Nanoscale-Textured Tantalum Surfaces for Mammalian Cell Alignment
Abstract
:1. Introduction
2. Materials and Methods
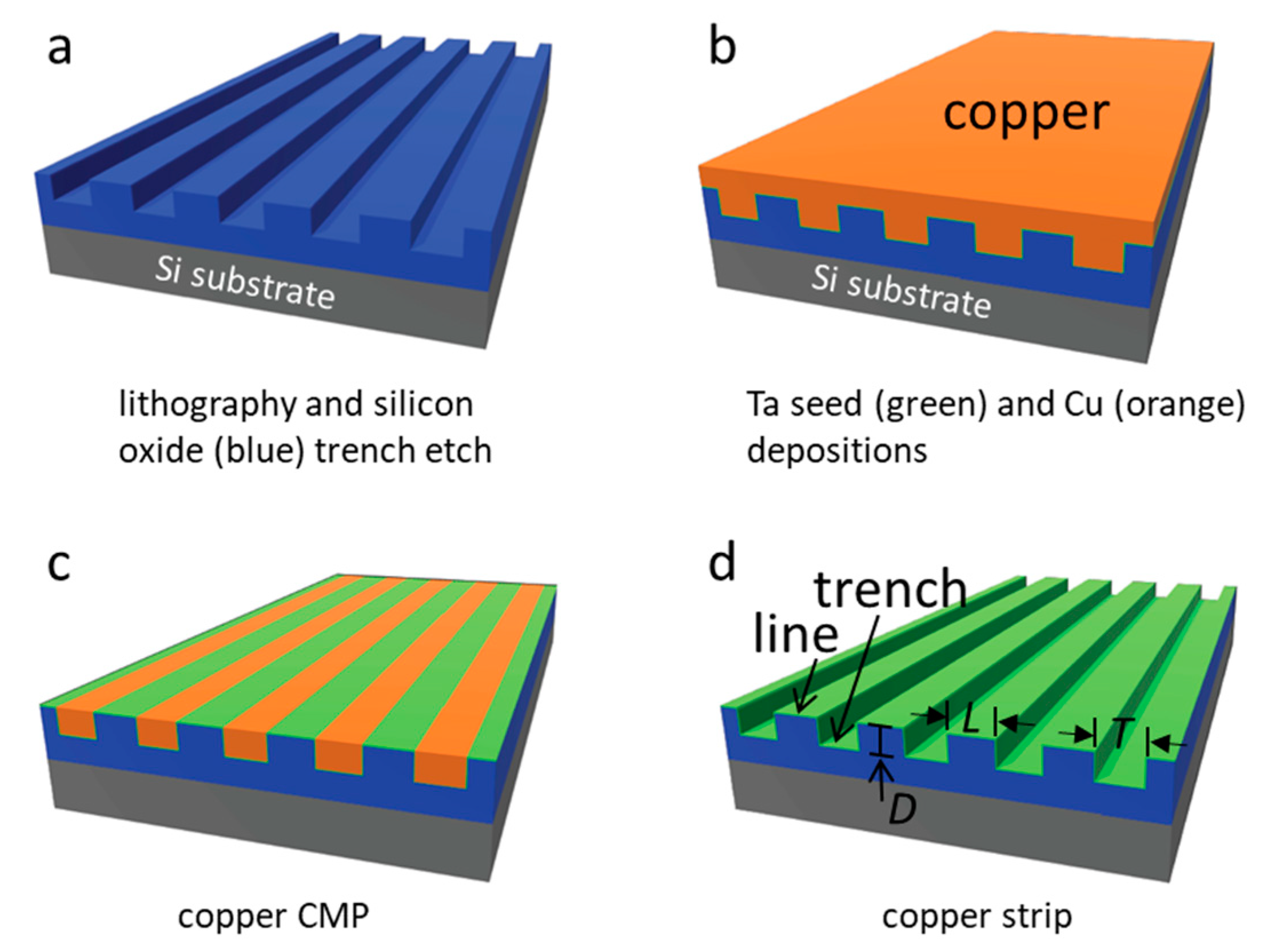
2.1. Tantalum Comb Structures
2.2. Cell Culture and Deposition
2.3. Cell Fixation and Staining Processes
2.4. Scanning Electron Microscopy
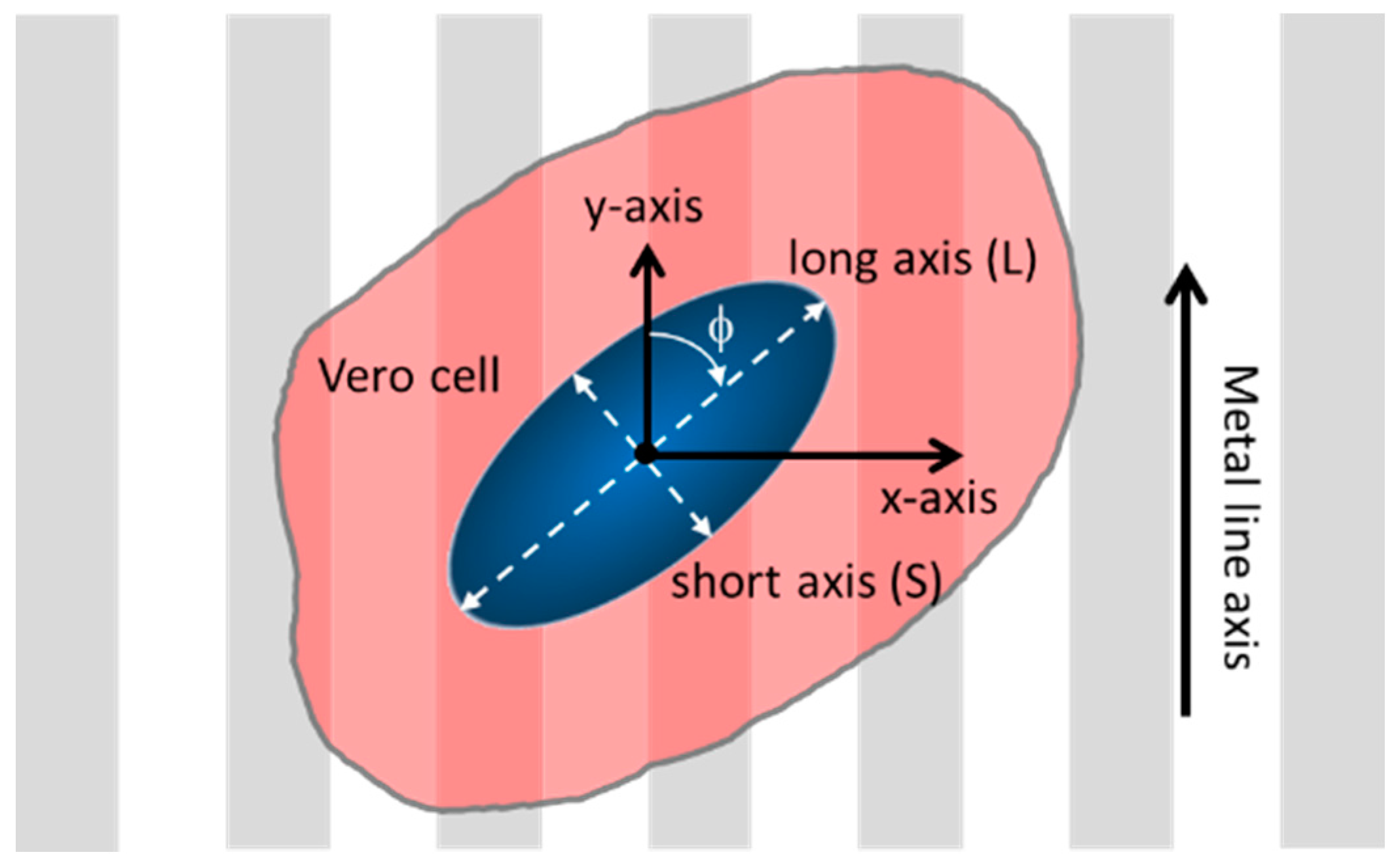
2.5. Adherent Cell Alignment and Elongation Characterizations
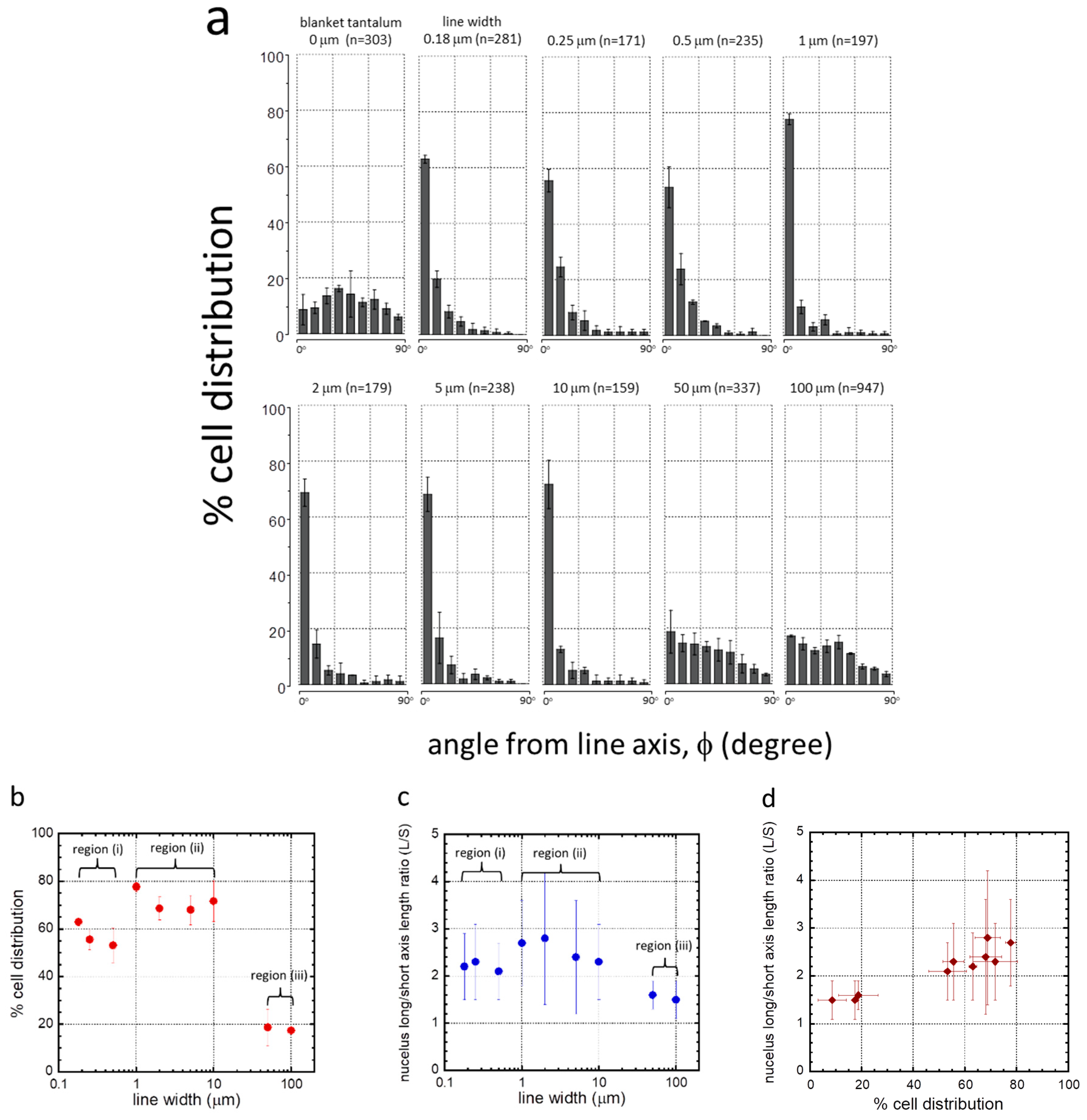
3. Results
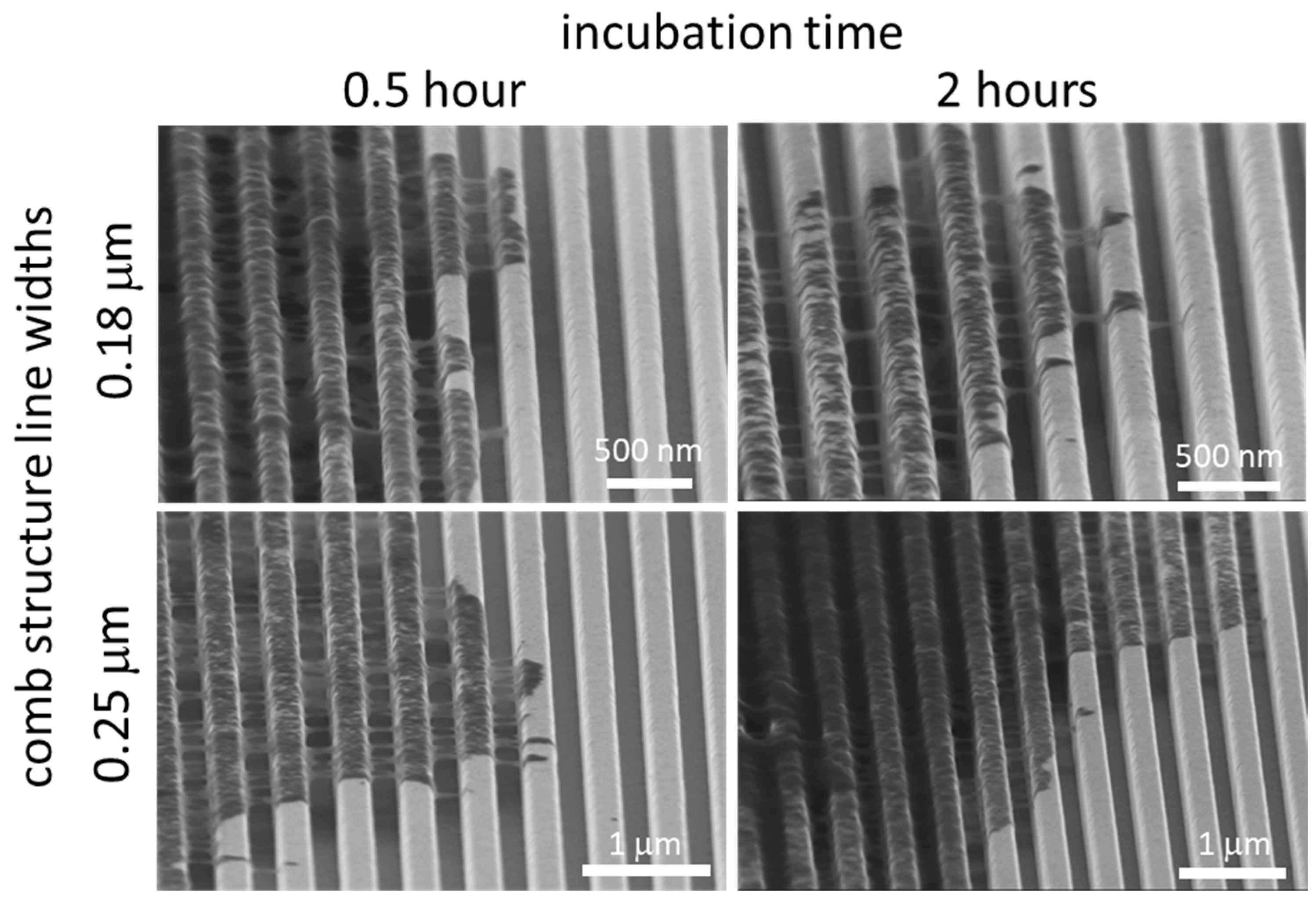
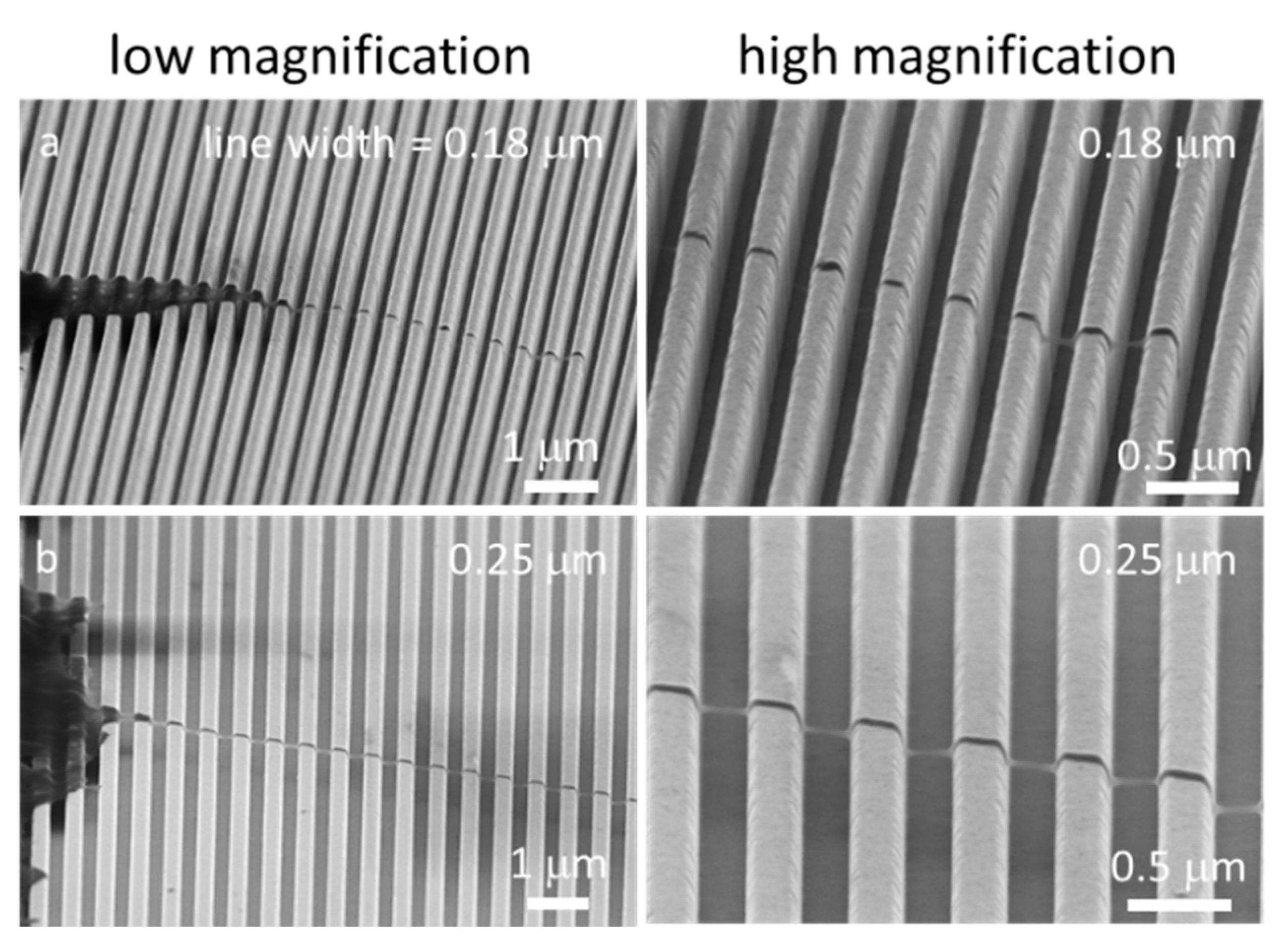
3.1. Test Structure Characterizations
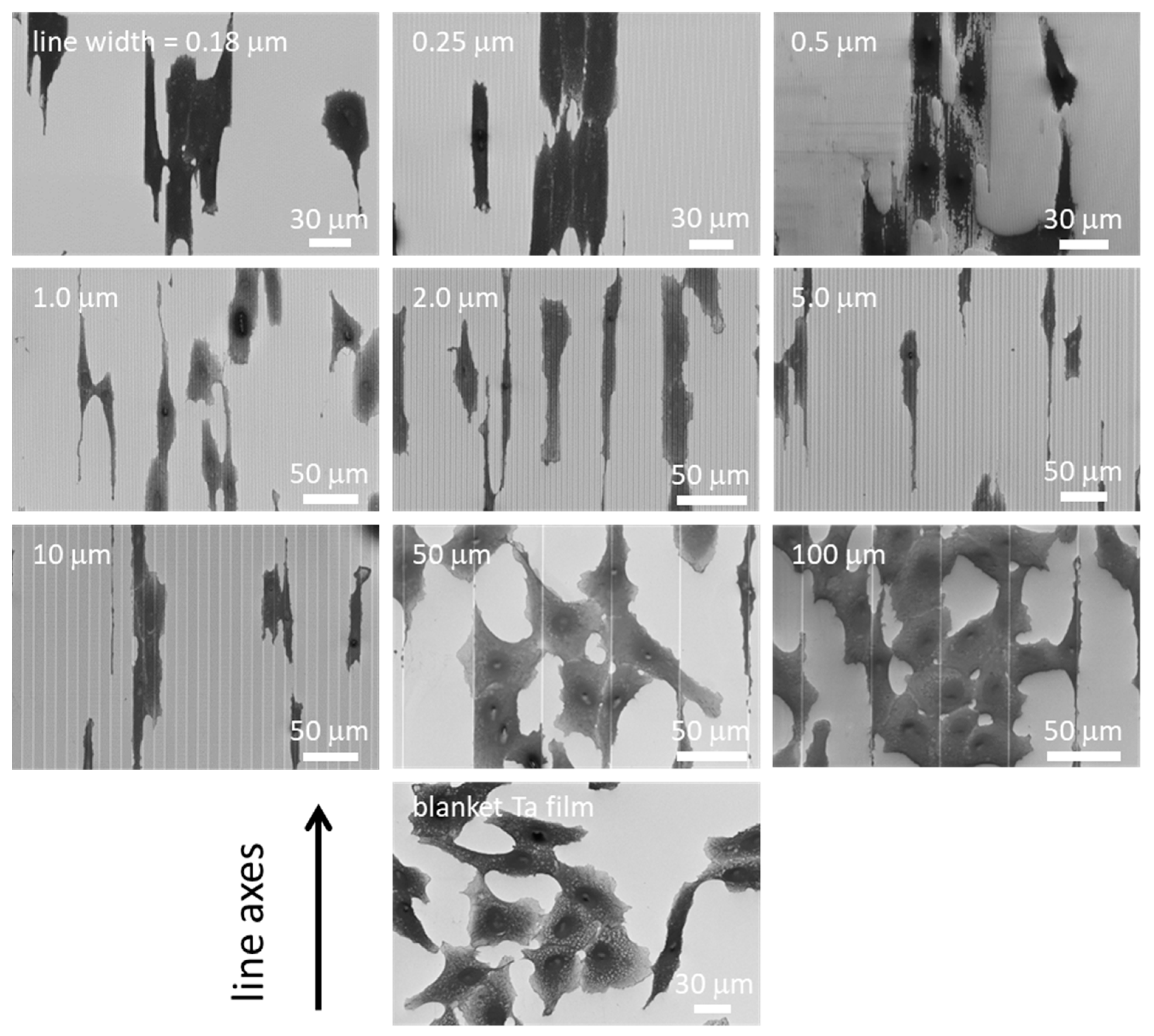
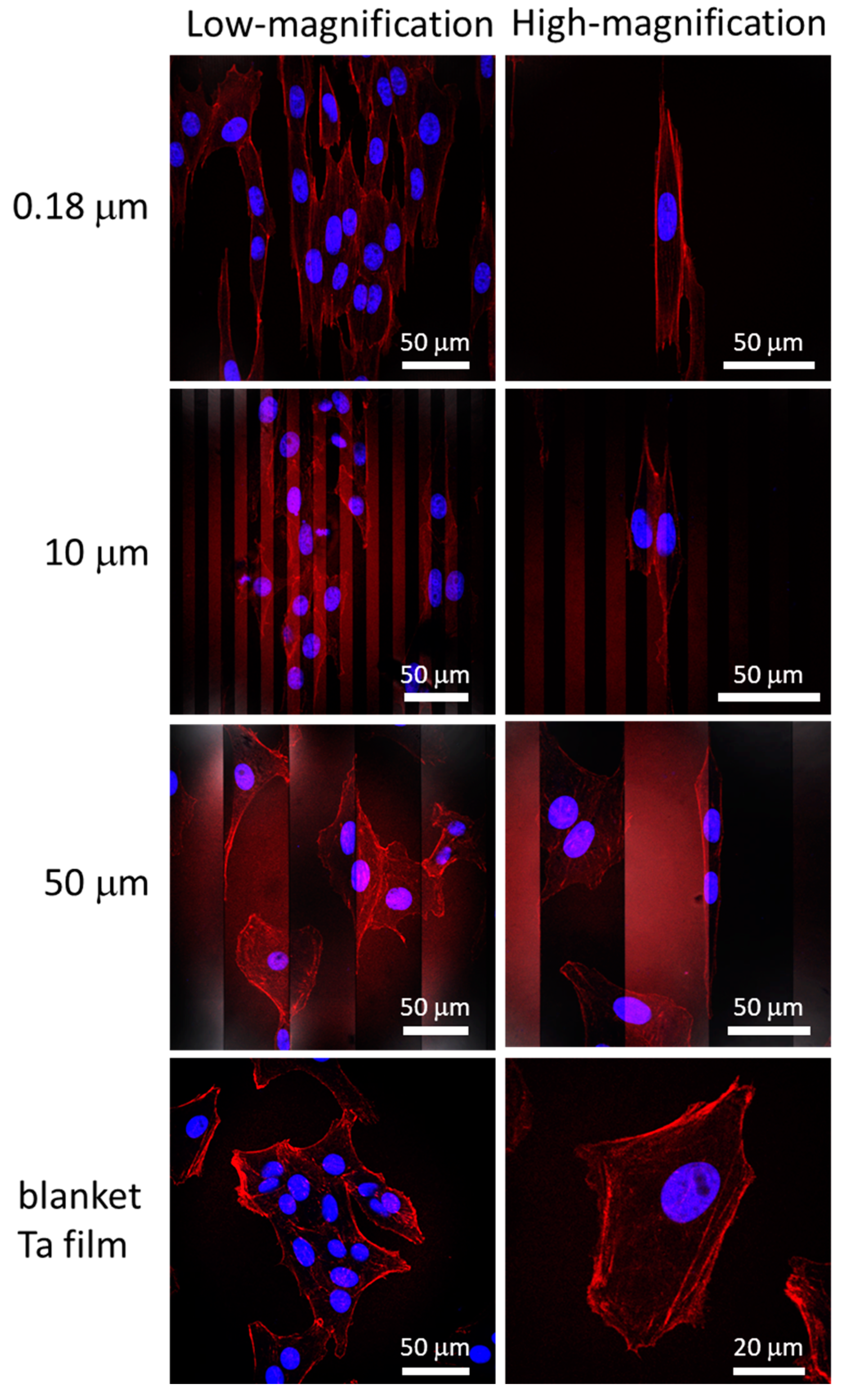
3.2. Cell Alignment and Elongation on Patterned Comb Structures
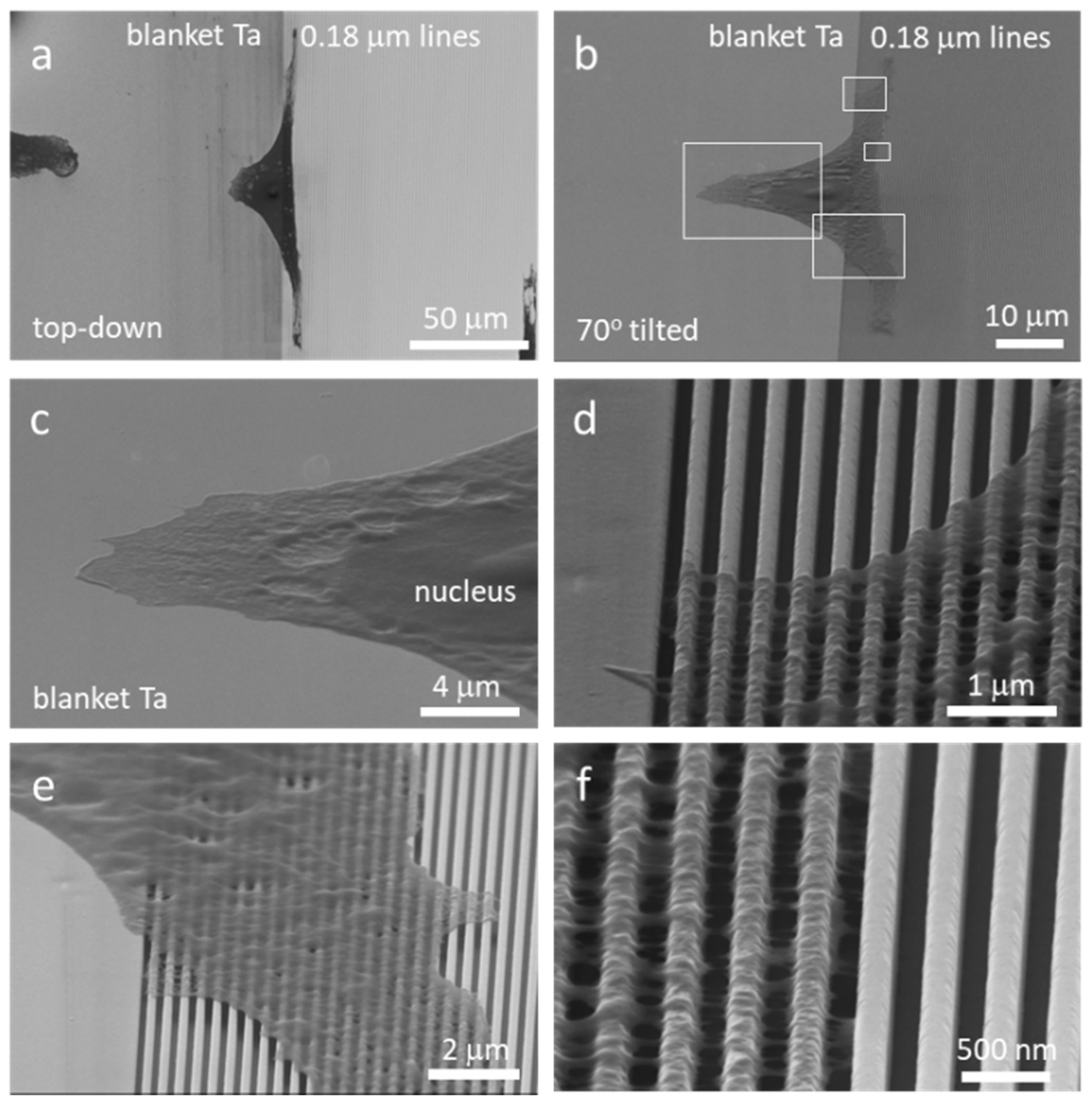
3.3. Nanometer Scale Morphology Analyses
3.3.1. Cells on Individual Comb Structures
3.3.2. A Single Cell Adhered on Multiply Structures Simultaneously
3.3.3. Long-Stranded Pseudopodia Structures
3.4. Possible Cell Alignment Mechanisms
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kaplan, R.B. Open Cell Tantalum Structures for Cancellous Bone Implants and Cell and Tissue Receptors. U.S. Patent 5,282,861, 1 February 1994. [Google Scholar]
- Black, J. Biological Performance of Tantalum. Clin. Mater. 1994, 16, 173–1994. [Google Scholar] [CrossRef]
- Balla, V.K.; Bose, S.; Davies, N.M.; Bandyopadhyay, A. Tantalum—A Bioactive Metal for Implants. JOM 2010, 62, 61–64. [Google Scholar] [CrossRef]
- Levine, B.R.; Sporer, S.; Poggie, R.A.; della Valle, C.J.; Jacobs, J.J. Experimental and clinical performance of porous tantalum in orthopedic surgery. Biomaterials 2006, 27, 4671–4681. [Google Scholar] [CrossRef] [PubMed]
- Miyazakia, T.; Kima, H.-M.; Kokuboa, T.; Ohtsuki, C.; Kato, H.; Nakamura, T. Mechanism of Apatite Formation on Bioactive Titanium Metal TEM-EDX study of mechanism of bonelike apatite formation on bioactive titanium metal in simulated body fluid. Biomaterials 2002, 23, 3–2002. [Google Scholar]
- Varitimidis, S.E.; Dimitroulias, A.P.; Karachalios, T.S.; Dailiana, Z.H.; Malizos, K.N. Outcome after tantalum rod implantation for treatment of femoral head osteonecrosis: 26 hips followed for an average of 3 years. Acta Orthop. 2009, 80, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Zhai, Z.; Guo, K.; Liu, Y.; Hou, W.; Zhu, Q.; Zhu, J. The application of porous tantalum cylinder to the repair of comminuted bone defects: A study of rabbit firearm injuries. Int. J. Clin. Exp. Med. 2015, 8, 5055–5064. [Google Scholar] [PubMed]
