KLF10 as a Tumor Suppressor Gene and Its TGF-β Signaling
Abstract
1. SP/KLF Family
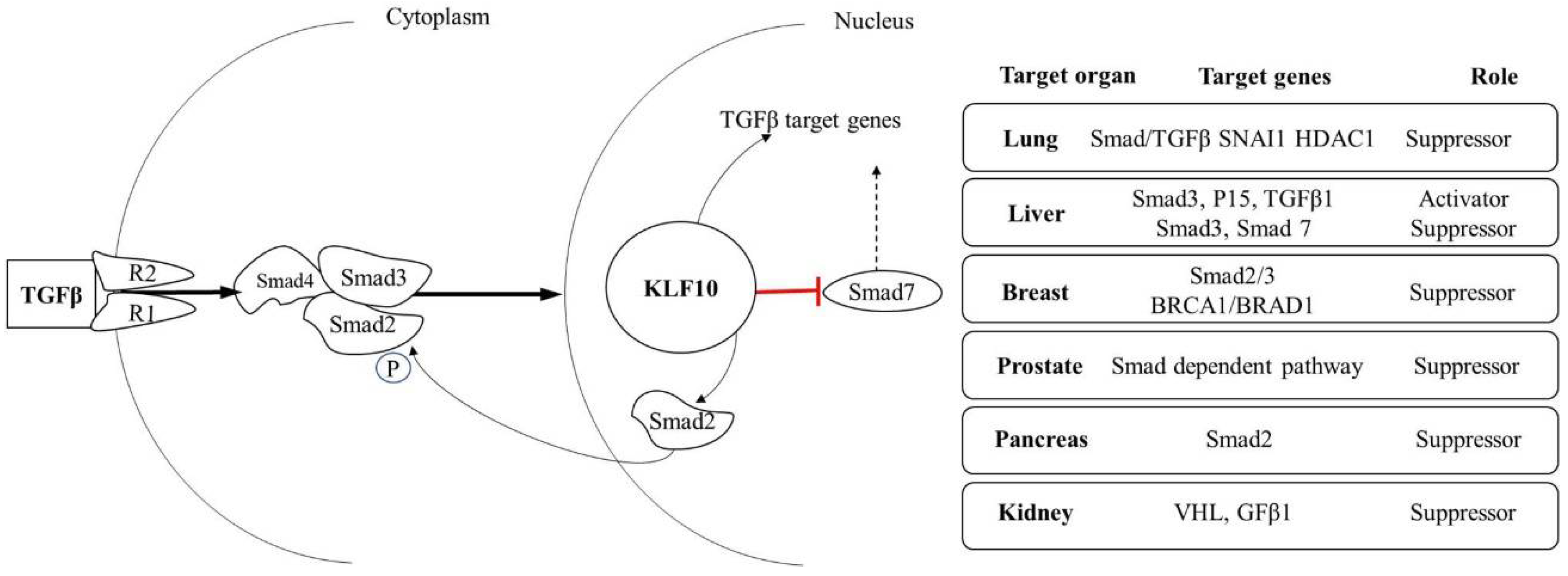
2. KLF10 Induced Mechanism of Gene Activation
3. KLF10 Role in Various Diseases
3.1. Diabetes
3.2. Bone Disease
3.3. Heart Hypertrophy
3.4. Other Diseases
3.5. Phenotype in KLF10 Deficient Models
4. Role of KLF10 as a Tumor Suppressor in Various Cancers
4.1. Liver Cancer
4.2. Pancreatic Cancer
4.3. Lung Cancer
4.4. Breast Cancer
4.5. Colon Cancer
4.6. Human Prostate Cancer
4.7. Metastatic Brain Tumors
4.8. Renal Cancer
4.9. Other Cancers
5. Concluding Remarks
Acknowledgments
Conflicts of Interest
References
- Turner, J.; Crossley, M. Mammalian Kruppel-like transcription factors: More than just a pretty finger. Trends Biochem. Sci. 1999, 24, 236–240. [Google Scholar] [CrossRef]
- Dynan, W.S.; Tjian, R. The promoter-specific transcription factor sp1 binds to upstream sequences in the SV40 early promoter. Cell 1983, 35, 79–87. [Google Scholar] [CrossRef]
- Pei, J.; Grishin, N.V. A new family of predicted Kruppel-like factor genes and pseudogenes in placental mammals. PLoS ONE 2013, 8, e81109. [Google Scholar] [CrossRef] [PubMed]
- Kaczynski, J.; Cook, T.; Urrutia, R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003, 4, 206. [Google Scholar] [CrossRef] [PubMed]
- Pearson, R.; Fleetwood, J.; Eaton, S.; Crossley, M.; Bao, S. Kruppel-like transcription factors: A functional family. Int. J. Biochem. Cell Biol. 2008, 40, 1996–2001. [Google Scholar] [CrossRef] [PubMed]
- Miller, I.J.; Bieker, J.J. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol. Cell. Biol. 1993, 13, 2776–2786. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.M.; Houzelstein, D.; Dunwoodie, S.L.; Beddington, R.S. Sp5, a new member of the Sp1 family, is dynamically expressed during development and genetically interacts with Brachyury. Dev. Biol. 2000, 227, 358–372. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, M.; Harris, S.A.; Oursler, M.J.; Rasmussen, K.; Riggs, B.L.; Spelsberg, T.C. Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res. 1995, 23, 4907–4912. [Google Scholar] [CrossRef] [PubMed]
- Cook, T.; Gebelein, B.; Mesa, K.; Mladek, A.; Urrutia, R. Molecular cloning and characterization of TIEG2 reveals a new subfamily of transforming growth factor-beta-inducible sp1-like zinc finger-encoding genes involved in the regulation of cell growth. J. Biol. Chem. 1998, 273, 25929–25936. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.P.; Kern, C.B.; Crable, S.C.; Lingrel, J.B. Isolation of a gene encoding a functional zinc finger protein homologous to erythroid Kruppel-like factor: Identification of a new multigene family. Mol. Cell. Biol. 1995, 15, 5957–5965. [Google Scholar] [CrossRef] [PubMed]
- Crossley, M.; Whitelaw, E.; Perkins, A.; Williams, G.; Fujiwara, Y.; Orkin, S.H. Isolation and characterization of the CDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol. Cell. Biol. 1996, 16, 1695–1705. [Google Scholar] [CrossRef] [PubMed]
- Shields, J.M.; Christy, R.J.; Yang, V.W. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J. Biol. Chem. 1996, 271, 20009–20017. [Google Scholar] [CrossRef] [PubMed]
- Imataka, H.; Sogawa, K.; Yasumoto, K.; Kikuchi, Y.; Sasano, K.; Kobayashi, A.; Hayami, M.; Fujii-Kuriyama, Y. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J. 1992, 11, 3663–3671. [Google Scholar] [PubMed]
- Conkright, M.D.; Wani, M.A.; Anderson, K.P.; Lingrel, J.B. A gene encoding an intestinal-enriched member of the Kruppel-like factor family expressed in intestinal epithelial cells. Nucleic Acids Res. 1999, 27, 1263–1270. [Google Scholar] [CrossRef] [PubMed]
- Suske, G.; Bruford, E.; Philipsen, S. Mammalian SP/KLF transcription factors: Bring in the family. Genomics 2005, 85, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Burns, K.H.; Ma, L.; Matzuk, M.M. Identification of Zfp393, a germ cell-specific gene encoding a novel zinc finger protein. Mech. Dev. 2002, 118, 233–239. [Google Scholar] [CrossRef]
- McConnell, B.B.; Yang, V.W. Mammalian Kruppel-like factors in health and diseases. Physiol. Rev. 2010, 90, 1337–1381. [Google Scholar] [CrossRef] [PubMed]
- White, J.A.; McAlpine, P.J.; Antonarakis, S.; Cann, H.; Eppig, J.T.; Frazer, K.; Frezal, J.; Lancet, D.; Nahmias, J.; Pearson, P.; et al. Guidelines for human gene nomenclature (1997). Hugo nomenclature committee. Genomics 1997, 45, 468–471. [Google Scholar] [CrossRef] [PubMed]
