancer-Testis Antigens and Class I Major Histocompatibility Complex-Encoded Molecules. Cancer Immunol. Immunother. 2009, 58, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A. Inhibiting DNA Methylation Causes an Interferon Response in Cancer Via dsRNA Including Endogenous Retroviruses. Cell 2015, 162, 974–986. [Google Scholar] [CrossRef] [PubMed]
- Korsunsky, I.; Parameswaran, J.; Shapira, I.; Lovecchio, J.; Menzin, A.; Whyte, J.; Dos Santos, L.; Liang, S.; Bhuiya, T.; Keogh, M.; et al. Two microRNA Signatures for Malignancy and Immune Infiltration Predict overall Survival in Advanced Epithelial Ovarian Cancer. J. Investig. Med. 2017, 65, 1068–1076. [Google Scholar] [CrossRef] [PubMed]
- McKenna, K.; Beignon, A.S.; Bhardwaj, N. Plasmacytoid Dendritic Cells: Linking Innate and Adaptive Immunity. J. Virol. 2005, 79, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Vermi, W.; Soncini, M.; Melocchi, L.; Sozzani, S.; Facchetti, F. Plasmacytoid Dendritic Cells and Cancer. J. Leukoc. Biol. 2011, 90, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wu, J.; Zhu, S.; Liu, Y.; Chen, J. Disease-Associated Plasmacytoid Dendritic Cells. Front. Immunol. 2017, 8, 1268. [Google Scholar] [CrossRef] [PubMed]
- Wertel, I.; Polak, G.; Bednarek, W.; Barczyński, B.; Roliński, J.; Kotarski, J. Dendritic Cell Subsets in the Peritoneal Fluid and Peripheral Blood of Women Suffering from Ovarian Cancer. Cytom. Part B Clin. Cytom. 2008, 74, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Curiel, T.J.; Cheng, P.; Mottram, P.; Alvarez, X.; Moons, L.; Evdemon-Hogan, M.; Wei, S.; Zou, L.; Kryczek, I.; Hoyle, G.; et al. Dendritic Cell Subsets Differentially Regulate Angiogenesis in Human Ovarian Cancer. Cancer Res. 2004, 64, 5535–5538. [Google Scholar] [CrossRef] [PubMed]
- Conejo-Garcia, J.R.; Benencia, F.; Courreges, M.; Kang, E.; Mohamed-Hadley, A.; Buckanovich, R.J.; Holtz, D.O.; Jenkins, A.; Na, H.; Zhang, L. Tumor-Infiltrating Dendritic Cell Precursors Recruited by a B-Defensin Contribute to Vasculogenesis Under the Influence of Vegf-A. Nat. Med. 2004, 10, 950–958. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Kryczek, I.; Zou, L.; Daniel, B.; Cheng, P.; Mottram, P.; Curiel, T.; Lange, A.; Zou, W. Plasmacytoid Dendritic Cells Induce CD8+ Regulatory T Cells in Human Ovarian Carcinoma. Cancer Res. 2005, 65, 5020–5026. [Google Scholar] [CrossRef] [PubMed]
- Labidi-Galy, S.I.; Treilleux, I.; Goddard-Leon, S.; Combes, J.; Blay, J.; Ray-Coquard, I.; Caux, C.; Bendriss-Vermare, N. Plasmacytoid Dendritic Cells Infiltrating Ovarian Cancer are Associated with Poor Prognosis. Oncoimmunology 2012, 1, 380–382. [Google Scholar] [CrossRef] [PubMed]