- Tang, Z.; Xie, Y.; Yang, F.; Huang, Y.; Wang, C.; Dai, K.; Zheng, X.; Zhang, X. Porous Tantalum Coatings Prepared by Vacuum Plasma Spraying Enhance BMSCs Osteogenic Differentiation and Bone Regeneration In Vitro and In Vivo. PLoS ONE 2013, 8, e66263. [Google Scholar] [CrossRef] [PubMed]
- Matassi, F.; Botti, A.; Sirleo, L.; Carulli, C.; Innocenti, M. Porous metal for orthopedics implants. Clin. Cases Miner. Bone Metab. 2013, 10, 111–115. [Google Scholar] [PubMed]
- Balla, V.K.; Bodhak, S.; Bose, S.; Bandyopadhyay, A. Porous Tantalum Structures for Bone Implants: Fabrication, Mechanical and In vitro Biological Properties. Acta Biomater. 2011, 6, 3349–3359. [Google Scholar] [CrossRef] [PubMed]
- Bobyn, J.D.; Stackpool, G.J.; Hacking, S.A.; Tanzer, M.; Krygier, J.J. Characteristics of bone ingrowth and interface mechanics of a new porous tantalum biomaterial. J. Bone Jt. Surg. 1999, 81-B, 907–914. [Google Scholar] [CrossRef]
- Guisbiers, G.; Herth, E.; Buchaillot, L.; Pardoen, T. Fracture toughness, hardness, and Young’s modulus of tantalum nanocrystalline films. Appl. Phys. Lett. 2010, 97, 143115. [Google Scholar] [CrossRef] [Green Version]
- Tran, R.; Xu, Z.; Radhakrishnan, B.; Winston, D.; Sun, W. Data Descriptor: Surface energies of elemental crystals. Sci. Data 2016, 3, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Qiao, Y.; Cheng, M.; Jiang, G.; He, G.; Chen, Y. Tantalum implanted entangled porous titanium promotes surface osseointegration and bone ingrowth. Nat. Publ. Gr. 2016, 6, 26248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, X.; Zhao, D.; Wang, B.; Wang, W.; Kang, K.; Xie, H.; Liu, B.; Zhang, X.; Zhang, J.; Yang, Z. Tantalum coating of porous carbon scaffold supplemented with autologous bone marrow stromal stem cells for bone regeneration in vitro and in vivo. Exp. Biol. Med. 2016, 241, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, A.I.; Abrams, G.A.; Bertics, P.J.; Murphy, C.J.; Nealey, P.F. Epithelial contact guidance on well-defined micro- and nanostructured substrates. J. Cell Sci. 2003, 116, 1892–2003. [Google Scholar] [CrossRef] [PubMed]