- Black, A.R.; Black, J.D.; Azizkhan-Clifford, J. Sp1 and Kruppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell. Physiol. 2001, 188, 143–160. [Google Scholar] [CrossRef] [PubMed]
- Presnell, J.S.; Schnitzler, C.E.; Browne, W.E. KLF/SP transcription factor family evolution: Expansion, diversification, and innovation in eukaryotes. Genome Biol. Evol. 2015, 7, 2289–2309. [Google Scholar] [CrossRef] [PubMed]
- Pollak, N.M.; Hoffman, M.; Goldberg, I.J.; Drosatos, K. Krüppel-like factors: Crippling and uncrippling metabolic pathways. JACC Basic Transl. Sci. 2018, 3, 132–156. [Google Scholar] [CrossRef]
- Cook, T.; Gebelein, B.; Urrutia, R. Sp1 and its likes: Biochemical and functional predictions for a growing family of zinc finger transcription factors. Ann. N. Y. Acad. Sci. 1999, 880, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Dang, D.T.; Pevsner, J.; Yang, V.W. The biology of the mammalian Kruppel-like family of transcription factors. Int. J. Biochem. Cell Biol. 2000, 32, 1103–1121. [Google Scholar] [CrossRef]
- Philipsen, S.; Suske, G. A tale of three fingers: The family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999, 27, 2991–3000. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.; Crossley, M. Basic Kruppel-like factor functions within a network of interacting haematopoietic transcription factors. Int. J. Biochem. Cell Biol. 1999, 31, 1169–1174. [Google Scholar] [CrossRef]
- Schmierer, B.; Hill, C.S. TGFbeta-SMAD signal transduction: Molecular specificity and functional flexibility. Nat. Rev. Mol. Cell Biol. 2007, 8, 970–982. [Google Scholar] [CrossRef] [PubMed]
- Wrighton, K.H.; Lin, X.; Feng, X.H. Phospho-control of TGF-beta superfamily signaling. Cell Res. 2009, 19, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Goumans, M.J.; Valdimarsdottir, G.; Itoh, S.; Lebrin, F.; Larsson, J.; Mummery, C.; Karlsson, S.; ten Dijke, P. Activin receptor-like kinase (ALK)1 is an antagonistic mediator of lateral TGFbeta/ALK5 signaling. Mol. Cell 2003, 12, 817–828. [Google Scholar] [CrossRef]
- Schiffer, M.; Von Gersdorff, G.; Bitzer, M.; Susztak, K.; Böttinger, E.P. SMAD proteins and transforming growth factor-β signaling. Kidney Int. 2000, 58, S45–S52. [Google Scholar] [CrossRef]
- Tachibana, I.; Imoto, M.; Adjei, P.N.; Gores, G.J.; Subramaniam, M.; Spelsberg, T.C.; Urrutia, R. Overexpression of the TGFbeta-regulated zinc finger encoding gene, TIEG, induces apoptosis in pancreatic epithelial cells. J. Clin. Investig. 1997, 99, 2365–2374. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Di, G.; Li, J.; Chen, Y.; Li, W.; Wu, J.; Cheng, T.; Yao, M.; Shao, Z. TIEG1 induces apoptosis through mitochondrial apoptotic pathway and promotes apoptosis induced by homoharringtonine and velcade. FEBS Lett. 2007, 581, 3826–3832. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.; Bronk, S.F.; Roberts, P.J.; Urrutia, R.; Gores, G.J. The transforming growth factor beta(1)-inducible transcription factor tieg1, mediates apoptosis through oxidative stress. Hepatology 1999, 30, 1490–1497. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, S.A.; Subramaniam, M.; Janknecht, R.; Spelsberg, T.C. TGFbeta inducible early gene enhances TGFbeta/SMAD-dependent transcriptional responses. Oncogene 2002, 21, 5783–5790. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, M.; Hawse, J.R.; Johnsen, S.A.; Spelsberg, T.C. Role of TIEG1 in biological processes and disease states. J. Cell. Biochem. 2007, 102, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Hawse, J.R.; Cicek, M.; Grygo, S.B.; Bruinsma, E.S.; Rajamannan, N.M.; van Wijnen, A.J.; Lian, J.B.; Stein, G.S.; Oursler, M.J.; Subramaniam, M.; et al. TIEG1/KLF10 modulates Runx2 expression and activity in osteoblasts. PLoS ONE 2011, 6, e19429. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Aguilar, R.; Benmezroua, Y.; Balkau, B.; Marre, M.; Helbecque, N.; Charpentier, G.; Polychronakos, C.; Sladek, R.; Froguel, P.; Neve, B. Minor contribution of SMAD7 and KLF10 variants to genetic susceptibility of type 2 diabetes. Diabetes Metab. 2007, 33, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Rajamannan, N.M.; Subramaniam, M.; Abraham, T.P.; Vasile, V.C.; Ackerman, M.J.; Monroe, D.G.; Chew, T.L.; Spelsberg, T.C. TGFbeta inducible early gene-1 (TIEG1) and cardiac hypertrophy: Discovery and characterization of a novel signaling pathway. J. Cell. Biochem. 2007, 100, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Wara, A.K.; Icli, B.; Sun, X.; Packard, R.R.; Esen, F.; Stapleton, C.J.; Subramaniam, M.; Kretschmer, K.; Apostolou, I.; et al. Kruppel-like factor KLF10 targets transforming growth factor-beta1 to regulate CD4(+)CD25(−) T cells and T regulatory cells. J. Biol. Chem. 2009, 284, 24914–24924. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Ding, J.; Marcoux, Y.; Iwashina, T.; Sakurai, H.; Tredget, E.E. Impaired cutaneous wound healing in transforming growth factor-beta inducible early gene1 knockout mice. Wound Repair Regen. 2012, 20, 166–177. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Lee, K.S.; Chang, H.Y.; Lee, W.K.; Lee, J.I. Progression of diet induced nonalcoholic steatohepatitis is accompanied by increased expression of Kruppel-like-factor 10 in mice. J. Transl. Med. 2014, 12, 186. [Google Scholar] [CrossRef] [PubMed]
- Papadakis, K.A.; Krempski, J.; Svingen, P.; Xiong, Y.; Sarmento, O.F.; Lomberk, G.A.; Urrutia, R.A.; Faubion, W.A. Kruppel-like factor KLF10 deficiency predisposes to colitis through colonic macrophage dysregulation. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 309, G900–G909. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Chen, J.; Zhang, H.; Wang, X.; Yao, H.; Peng, Y.; Zhang, W. LncRNA OIP5-AS1 loss-induced microRNA-410 accumulation regulates cell proliferation and apoptosis by targeting KLF10 via activating PTEN/PI3K/AKT pathway in multiple myeloma. Cell Death Dis. 2017, 8, e2975. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, K.; Takeda, J.; Horikawa, Y. Kruppel-like factor-10 is directly regulated by carbohydrate response element-binding protein in rat primary hepatocytes. Biochem. Biophys. Res. Commun. 2011, 412, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Guillaumond, F.; Grechez-Cassiau, A.; Subramaniam, M.; Brangolo, S.; Peteri-Brunback, B.; Staels, B.; Fievet, C.; Spelsberg, T.C.; Delaunay, F.; Teboul, M. Kruppel-like factor kLF10 is a link between the circadian clock and metabolism in liver. Mol. Cell. Biol. 2010, 30, 3059–3070. [Google Scholar] [CrossRef] [PubMed]
- Song, C.Z.; Gavriilidis, G.; Asano, H.; Stamatoyannopoulos, G. Functional study of transcription factor KLF11 by targeted gene inactivation. Blood Cells Mol. Dis. 2005, 34, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, M.; Gorny, G.; Johnsen, S.A.; Monroe, D.G.; Evans, G.L.; Fraser, D.G.; Rickard, D.J.; Rasmussen, K.; van Deursen, J.M.; Turner, R.T.; et al. TIEG1 null mouse-derived osteoblasts are defective in mineralization and in support of osteoclast differentiation in vitro. Mol. Cell. Biol. 2005, 25, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Zitman-Gal, T.; Green, J.; Pasmanik-Chor, M.; Golan, E.; Bernheim, J.; Benchetrit, S. Vitamin D manipulates miR-181c, miR-20b and miR-15a in human umbilical vein endothelial cells exposed to a diabetic-like environment. Cardiovasc. Diabetol. 2014, 13, 8. [Google Scholar] [CrossRef] [PubMed]
- Janssens, K.; ten Dijke, P.; Janssens, S.; Van Hul, W. Transforming growth factor-beta1 to the bone. Endocr. Rev. 2005, 26, 743–774. [Google Scholar] [CrossRef] [PubMed]
- Prashar, P.; Yadav, P.S.; Samarjeet, F.; Bandyopadhyay, A. Microarray meta-analysis identifies evolutionarily conserved bmp signaling targets in developing long bones. Dev. Biol. 2014, 389, 192–207. [Google Scholar] [CrossRef] [PubMed]
- Cicek, M.; Vrabel, A.; Sturchio, C.; Pederson, L.; Hawse, J.R.; Subramaniam, M.; Spelsberg, T.C.; Oursler, M.J. TGF-beta inducible early gene 1 regulates osteoclast differentiation and survival by mediating the NFATc1, Akt, and MEK/ERK signaling pathways. PLoS ONE 2011, 6, e17522. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Shen, P.; Zeng, S.; Liu, P. TIEG1 inhibits angiotensin II-induced cardiomyocyte hypertrophy by inhibiting transcription factor GATA4. J. Cardiovasc. Pharmacol. 2015, 66, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Heaney, A.P.; Horwitz, G.A.; Wang, Z.; Singson, R.; Melmed, S. Early involvement of estrogen-induced pituitary tumor transforming gene and fibroblast growth factor expression in prolactinoma pathogenesis. Nat. Med. 1999, 5, 1317–1321. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Svingen, P.A.; Sarmento, O.O.; Smyrk, T.C.; Dave, M.; Khanna, S.; Lomberk, G.A.; Urrutia, R.A.; Faubion, W.A., Jr. Differential coupling of kLF10 to Sin3-HDAC and PCAF regulates the inducibility of the FOXP3 gene. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014, 307, R608–R620. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Khanna, S.; Grzenda, A.L.; Sarmento, O.F.; Svingen, P.A.; Lomberk, G.A.; Urrutia, R.A.; Faubion, W.A., Jr. Polycomb antagonizes p300/CREB-binding protein-associated factor to silence FOXP3 in a Kruppel-like factor-dependent manner. J. Biol. Chem. 2012, 287, 34372–34385. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Chen, Q.; Sun, L.; Zhang, H.; Yao, L.; Cui, X.; Gao, Y.; Fang, F.; Chang, Y. KLF10 transcription factor regulates hepatic glucose metabolism in mice. Diabetologia 2017, 60, 2443–2452. [Google Scholar] [CrossRef] [PubMed]