- Huarte, E.; Cubillos-Ruiz, J.R.; Nesbeth, Y.C.; Scarlett, U.K.; Martinez, D.G.; Buckanovich, R.J.; Benencia, F.; Stan, R.V.; Keler, T.; Sarobe, P.; et al. Depletion of Dendritic Cells Delays Ovarian Cancer Progression by Boosting Antitumor Immunity. Cancer Res. 2008, 68, 7684–7691. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Meng, L.; Zhang, Y. Epigenetic Regulation of Dendritic Cell Development and Function. Cancer J. 2017, 23, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Paul, F.; Amit, I. Plasticity in the Transcriptional and Epigenetic Circuits Regulating Dendritic Cell Lineage Specification and Function. Curr. Opin. Immunol. 2014, 30, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, Y.; Park, J.H.; Kim, Y. Transcriptional and Epigenetic Networks in the Development and Maturation of Dendritic Cells. Epigenomics 2013, 5, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Smyth, L.A.; Boardman, D.A.; Tung, S.L.; Lechler, R.; Lombardi, G. MicroRNAs Affect Dendritic Cell Function and Phenotype. Immunology 2015, 144, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Valadkhan, S.; Gunawardane, L.S. lncRNA-Mediated Regulation of the Interferon Response. Virus Res. 2016, 212, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Cubillos-Ruiz, J.R.; Baird, J.R.; Tesone, A.J.; Rutkowski, M.R.; Scarlett, U.K.; Camposeco-Jacobs, A.L.; Anadon-Arnillas, J.; Harwood, N.M.; Korc, M.; Fiering, S.N.; et al. Reprogramming Tumor-Associated Dendritic Cells in Vivo using miRNA Mimetics Triggers Protective Immunity Against Ovarian Cancer. Cancer Res. 2012, 72, 1683–1693. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Ubreva, J.; Català-Moll, F.; Obermajer, N.; Álvarez-Errico, D.; Ramirez, R.N.; Vento-Tormo, R.; Moreno-Bueno, G.; Edwards, R.P.; Mortazavi, A.; Kalinski, P. Prostaglandin E2 Leads to the Acquisition of DNMT3A-Dependent Tolerogenic Functions in Human Myeloid-Derived Suppressor Cells. Cell Rep. 2017, 21, 154–167. [Google Scholar] [CrossRef] [PubMed]
- Davidowitz, R.A.; Iwanicki, M.P.; Brugge, J.S. In Vitro Mesothelial Clearance Assay that Models the Early Steps of Ovarian Cancer Metastasis. J. Vis. Exp. 2012, 60, 3888. [Google Scholar] [CrossRef] [PubMed]
- Klymenko, Y.; Kim, O.; Loughran, E.; Yang, J.; Lombard, R.; Alber, M.; Stack, M. Cadherin Composition and Multicellular Aggregate Invasion in Organotypic Models of Epithelial Ovarian Cancer Intraperitoneal Metastasis. Oncogene 2017, 19, 5840–5851. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Xiao, Y.J.; Singh, L.S.; Zhao, X.; Zhao, Z.; Feng, L.; Rose, T.M.; Prestwich, G.D.; Xu, Y. Lysophosphatidic Acid is Constitutively Produced by Human Peritoneal Mesothelial Cells and Enhances Adhesion, Migration, and Invasion of Ovarian Cancer Cells. Cancer Res. 2006, 66, 3006–3014. [Google Scholar] [CrossRef] [PubMed]
- Stadlmann, S.; Amberger, A.; Pollheimer, J.; Gastl, G.; Offner, F.A.; Margreiter, R.; Zeimet, A.G. Ovarian Carcinoma Cells and IL-1beta-Activated Human Peritoneal Mesothelial Cells are Possible Sources of Vascular Endothelial Growth Factor in Inflammatory and Malignant Peritoneal Effusions. Gynecol. Oncol. 2005, 97, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Kassim, S.K.; El-Salahy, E.M.; Fayed, S.T.; Helal, S.A.; Helal, T.; Azzam, E.E.; Khalifa, A. Vascular Endothelial Growth Factor and Interleukin-8 are Associated with Poor Prognosis in Epithelial Ovarian Cancer Patients. Clin. Biochem. 