- Moussa, H.I.; Logan, M.; Siow, G.C.; Phann, D.L.; Rao, Z.; Aucoin, M.G.; Tsui, T.Y. Manipulating mammalian cell morphologies using chemical-mechanical polished integrated circuit chips. Sci. Technol. Adv. Mater. 2017, 18, 839–856. [Google Scholar] [CrossRef] [PubMed]
- Moussa, H.; Logan, M.; Chan, W.; Wong, K.; Rao, Z.; Aucoin, M.; Tsui, T. Pattern-Dependent Mammalian Cell (Vero) Morphology on Tantalum/Silicon Oxide 3D Nanocomposites. Materials 2018, 11, 1306. [Google Scholar] [CrossRef] [PubMed]
- Le Digabel, J.; Richert, A.; Hersen, P.; Ghibaudo, M. Substrate Topography Induces a Crossover from 2D to 3D Behavior in Fibroblast Migration. Biophys. J. 2009, 97, 357–368. [Google Scholar]
- Seo, B.B.; Jahed, Z.; Coggan, J.A.; Chau, Y.Y.; Rogowski, J.L.; Gu, F.X.; Wen, W.; Mofrad, M.R.K.; Tsui, T.Y. Mechanical contact characteristics of pc3 human prostate cancer cells on complex-shaped silicon micropillars. Materials 2017, 10, 892. [Google Scholar] [CrossRef] [PubMed]
- Jahed, Z.; Molladavoodi, S.; Seo, B.B.; Gorbet, M.; Tsui, T.Y.; Mofrad, M.R.K. Cell responses to metallic nanostructure arrays with complex geometries. Biomaterials 2014, 35, 9363–9371. [Google Scholar] [CrossRef] [PubMed]
- Gentleman, M.M.; Gentleman, E. The role of surface free energy in osteoblast—Biomaterial interactions The role of surface free energy in osteoblast—Biomaterial interactions. Int. Mater. Rev. 2014, 59, 417–429. [Google Scholar] [CrossRef]
- Deligianni, D.D.; Katsala, N.; Ladas, S.; Sotiropoulou, D.; Amedee, J.; Missirlis, Y.F. Effect of surface roughness of the titanium alloy Ti-6Al-4V on human bone marrow cell response and on protein adsorption. Biomaterials 2001, 22, 1251–2001. [Google Scholar] [CrossRef]
- Khalili, A.A.; Ahmad, M.R. A Review of Cell Adhesion Studies for Biomedical and Biological Applications. Int. J. Mol. Sci. 2015, 16, 18149–18184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zareidoost, A.; Yousefpour, M.; Ghaseme, B.; Amanzadeh, A. The relationship of surface roughness and cell response of chemical surface modification of titanium. J. Mater. Sci. Mater. Med. 2012, 23, 1479–1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolatshahi-Pirouz, A.; Jensen, T.; Kraft, D.C.; Foss, Ќ.M.; Kingshott, P.; Hansen, J.L.; Larsen, A.N.; Chevallier, J.; Besenbacher, F. Fibronectin Adsorption, Cell Adhesion, and Proliferation on Nanostructured Tantalum Surfaces. ACS Nano 2010, 4, 2874–2882. [Google Scholar] [CrossRef] [PubMed]