- Koczulla, A.R.; Jonigk, D.; Wolf, T.; Herr, C.; Noeske, S.; Klepetko, W.; Vogelmeier, C.; von Neuhoff, N.; Rische, J.; Wrenger, S.; et al. Kruppel-like zinc finger proteins in end-stage COPD lungs with and without severe alpha1-antitrypsin deficiency. Orphanet J. Rare Dis. 2012, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Tsubone, T.; Moran, S.L.; Subramaniam, M.; Amadio, P.C.; Spelsberg, T.C.; An, K.N. Effect of TGF-beta inducible early gene deficiency on flexor tendon healing. J. Orthop. Res. 2006, 24, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Koltsova, S.V.; Shilov, B.; Birulina, J.G.; Akimova, O.A.; Haloui, M.; Kapilevich, L.V.; Gusakova, S.V.; Tremblay, J.; Hamet, P.; Orlov, S.N. Transcriptomic changes triggered by hypoxia: Evidence for HIF-1alpha-independent, [Na+]i/[K+]i-mediated, excitation-transcription coupling. PLoS ONE 2014, 9, e110597. [Google Scholar] [CrossRef] [PubMed]
- Bensamoun, S.F.; Tsubone, T.; Subramaniam, M.; Hawse, J.R.; Boumediene, E.; Spelsberg, T.C.; An, K.N.; Amadio, P.C. Age-dependent changes in the mechanical properties of tail tendons in TGF-beta inducible early gene-1 knockout mice. J. Appl. Physiol. 2006, 101, 1419–1424. [Google Scholar] [CrossRef] [PubMed]
- Bensamoun, S.F.; Hawse, J.R.; Subramaniam, M.; Ilharreborde, B.; Bassillais, A.; Benhamou, C.L.; Fraser, D.G.; Oursler, M.J.; Amadio, P.C.; An, K.N.; et al. TGFbeta inducible early gene-1 knockout mice display defects in bone strength and microarchitecture. Bone 2006, 39, 1244–1251. [Google Scholar] [CrossRef] [PubMed]
- Haddad, O.; Hawse, J.R.; Subramaniam, M.; Spelsberg, T.C.; Bensamoun, S.F. TIEG1-null osteocytes display defects in their morphology, density and surrounding bone matrix. J. Musculoskelet. Res. 2009, 12, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Gumez, L.; Bensamoun, S.F.; Doucet, J.; Haddad, O.; Hawse, J.R.; Subramaniam, M.; Spelsberg, T.C.; Pichon, C. Molecular structure of tail tendon fibers in TIEG1 knockout mice using synchrotron diffraction technology. J. Appl. Physiol. 2010, 108, 1706–1710. [Google Scholar] [CrossRef] [PubMed]
- Hawse, J.R.; Iwaniec, U.T.; Bensamoun, S.F.; Monroe, D.G.; Peters, K.D.; Ilharreborde, B.; Rajamannan, N.M.; Oursler, M.J.; Turner, R.T.; Spelsberg, T.C.; et al. TIEG-null mice display an osteopenic gender-specific phenotype. Bone 2008, 42, 1025–1031. [Google Scholar] [CrossRef] [PubMed]
- Hawse, J.R.; Pitel, K.S.; Cicek, M.; Philbrick, K.A.; Gingery, A.; Peters, K.D.; Syed, F.A.; Ingle, J.N.; Suman, V.J.; Iwaniec, U.T.; et al. TGFbeta inducible early gene-1 plays an important role in mediating estrogen signaling in the skeleton. J. Bone Miner. Res. 2014, 29, 1206–1216. [Google Scholar] [CrossRef] [PubMed]
- Kammoun, M.; Meme, S.; Meme, W.; Subramaniam, M.; Hawse, J.R.; Canon, F.; Bensamoun, S.F. Impact of tieg1 on the structural properties of fast- and slow-twitch skeletal muscle. Muscle Nerve 2017, 55, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Sartorelli, V.; Huang, J.; Hamamori, Y.; Kedes, L. Molecular mechanisms of myogenic coactivation by p300: Direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol. Cell. Biol. 1997, 17, 1010–1026. [Google Scholar] [CrossRef] [PubMed]
- Wara, A.K.; Foo, S.; Croce, K.; Sun, X.; Icli, B.; Tesmenitsky, Y.; Esen, F.; Lee, J.S.; Subramaniam, M.; Spelsberg, T.C.; et al. TGF-beta1 signaling and Kruppel-like factor 10 regulate bone marrow-derived proangiogenic cell differentiation, function, and neovascularization. Blood 2011, 118, 6450–6460. [Google Scholar] [CrossRef] [PubMed]
- Wara, A.K.; Manica, A.; Marchini, J.F.; Sun, X.; Icli, B.; Tesmenitsky, Y.; Croce, K.; Feinberg, M.W. Bone marrow-derived Kruppel-like factor 10 controls reendothelialization in response to arterial injury. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1552–1560. [Google Scholar] [CrossRef] [PubMed]
- Hefferan, T.E.; Reinholz, G.G.; Rickard, D.J.; Johnsen, S.A.; Waters, K.M.; Subramaniam, M.; Spelsberg, T.C. Overexpression of a nuclear protein, TIEG, mimics transforming growth factor-beta action in human osteoblast cells. J. Biol. Chem. 2000, 275, 20255–20259. [Google Scholar] [CrossRef] [PubMed]