2004, 37, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, S.M.; Amini, A.; Morris, D.L.; Pourgholami, M.H. Significance of Vascular Endothelial Growth Factor in Growth and Peritoneal Dissemination of Ovarian Cancer. Cancer Metastasis Rev. 2012, 31, 143–162. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, D.; Zhang, H.; Kirmani, K.; Zhao, Z.; Steinmetz, R.; Xu, Y. Lysophosphatidic Acid Stimulates Cell Migration, Invasion, and Colony Formation as Well as Tumorigenesis/Metastasis of Mouse Ovarian Cancer in Immunocompetent Mice. Mol. Cancer Ther. 2009, 8, 1692–1701. [Google Scholar] [CrossRef] [PubMed]
- Zeimet, A.G.; Stadlmann, S.; Natoli, C.; Widschwendter, M.; Hermann, M.; Abendstein, B.; Daxenbichler, G.; Offner, F.A.; Iacobelli, S.; Marth, C. Peritoneal Mesothelial Cells as a Significant Source of Ascitic Immunostimulatory Protein 90K. Anticancer Res. 2000, 20, 4507–4511. [Google Scholar] [PubMed]
- Zeimet, A.G.; Natoli, C.; Herold, M.; Fuchs, D.; Windbichler, G.; Daxenbichler, G.; Iacobelli, S.; Dapunt, O.; Marth, C. Circulating Immunostimulatory Protein 90K and Soluble Interleukin-2-receptor in Human Ovarian Cancer. Int. J. Cancer 1996, 68, 34–38. [Google Scholar] [CrossRef]
- Iacobelli, S.; Sismondi, P.; Giai, M.; d’Egidio, M.; Tinari, N.; Amatetti, C.; Di Stefano, P.; Natoli, C. Prognostic Value of a Novel Circulating Serum 90K Antigen in Breast Cancer. Br. J. Cancer 1994, 69, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Fornarini, B.; D’Ambrosio, C.; Natoli, C.; Tinari, N.; Silingardi, V.; Iacobelli, S. Adhesion to 90K (Mac-2 BP) as a Mechanism for Lymphoma Drug Resistance in Vivo. Blood 2000, 96, 3282–3285. [Google Scholar] [PubMed]
- Matte, I.; Lane, D.; Bachvarov, D.; Rancourt, C.; Piché, A. Role of Malignant Ascites on Human Mesothelial Cells and their Gene Expression Profiles. BMC Cancer 2014, 14, 288. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Munck, J.; Tang, J.; Taverna, P.; Wang, Y.; Miller, D.F.; Pilrose, J.; Choy, G.; Azab, M.; Pawelczak, K.S.; et al. The Novel, Small-Molecule DNA Methylation Inhibitor SGI-110 as an Ovarian Cancer Chemosensitizer. Clin. Cancer Res. 2014, 20, 6504–6516. [Google Scholar]
- Fang, F.; Cardenas, H.; Huang, H.; Jiang, G.; Perkins, S.M.; Zhang, C.; Keer, H.N.; Liu, Y.; Nephew, K.P.; Matei, D. Genomic and Epigenomic Signatures in Ovarian Cancer Associated with Resensitization to Platinum Drugs. Cancer Res. 2018, 78, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Balch, C.; Schilder, J.; Breen, T.; Zhang, S.; Shen, C.; Li, L.; Kulesavage, C.; Snyder, A.J.; Nephew, K.P. A Phase 1 and Pharmacodynamic Study of Decitabine in Combination with Carboplatin in Patients with Recurrent, Platinum-resistant, Epithelial Ovarian Cancer. Cancer 2010, 116, 4043–4053. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cardenas, H.; Fang, F.; Condello, S.; Taverna, P.; Segar, M.; Liu, Y.; Nephew, K.P.; Matei, D. Epigenetic Targeting of Ovarian Cancer Stem Cells. Cancer Res. 2014, 74, 4922–4936. [Google Scholar] [CrossRef] [PubMed]