- Dolatshahi-Pirouz, A.; Pennisi, C.P.; Skeldal, S.; Foss, M.; Chevallier, J.; Zachar, V.; Andreasen, P.; Yoshida, K.; Besenbacher, F. The influence of glancing angle deposited nano-rough platinum surfaces on the adsorption of fibrinogen and the proliferation of primary human fibroblasts. Nanotechnology 2009, 20, 095101. [Google Scholar] [CrossRef] [PubMed]
- Hallab, N.J.; Bundy, K.J.; Connor, K.O.; Moses, R.L.; Jacobs, J.J. Evaluation of Metallic and Polymeric Biomaterial Surface Energy and Surface Roughness Characteristics for Directed Cell Adhesion. Tissue Eng. 2001, 7, 55–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchesano, V.; Gennari, O.; Mecozzi, L.; Grilli, S.; Ferraro, P. Effects of Lithium Niobate Polarization on Cell Adhesion and Morphology. ACS Appl. Mater. Interfaces 2015, 7, 18113–18119. [Google Scholar] [CrossRef] [PubMed]
- Christophi, C.; Cavalcanti-Adam, E.A.; Hanke, M.; Kitamura, K.; Gruverman, A.; Grunze, M.; Dowben, P.A.; Rosenhahn, A. Adherent cells avoid polarization gradients on periodically poled LiTaO3ferroelectrics. Biointerphases 2013, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Carville, N.C.; Collins, L.; Manzo, M.; Gallo, K.; Lukasz, B.I.; McKayed, K.K.; Simpson, J.C.; Rodriguez, B.J. Biocompatibility of ferroelectric lithium niobate and the influence of polarization charge on osteoblast proliferation and function. J. Biomed. Mater. Res. Part A 2015, 103, 2540–2548. [Google Scholar] [CrossRef] [PubMed]
- Vilarinho, P.M.; Barroca, N.; Zlotnik, S.; Félix, P.; Fernandes, M.H. Are lithium niobate (LiNbO3) and lithium tantalate (LiTaO3) ferroelectrics bioactive? Mater. Sci. Eng. C 2014, 39, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Mandracchia, B.; Gennari, O.; Marchesano, V.; Paturzo, M.; Ferraro, P. Label free imaging of cell-substrate contacts by holographic total internal reflection microscopy. J. Biophotonics 2017, 10, 1163–1170. [Google Scholar] [CrossRef] [PubMed]