- Engelmann, D.; Knoll, S.; Ewerth, D.; Steder, M.; Stoll, A.; Putzer, B.M. Functional interplay between E2F1 and chemotherapeutic drugs defines immediate E2F1 target genes crucial for cancer cell death. Cell. Mol. Life Sci. 2010, 67, 931–948. [Google Scholar] [CrossRef] [PubMed]
- Partin, J.V.; Anglin, I.E.; Kyprianou, N. Quinazoline-based alpha 1-adrenoceptor antagonists induce prostate cancer cell apoptosis via TGF-beta signalling and I kappa B alpha induction. Br. J. Cancer 2003, 88, 1615–1621. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.G.; Xu, H.; Lee, J.F.; Subramaniam, M.; Leung, K.L.; Wang, S.H.; Chan, U.P.; Spelsberg, T.C. 15-hydroxy-eicosatetraenoic acid arrests growth of colorectal cancer cells via a peroxisome proliferator-activated receptor gamma-dependent pathway. Int. J. Cancer 2003, 107, 837–843. [Google Scholar] [CrossRef] [PubMed]
- Reinholz, M.M.; An, M.W.; Johnsen, S.A.; Subramaniam, M.; Suman, V.J.; Ingle, J.N.; Roche, P.C.; Spelsberg, T.C. Differential gene expression of TGF beta inducible early gene (TIEG), SMAD7, SMAD2 and Bard1 in normal and malignant breast tissue. Breast Cancer Res. Treat. 2004, 86, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Sebestyen, A.; Barna, G.; Nagy, K.; Janosi, J.; Paku, S.; Kohut, E.; Berczi, L.; Mihalik, R.; Kopper, L. SMAD signal and TGFbeta induced apoptosis in human lymphoma cells. Cytokine 2005, 30, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Zohrabian, V.M.; Nandu, H.; Gulati, N.; Khitrov, G.; Zhao, C.; Mohan, A.; Demattia, J.; Braun, A.; Das, K.; Murali, R.; et al. Gene expression profiling of metastatic brain cancer. Oncol. Rep. 2007, 18, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, S.V.; Ivanova, A.V.; Salnikow, K.; Timofeeva, O.; Subramaniam, M.; Lerman, M.I. Two novel VHL targets, TGFBI (BIGH3) and its transactivator KLF10, are up-regulated in renal clear cell carcinoma and other tumors. Biochem. Biophys. Res. Commun. 2008, 370, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, F.; Lin, F.; Gao, S.M.; Tan, Y.; Han, Y.; Chen, C.; Wu, J. Lentivirus-mediated overexpression of TGF-beta inducible early gene 1 inhibits SW1990 pancreatic cancer cell growth. Cell Biol. Int. 2011, 35, 891–896. [Google Scholar] [CrossRef] [PubMed]
- Antonello, D.; Moore, P.S.; Zamboni, G.; Falconi, M.; Scarpa, A. Absence of mutations in the transforming growth factor-beta inducible early gene 1, TIEG1, in pancreatic cancer. Cancer Lett. 2002, 183, 179–183. [Google Scholar] [CrossRef]
- Jiang, L.; Lai, Y.K.; Zhang, J.F.; Chan, C.Y.; Lu, G.; Lin, M.C.; He, M.L.; Li, J.C.; Kung, H.F. Transactivation of the TIEG1 confers growth inhibition of transforming growth factor-beta-susceptible hepatocellular carcinoma cells. World J. Gastroenterol. 2012, 18, 2035–2042. [Google Scholar] [CrossRef] [PubMed]
- Heo, S.H.; Jeong, E.S.; Lee, K.S.; Seo, J.H.; Lee, W.K.; Choi, Y.K. Kruppel-like factor 10 null mice exhibit lower tumor incidence and suppressed cellular proliferation activity following chemically induced liver tumorigenesis. Oncol. Rep. 2015, 33, 2037–2044. [Google Scholar] [CrossRef] [PubMed]
- Jim, H.S.; Lin, H.Y.; Tyrer, J.P.; Lawrenson, K.; Dennis, J.; Chornokur, G.; Chen, Z.; Chen, A.Y.; Permuth-Wey, J.; Aben, K.K.; et al. Common genetic variation in circadian rhythm genes and risk of Epithelial Ovarian Cancer (EOC). J. Genet. Genome Res. 2015, 2, 17. [Google Scholar] [CrossRef]
- Mishra, V.K.; Subramaniam, M.; Kari, V.; Pitel, K.S.; Baumgart, S.J.; Naylor, R.M.; Nagarajan, S.; Wegwitz, F.; Ellenrieder, V.; Hawse, J.R.; et al. Kruppel-like transcription factor KLF10 suppresses TGFbeta-induced epithelial-to-mesenchymal transition via a negative feedback mechanism. Cancer Res. 2017, 77, 2387–2400. [Google Scholar] [CrossRef] [PubMed]
- Song, K.D.; Kim, D.J.; Lee, J.E.; Yun, C.H.; Lee, W.K. KLF10, transforming growth factor-beta-inducible early gene 1, acts as a tumor suppressor. Biochem. Biophys. Res. Commun. 2012, 419, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Mouzaki, M.; You, H.; Laird, J.C.; Mato, J.; Lu, S.C.; Rountree, C.B. CD133+ liver cancer stem cells from methionine adenosyl transferase 1A-deficient mice demonstrate resistance to transforming growth factor (TGF)-beta-induced apoptosis. Hepatology 2009, 49, 1277–1286. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Kitisin, K.; Jogunoori, W.; Li, C.; Deng, C.X.; Mueller, S.C.; Ressom, H.W.; Rashid, A.; He, A.R.; Mendelson, J.S.; et al. Progenitor/stem cells give rise to liver cancer due to aberrant TGF-beta and IL-6 signaling. Proc. Natl. Acad. Sci. USA 2008, 105, 2445–2450. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, A.; Kurita, M.; Oshima, K.; Osada, S.; Nishihara, T.; Imagawa, M. Functional analysis of zinc finger proteins that bind to the silencer element in the glutathione transferase P gene. Biol. Pharm. Bull. 2002, 25, 970–974. [Google Scholar] [CrossRef] [PubMed]