- Matei, D.; Ghamande, S.; Roman, L.; Alvarez Secord, A.; Nemunaitis, J.; Markham, M.J.; Nephew, K.P.; Jueliger, S.; Oganesian, A.; Naim, S.; et al. A Phase I Clinical Trial of Guadecitabine and Carboplatin in Platinum-Resistant, Recurrent Ovarian Cancer: Clinical, Pharmacokinetic, and Pharmacodynamic Analyses. Clin. Cancer Res. 2018, 24, 2285–2293. [Google Scholar] [CrossRef] [PubMed]
- Modesitt, S.C.; Sill, M.; Hoffman, J.S.; Bender, D.P. A Phase II Study of Vorinostat in the Treatment of Persistent or Recurrent Epithelial Ovarian Or Primary Peritoneal Carcinoma: A Gynecologic Oncology Group Study. Gynecol. Oncol. 2008, 109, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; LaRochelle, W.J.; Ara, G.; Wu, F.; Petersen, K.D.; Thougaard, A.; Sehested, M.; Lichenstein, H.S.; Jeffers, M. Activity of PXD101, a Histone Deacetylase Inhibitor, in Preclinical Ovarian Cancer Studies. Mol. Cancer. Ther. 2006, 5, 2086–2095. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.T.; Balch, C.; Kulp, S.K.; Mand, M.R.; Nephew, K.P.; Chen, C.S. A Rationally Designed Histone Deacetylase Inhibitor with Distinct Antitumor Activity Against Ovarian Cancer. Neoplasia 2009, 11, 552–563, 3 p following 563. [Google Scholar] [CrossRef] [PubMed]
- Halsall, J.A.; Turan, N.; Wiersma, M.; Turner, B.M. Cells Adapt to the Epigenomic Disruption Caused by Histone Deacetylase Inhibitors through a Coordinated, Chromatin-Mediated Transcriptional Response. Epigenet. Chromatin 2015, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Adler, J.T.; Hottinger, D.G.; Kunnimalaiyaan, M.; Chen, H. Combination Therapy with Histone Deacetylase Inhibitors and Lithium Chloride: A Novel Treatment for Carcinoid Tumors. Ann. Surg. Oncol. 2009, 16, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Halsall, J.A.; Turner, B.M. Histone Deacetylase Inhibitors for Cancer Therapy: An Evolutionarily Ancient Resistance Response May Explain Their Limited Success. Bioessays 2016, 38, 1102–1110. [Google Scholar] [CrossRef] [PubMed]
- Meseure, D.; Drak Alsibai, K.; Nicolas, A.; Bieche, I.; Morillon, A. Long Noncoding RNAs as New Architects in Cancer Epigenetics, Prognostic Biomarkers, and Potential Therapeutic Targets. BioMed Res. Int. 2015, 2015, 320214. [Google Scholar] [CrossRef] [PubMed]
- Labiche, A.; Heutte, N.; Herlin, P.; Chasle, J.; Gauduchon, P.; Elie, N. Stromal Compartment as a Survival Prognostic Factor in Advanced Ovarian Carcinoma. Int. J. Gynecol. Cancer 2010, 20, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Steele, N.; Finn, P.; Brown, R.; Plumb, J. Combined Inhibition of DNA Methylation and Histone Acetylation Enhances Gene Re-Expression and Drug Sensitivity in Vivo. Br. J. Cancer 2009, 100, 758–763. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, A.; Nolan, L.; Crabb, S.; Packham, G. Epigenetic Therapy: Histone Acetylation, DNA Methylation and Anti-Cancer Drug Discovery. Curr. Cancer Drug Targets 2009, 9, 963–981. [Google Scholar] [CrossRef] [PubMed]
- Yeung, T.; Leung, C.S.; Mok, S.C. CAF Reprogramming Inhibits Ovarian Cancer Progression. Cell Cycle 2014, 13, 3783–3784. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Wu, S.; Zhang, K.; Xu, T. A Comprehensive Overview of Exosomes in Ovarian Cancer: Emerging Biomarkers and Therapeutic Strategies. J. Ovar. Res. 2017, 10, 73. [Google Scholar] [CrossRef] [PubMed]