- Mandracchia, B.; Gennari, O.; Bramanti, A.; Grilli, S.; Ferraro, P. Label-free quantification of the effects of lithium niobate polarization on cell adhesion via holographic microscopy. J. Biophotonics 2018, 11, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Rega, R.; Gennari, O.; Mecozzi, L.; Grilli, S.; Pagliarulo, V.; Ferraro, P. Bipolar Patterning of Polymer Membranes by Pyroelectrification. Adv. Mater. 2016, 28, 454–459. [Google Scholar] [CrossRef] [PubMed]
- English, A.; Azeem, A.; Spanoudes, K.; Jones, E.; Tripathi, B.; Basu, N.; Mcnamara, K.; Tofail, S.A.M.; Rooney, N.; Riley, G.; et al. Acta Biomaterialia Substrate topography: A valuable in vitro tool, but a clinical red herring for in vivo tenogenesis. Acta Biomater. 2015, 27, 12–2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamoto, T.; Wang, X.; Kawazoe, N.; Chen, G. Biointerfaces Influence of micropattern width on differentiation of human mesenchymal stem cells to vascular smooth muscle cells. Colloids Surf. B Biointerfaces 2014, 122, 323–2014. [Google Scholar] [CrossRef] [PubMed]
- Béduer, A.; Vieu, C.; Arnauduc, F.; Sol, J.; Loubinoux, I.; Vaysse, L. Biomaterials Engineering of adult human neural stem cells differentiation through surface micropatterning. Biomaterials 2012, 33, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, A.; Cecchini, M.; Serresi, M.; Faraci, P.; Pisignano, D.; Beltram, F. Biomaterials Neuronal polarity selection by topography-induced focal adhesion control. Biomaterials 2010, 31, 4682–4694. [Google Scholar] [CrossRef] [PubMed]
- Joo, S.; Kim, J.Y.; Lee, E.; Hong, N.; Sun, W.; Nam, Y. Effects of ECM protein micropatterns on the migration and differentiation of adult neural stem cells. Sci. Rep. 2015, 5, 13043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Provenzano, P.P.; Smith, C.L.; Levchenko, A. Matrix nanotopography as a regulator of cell function. J. Cell Biol. 2012, 197, 351–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yim, E.K.F.; Reano, R.M.; Pang, S.W.; Yee, A.F.; Chen, C.S.; Leong, K.W. Nanopattern-induced changes in morphology and motility of smooth muscle cells. Biomaterials 2005, 26, 5405–5413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Lipke, E.A.; Kim, P.; Cheong, R.; Thompson, S.; Delannoy, M. Nanoscale cues regulate the structure and function of macroscopic cardiac tissue constructs. Proc. Natl. Acad. Sci. USA 