- Chang, V.H.; Chu, P.Y.; Peng, S.L.; Mao, T.L.; Shan, Y.S.; Hsu, C.F.; Lin, C.Y.; Tsai, K.K.; Yu, W.C.; Ch’ang, H.J. Kruppel-like factor 10 expression as a prognostic indicator for pancreatic adenocarcinoma. Am. J. Pathol. 2012, 181, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.J.; Wu, W.C.; Chang, H.W.; Lai, Y.T.; Lin, C.H.; Yu, W.C.; Chang, V.H. KLF10 affects pancreatic function via the SEI-1/p21Cip1 pathway. Int. J. Biochem. Cell Biol. 2015, 60, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Chen, Y.; Chan, C.Y.; Wang, X.; Lin, L.; He, M.L.; Lin, M.C.; Yew, D.T.; Sung, J.J.; Li, J.C.; et al. Down-regulation of stathmin is required for TGF-beta inducible early gene 1 induced growth inhibition of pancreatic cancer cells. Cancer Lett. 2009, 274, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Mizumoto, K.; Maehara, N.; Shono, M.; Nagai, E.; Tanaka, M. Instability of chromosome 8 as an indicator of aggressive tumor phenotype in pancreatic cancer. J. Surg. Oncol. 2001, 76, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Venkov, C.; Plieth, D.; Ni, T.; Karmaker, A.; Bian, A.; George, A.L., Jr.; Neilson, E.G. Transcriptional networks in epithelial-mesenchymal transition. PLoS ONE 2011, 6, e25354. [Google Scholar] [CrossRef] [PubMed]
- Hung, M.S.; Chen, I.C.; You, L.; Jablons, D.M.; Li, Y.C.; Mao, J.H.; Xu, Z.; Lung, J.H.; Yang, C.T.; Liu, S.T. Knockdown of cullin 4A inhibits growth and increases chemosensitivity in lung cancer cells. J. Cell. Mol. Med. 2016, 20, 1295–1306. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Chen, B.B.; Li, J.Y.; Zhu, H.; Huang, M.; Gu, S.M.; Wang, Q.Q.; Chen, J.Y.; Yu, S.; Wu, J.; et al. TIEG1 inhibits breast cancer invasion and metastasis by inhibition of epidermal growth factor receptor (EGFR) transcription and the EGFR signaling pathway. Mol. Cell. Biol. 2012, 32, 50–63. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.F.; Sui, C.L.; Wu, W.C.; Wang, J.J.; Yang, D.H.; Chen, Y.C.; Yu, W.C.; Chang, H.S. KLF10 induces cell apoptosis through modulation of BI-1 expression and Ca2+ homeostasis in estrogen-responding adenocarcinoma cells. Int. J. Biochem. Cell Biol. 2011, 43, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Nagy, L.; Tontonoz, P.; Alvarez, J.G.; Chen, H.; Evans, R.M. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARgamma. Cell 1998, 93, 229–240. [Google Scholar] [CrossRef]
- Eid, M.A.; Kumar, M.V.; Iczkowski, K.A.; Bostwick, D.G.; Tindall, D.J. Expression of early growth response genes in human prostate cancer. Cancer Res. 1998, 58, 2461–2468. [Google Scholar] [PubMed]
- Yono, M.; Mane, S.M.; Lin, A.; Weiss, R.M.; Latifpour, J. Differential effects of diabetes induced by streptozotocin and that develops spontaneously on prostate growth in Bio Breeding (BB) rats. Life Sci. 2008, 83, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Barna, G.; Sebestyen, A.; Chinopoulos, C.C.; Nagy, K.; Mihalik, R.; Paku, S.; Kopper, L. TGF beta 1 kills lymphoma cells using mitochondrial apoptotic pathway with the help of caspase-8. Anticancer Res. 2002, 22, 3867–3872. [Google Scholar] [PubMed]
- Zhou, F.L.; Zhang, W.G.; Chen, G.; Zhao, W.H.; Cao, X.M.; Chen, Y.X.; Tian, W.; Liu, J.; Liu, S.H. Serological identification and bioinformatics analysis of immunogenic antigens in multiple myeloma. Cancer Immunol. Immunother. 2006, 55, 910–917. [Google Scholar] [CrossRef] [PubMed]
- Noti, J.D.; Johnson, A.K.; Dillon, J.D. The zinc finger transcription factor transforming growth factor beta-inducible early gene-1 confers myeloid-specific activation of the leukocyte integrin CD11d promoter. J. Biol. Chem. 2004, 279, 26948–26958. [Google Scholar] [CrossRef] [PubMed]
- Noti, J.D.; Johnson, A.K.; Dillon, J.D. The leukocyte integrin gene CD11d is repressed by gut-enriched Kruppel-like factor 4 in myeloid cells. J. Biol. Chem. 2005, 280, 3449–3457. [Google Scholar] [CrossRef] [PubMed]
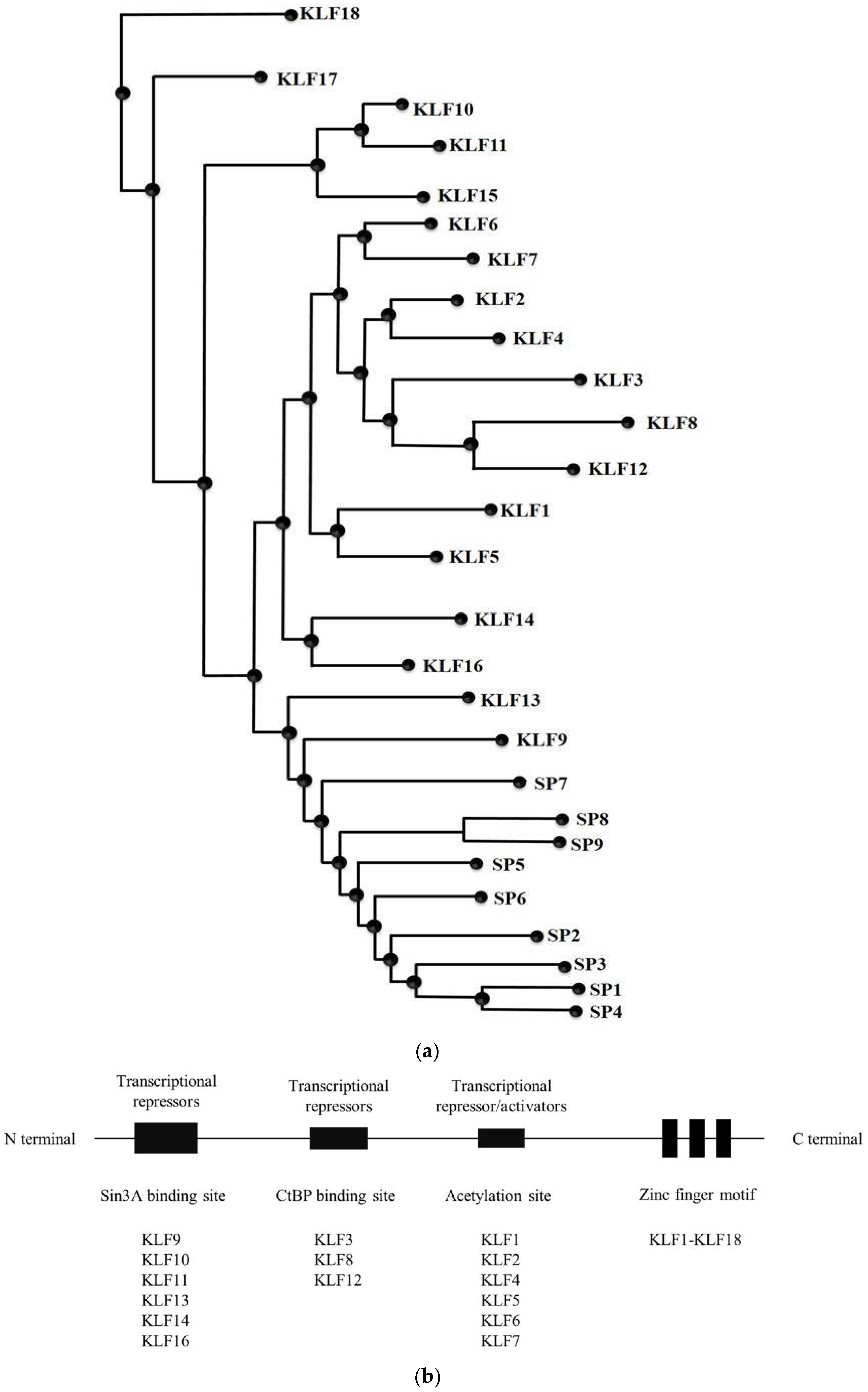

| Name | Previous Name | Transcription Activity | Expression Pattern | Disease | TGFβ Signaling |
|---|---|---|---|---|---|