- Chiriva-Internati, M.; Mirandola, L.; Kast, W.M.; Jenkins, M.R.; Cobos, E.; Cannon, M.J. Understanding the Cross-Talk between Ovarian Tumors and Immune Cells: Mechanisms for Effective Immunotherapies. Int. Rev. Immunol. 2011, 30, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Chae, C.; Teran-Cabanillas, E.; Cubillos-Ruiz, J.R. Dendritic Cell Rehab: New Strategies to Unleash Therapeutic Immunity in Ovarian Cancer. Cancer Immunol. Immunother. 2017, 66, 969–977. [Google Scholar] [CrossRef] [PubMed]
- Drakes, M.L.; Stiff, P.J. Understanding Dendritic Cell Immunotherapy in Ovarian Cancer. Expert Rev. Anticancer Ther. 2016, 16, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Wefers, C.; Lambert, L.J.; Torensma, R.; Hato, S.V. Cellular Immunotherapy in Ovarian Cancer: Targeting the Stem of Recurrence. Gynecol. Oncol. 2015, 137, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Odunsi, K. Immunotherapy in Ovarian Cancer. Ann. Oncol. 2017, 28, viii1–viii7. [Google Scholar] [CrossRef] [PubMed]
- Barrero, M.J. Epigenetic Strategies to Boost Cancer Immunotherapies. Int. J. Mol. Sci. 2017, 18, 1108. [Google Scholar] [CrossRef] [PubMed]
- Terranova-Barberio, M.; Thomas, S.; Munster, P.N. Epigenetic Modifiers in Immunotherapy: A Focus on Checkpoint Inhibitors. Immunotherapy 2016, 8, 705–719. [Google Scholar] [CrossRef] [PubMed]
- Strick, R.; Strissel, P.L.; Baylin, S.B.; Chiappinelli, K.B. Unraveling the Molecular Pathways of DNA-Methylation Inhibitors: Human Endogenous Retroviruses Induce the Innate Immune Response in Tumors. Oncoimmunology 2016, 5, e1122160. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Ye, Q.; Santoro, S.; Fang, C.; Best, A.; Powell, D.J., Jr. Chimeric NKG2D CAR-Expressing T Cell-Mediated Attack of Human Ovarian Cancer is Enhanced by Histone Deacetylase Inhibition. Hum. Gene Ther. 2013, 24, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Wargo, J.A.; Robbins, P.F.; Li, Y.; Zhao, Y.; El-Gamil, M.; Caragacianu, D.; Zheng, Z.; Hong, J.A.; Downey, S.; Schrump, D.S. Recognition of NY-ESO-1 Tumor Cells by Engineered Lymphocytes is Enhanced by Improved Vector Design and Epigenetic Modulation of Tumor Antigen Expression. Cancer Immunol. Immunother. 2009, 58, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Scanlan, M.J.; Gure, A.O.; Jungbluth, A.A.; Old, L.J.; Chen, Y. Cancer/Testis Antigens: An Expanding Family of Targets for Cancer Immunotherapy. Immunol. Rev. 2002, 188, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Odunsi, K.; Matsuzaki, J.; James, S.R.; Mhawech-Fauceglia, P.; Tsuji, T.; Miller, A.; Zhang, W.; Akers, S.N.; Griffiths, E.A.; Miliotto, A.; et al. Epigenetic Potentiation of NY-ESO-1 Vaccine Therapy in Human Ovarian Cancer. Cancer. Immunol. Res. 2014, 2, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Quiros, J.; Mahuron, K.; Pai, C.; Ranzani, V.; Young, A.; Silveria, S.; Harwin, T.; Abnousian, A.; Pagani, M. Targeting EZH2 Reprograms Intratumoral Regulatory T Cells to Enhance Cancer Immunity. Cell Rep. 2018, 23, 3262–3274. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).