2009, 107, 565–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teixeira, A.I.; McKie, G.A.; Foley, J.D.; Bertics, P.J.; Nealey, P.F.; Murphy, C.J. The effect of environmental factors on the response of human corneal epithelial cells to nanoscale substrate topography. Biomaterials 2006, 27, 3945–3954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zahor, D.; Radko, A.; Vago, R.; Gheber, L.A. Organization of mesenchymal stem cells is controlled by micropatterned silicon substrates. Mater. Sci. Eng. C 2007, 27, 121–2007. [Google Scholar] [CrossRef]
- Kaiser, J.; Reinmann, A.; Bruinink, A. The effect of topographic characteristics on cell migration velocity. Biomaterials 2006, 27, 5230–5241. [Google Scholar] [CrossRef] [PubMed]
- Fujita, S.; Ohshima, M.; Iwata, H. Time-lapse observation of cell alignment on nanogrooved patterns. J. R. Soc. Interface 2009, 6, S269–S277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Shi, J.; Hu, J.; Chen, Y. Cells cultured on microgrooves with or without surface coating: Correlation between cell alignment, spreading and local membrane deformation. Mater. Sci. Eng. C 2013, 33, 855–863. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.Y.; Tong, W.Y.; Shi, J.; Shi, P. Influence of engineered surface on cell directionality and motility. Biofabrication 2014, 6, 15011. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.Z.; Tsuru, K.; Hayakawa, S.; Osaka, A. Modification of Ti implant surface for cell proliferation and cell alignment. J. Biomed. Mater. Res. Part A 2008, 84, 988–993. [Google Scholar]
- Chehroudi, B.; Ratkay, J.; Brunette, D.M. The role of implant surface geometry on mineralization in vivo and in vitro: A transmission and scanning electron microscopic study. Cells Mater. 1992, 2, 89–104. [Google Scholar]
- Owen, G.R.; Jackson, J.; Chehroudi, B.; Burt, H.; Brunette, D.M. A PLGA membrane controlling cell behaviour for promoting tissue regeneration. Biomaterials 2005, 26, 7447–7456. [Google Scholar] [CrossRef] [PubMed]
- Barr, S.; Hill, E.; Bayat, A. Current implant surface technology: An examination of their nanostructure and their influence on fibroblast alignment and biocompatibility. Eplasty 2009, 9, e22. [Google Scholar] [PubMed]