| Sp1 | TFSP1 | Activator | Ubiquitous | Alzheimer’s disease (AD) | Co-activator of Smad-dependent transduction pathway in AD |
| Sp2 | KIAA0048 | Activator/Repressor | Unknown | Unknown | Unknown |
| Sp3 | SPR-2 | Activator/Repressor | Ubiquitous | Pathogenesis of keratoconus | SP1/Sp3 activities control TGFβRII gene |
| Sp4 | SPR-1, HF1B | Activator/Repressor | Brain enriched | Unknown | Unknown |
| Sp5 | Unknown | Ubiquitous | Unknown | Unknown | |
| Sp6 | Activator | Ubiquitous | Unknown | Unknown | |
| Sp7 | OSX | Unknown | Osteoblastic cells | Bone cell differentiation | Unknown |
| Sp8 | BTD | Unknown | Neurogenic regions | Neural tube formation | Unknown |
| Sp9 | ZNF990 | Unknown | Unknown | Unknown | Embryonic limb morphogenesis |
| KLF1 | E-KLF | Activator | Erythropoietic tissues (fetal liver and adult bone marrow) | Anemia β-thalassemia | Unknown |
| KLF2 | L-KLF | Activator | Ubiquitous | Glomerular disease, atherosclerosis, vascular inflammation, cancers (leukemia, breast, colon, intestine, prostate) | Inhibits TGF-β signaling in atherosclerosis |
| KLF3 | BKLF, TEF-2 | Activator/Repressor | Ubiquitous | Cancer (leukemia, cervix) | Unknown |
| KLF4 | G-KLF, EZF | Activator/Repressor | Ubiquitous | Glomerular disease, IBD, acute kidney injury, liver fibrosis, heart failure, axon regeneration, different types of cancers (bladder, brain, breast, cervix, colon, intestine, esophagus, head and neck, liver, leukemia, lung, lymphoma, prostate, skin stomach, melanoma, pancreas) | Cell proliferation and differentiation, important target in macrophages |
| KLF5 | I-KLF, C-KLF, BTEB2 | Activator/Repressor | Gut and epithelial tissue, Placenta | IBD, kidney fibrosis, different types of cancers (leukemia, breast, colon, intestine, esophagus, head and neck, gastrointestinal stromal tumor, lung) pancreas, melanoma, prostate, stomach) | Proliferation, TGFβ induced growth arrest |
| KLF6 | BCD1, COPEB, CPBP, GBF, PAC1, ST12, Zf9 | Activator | Ubiquitous | Cardiac fibrosis, kidney fibrosis, different types of cancers (leukemia, bone, breast, brain, colon, intestine, head and neck, liver, lung, ovary, pancreas, pituitary, prostate, stomach) | Cell proliferation in skeletal myoblasts |
| KLF7 | U-KLF | Activator | Ubiquitous | Type 2 diabetes | Satellite cell quiescence |
| KLF8 | BKLF3, ZNF741 | Repressor | Ubiquitous | Cancers (breast, kidney, liver, ovary, prostate, stomach) | EMT |
| KLF9 | BTEB, BTEB1 | Activator | Ubiquitous | Demyelinating disorders, different types of cancers (brain, colon, intestine, multiple myeloma, uterus) | Thyroid hormone regulation |
| KLF10 | TIEG, TIEG1, EGRα | Activator/Repressor | Ubiquitous | Angiogenesis, cardiac hypertrophy, different types of cancers (breast, kidney, pancreas, prostate) | TGFβ induced growth inhibition |
| KLF11 | F-KLF, TIEG2, MODY7 | Activator/Repressor | Ubiquitous | Liver fibrosis, type 2 diabetes, different types of cancers (leukemia, breast, colon, intestine, kidney, lung, ovary, pancreas, stomach) | TGFβ induced growth inhibition |
| KLF12 | AP-2rep, AP2REP, HSPC122 | Repressor | Brain, kidney, liver, lung | Head and neck cancer, stomach progression of gastric cancer, salivary gland tumors, autosomal dominant polycystic kidney disease (ADPKD) | Unknown |
| KLF13 | BTEB3, NSLP1, RFLAT-1 | Activator/Repressor | Ubiquitous | Head and neck Cancer | Unknown |
| KLF14 | BTEB5, SP6, EPFN | Activator/Repressor | Ubiquitous | Type 2 diabetes | Transcription of TGFβRII |
| KLF15 | K-KLF | Repressor | Ubiquitous | Glomerular disease, cardiovascular disease, kidney fibrosis | Cardiac fibrosis |
| KLF16 | BTEB4, NSLP2, DRRF | Repressor | Ubiquitous | Adipose tissue expansion | Growth control mechanisms in NHK cells |
| KLF17 | ZNF393 | Repressor | Testis, brain, and bone | Cancers (metastasis in breast cancer, lung, hepatocellular carcinoma (HCC), gastric cancer, papillary thyroid carcinoma) | Downstream mediator of the TGF-β signaling pathway, anti-metastasis |
| KLF18 | Unknown | Unknown | Unknown | Unknown | Unknown |
| Disease | TGFβ Signaling | Comments | Reference |
|---|---|---|---|
| Bone diseases | RANKL RUNX2 Smad2 ↓ Smad7 ↑ | Osteopenia: KLF10plays a critical role in osteoblast-mediated mineralization and osteoblast support of osteoclast differentiation | [34] |
| TGF-β1, BMP2, EGF | Osteoblast: KLF10 plays an active role in mediating Runx2 responses following TGFβ1 and BMP2. | [35] | |
| Type 2 diabetes | KLF10, smad7 (weakly contributes) | KLF10 variants make minor contributions to a particular genetic background that increases susceptibility to the development of T2D. | [36] |
| Hypertrophy | Pttg1 ↑ (via Sp1 binding sites) | KLF10−/− mice develop a cardiac hypertrophic phenotype with asymmetric hypertrophy, interstitial fibrosis, and myocyte disarray. | [37] |
| Immune system | TGF-β1 and Foxp3 ↑ | Loss of KLF10 enhanced CD4+ CD25 T cell activity, which stimulated inflammation and atherosclerosis and increased peripheral proinflammatory cytokines | [38] |
| Wound healing | Smad 7 ↑ Smad 2, 3 ↓ | KLF10−/− mice delay wound healing. KLF10 may play a role in dermal wound healing via the TGFβ/Smad pathway. | [39] |
| NASH | TGFβ ↑ ChREBP ↓ | Expression of KLF10 significantly increases in diet-induced NASH and ECM producing activated HSCs. | [40] |
| Colitis | KLF10, smad2, TGFβRII ↓ | KLF10 regulates TGFβRII expression in murine macrophages via histone H3 modification. | [41] |
| Hyperglycemia | KLF10, Pgc-1α, Blood glucose ↑ | KLF10 is an important regulator of hepatic glucose metabolism in mice. | [42] |
| Cancer Type | Role | Comments | TGFβ Signaling | Reference |
|---|---|---|---|---|
| Prostate cancer | Suppressor | Doxazosin-mediated apoptosis in prostate cancer cells involves activation of KLF10 and Smad4 mRNA levels, as well as a decrease in Smad7 mRNA expression | Smad dependent pathway | [71] |
| Colorectal cancer | Suppressor | KLF10 is one of the members of the signal transduction of PPARγ pathway | Bcl2 | [72] |
| Breast cancer | Suppressor | KLF10 plays an inhibitory role in the proliferation of breast cancer. KLF10 and Smad7 in breast cancers are inversely correlated | Smad7 ↑,KLF10, Smad2, and Bard1 ↓ | [73] |
| Lymphoma cells | Suppressor | The participation of Smads in TGFβ induced apoptosis is supported by the increased expression of KLF10, which can activate the mitochondrial apoptotic pathway by increasing the intracellular level of ROS | KLF10 ↑Smad 2,3 ↓ | [74] |
| Brain cancer | Suppressor | KLF10 is involved in apoptosis and was expressed at low levels in metastatic brain tumors. | Transactivator of TGFβ | [75] |
| Leukemia cells | Suppressor | KLF10 promotes apoptosis through the mitochondrial apoptotic pathway. | BimBax ↑Bcl2, Bcl-xl ↓ | [31] |
| Renal cell carcinoma | Suppressor | KLF10 up-regulates the expression of TGFβI in von Hippel-Lindau gene (VHL) deficient tumors. KLF10 is a target of VHL | TGFβ1 ↑ | [76] |
| Pancreatic cancer | Suppressor | Overexpression of KLF10 induced by the lentivirus system inhibited pancreatic cancer cell growth in vitro and in vivo. | G1-phase arrest in vitro | [77] |
| Activator | Mutational screening of KLF10 in 22 pancreatic cancer cell lines revealed no alterations in expression. | No change in KLF10 expression | [78] | |
| Hepatocellular carcinoma (HCC) | Suppressor | Upregulation of KLF10 in the HCC cell line induces inhibition of cellular proliferation | Smad3 Smad 7 | [79] |
| Activator | Deficiency of KLF10 suppresses cellular proliferation of hepatocytes during liver tumorigenesis through the TGF-β/Smad pathway | Smad 3 TGFβ1, TGFβ R1 ↑ | [80] | |
| Ovarian cancer | Suppressor | KLF10 displays strong BMAL1-dependent circadian expression; the KLF10 promoter recruits BMAL1 and is transactivated by the CLOCK/BMAL1 dimer through a conserved E-box response element. | An interruption in Circadian genes | [81] |
| Non–small cell lung carcinoma (NSCLC) | Suppressor | KLF10 suppresses TGFβ-induced EMT in conjunction with SNAI2 and HDAC1. | KLF10 ↓ SNAI1 ↑TGFβ/SMAD signaling ↑ | [82] |
| Skin | Suppressor | Loss of KLF10 leads to enhanced tumor formation and progression. | P21 ↑ transcriptional activation in a p53 independent manner. | [83] |
| Multiple myelomas | Suppressor | MicroRNA-410 accumulation regulates cell proliferation and apoptosis by targeting KLF10 via activation of the PTEN/PI3K/AKT pathway in multiple myeloma. | PTEN/PI3K/AKT pathway | [42] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Memon, A.; Lee, W.K. KLF10 as a Tumor Suppressor Gene and Its TGF-β Signaling. Cancers 2018, 10, 161. https://doi.org/10.3390/cancers10060161
Memon A, Lee WK. KLF10 as a Tumor Suppressor Gene and Its TGF-β Signaling. Cancers. 2018; 10(6):161. https://doi.org/10.3390/cancers10060161
Chicago/Turabian StyleMemon, Azra, and Woon Kyu Lee. 2018. "KLF10 as a Tumor Suppressor Gene and Its TGF-β Signaling" Cancers 10, no. 6: 161. https://doi.org/10.3390/cancers10060161
APA StyleMemon, A., & Lee, W. K. (2018). KLF10 as a Tumor Suppressor Gene and Its TGF-β Signaling. Cancers, 10(6), 161. https://doi.org/10.3390/cancers10060161