- Yang, J.-Y.; Ting, Y.-C.; Lai, J.-Y.; Liu, H.-L.; Fang, H.-W.; Tsai, W.-B. Quantitative analysis of osteoblast-like cells (MG63) morphology on nanogrooved substrata with various groove and ridge dimensions. J. Biomed. Mater. Res. A 2009, 90, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Nobes, C.D.; Hall, A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 1995, 81, 53–62. [Google Scholar] [CrossRef] [Green Version]
- Ridley, A.J.; Hall, A. The small GTP-binding protein rho regulates the assembly of focal adhesions and stress fibres in response to growth factors. Cell 1992, 70, 399–1992. [Google Scholar]
- Huo, W.T.; Zhao, L.Z.; Yu, S.; Yu, Z.T.; Zhang, P.X.; Zhang, Y.S. Significantly enhanced osteoblast response to nano-grained pure tantalum. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Doering, R.; Nishi, Y. Handbook of Semiconductor Manufacturing Technology, 2nd ed.; CRC Press, Taylor & Francis Group: New York, NY, USA, 2007. [Google Scholar]
- Chen, W.-K. The VLSI Handbook, 2nd ed.; CRC Press, Taylor & Francis Group: New York, NY, USA, 2007. [Google Scholar]
- Li, Y. Microelectronic Applications of Chemical Mechanical Planarization; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2007. [Google Scholar]
- Van Zant, P. Microchip Fabrication: A Practical Guide to Semiconductor Processing, 6th ed.; McGraw Hilll Education: New York, NY, USA, 2014. [Google Scholar]
- Shi, X.; Murella, K.; Schlueter, J.A.; Choo, J.O. Chemical Mechanical Polishing Slurry Compositions and Method Using the Same for Copper and Through-Silicon via Applications. U.S. Patent 8,974,692 B2, 10 March 2015. [Google Scholar]
- Shi, X.; Palmer, B.J.; Sawayda, R.A.; Coder, F.A.; Perez, V. Method and Composition for Chemical Mechanical Planarization of a Metal. U.S. Patent 8,414,789 B2, 23 September 2013. [Google Scholar]
- Ventre, M.; Causa, F.; Netti, P.A. Determinants of cell-material crosstalk at the interface: Towards engineering of cell instructive materials. J. R. Soc. Interface 2012, 9, 2017–2032. [Google Scholar] [CrossRef] [PubMed]
- Lamers, E.; van Horssen, R.; te Riet, J.; van Delft, F.C.; Luttge, R.; Walboomers, X.F.; Jansen, J.A. The influence of nanoscale topographical cues on initial osteoblast morphology and migration. Eur. Cell Mater. 2010, 20, 329–343. [Google Scholar] [CrossRef] [PubMed]
- Loesberg, W.A.; te Riet, J.; van Delft, F.C.; Schön, P.; Figdor, C.G.; Speller, S.; van Loon, J.J.W.A.; Walboomers, X.F.; Jansen, J.A. The threshold at which substrate nanogroove dimensions may influence fibroblast alignment and adhesion. Biomaterials 2007, 28, 3944–3951. [Google Scholar] [CrossRef] [PubMed]
- Toworfe, G.K.; Composto, R.J.; Adams, C.S.; Shapiro, I.M.; Ducheyne, P. Fibronectin adsorption on surface-activated poly(dimethylsiloxane) and its effect on cellular function. J. Biomed. Mater. Res. Part A 2004, 71, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Lamers, E.; Walboomers, X.F.; Domanski, M.; te Riet, J.; van Delft, F.C.; Luttge, R.; Winnubst, L.A.J.A.; Gardeniers, H.J.G.E.; Jansen, J.A. The influence of nanoscale grooved substrates on osteoblast behavior and extracellular matrix deposition. Biomaterials 2010, 31, 3307–3316. [Google Scholar] [CrossRef] [PubMed]
- Yim, E.K.F.; Darling, E.M.; Kulangara, K.; Guilak, F.; Leong, K.W. Biomaterials Nanotopography-induced changes in focal adhesions, cytoskeletal organization, and mechanical properties of human mesenchymal stem cells. Biomaterials 2010, 31, 1299–1306. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Li, N.; Huang, R.; Song, Q.; Wang, L.; Zhang, Q.; Su, R.; Kong, T.; Tang, M.; Cheng, G. The Effects of Topographical Patterns and Sizes on Neural Stem Cell Behavior. PLoS ONE 2013, 8, e59022. [Google Scholar] [CrossRef] [PubMed]
- Sung, C.; Yang, C.; Yeh, J.A. Integrated Circuit-Based Biofabrication with Common Biomaterials for Probing Cellular Biomechanics. Trends Biotechnol. 2016, 34, 171–186. [Google Scholar] [CrossRef] [PubMed]


| Reference | Cell Type | Substrate | Line Width Range (μm) | Trench Width Range (μm) | Maximum Alignment Line/Trench Width (μm) |
|---|---|---|---|---|---|
| [44] | Human corneal epithelial cells | Silicon oxide | 0.07–1.9 | 0.3–2.1 | 0.85/1.15 |
| [54] | Osteoblast-like cells (MG63) | Silicon | 0.09–0.5 | 0.09–0.5 | 0.15/0.15 |
| [48] | HeLa cells | Polydimethylsiloxane | 2–30 | 1.5–3.0 | 2/2 |
| [38] | Human neural stem cells | Polydimethylsiloxane | 5–20 | 5–60 | 5/5 |
| [37] | Human mesenchymal stem cells | Polystyrene stripes | 5–1000 | 5–1000 | 20/20 |
| [40] | Adult neural stem cells | Poly-d-lysine with Printed laminin strips | 30 | 30–170 | 30/30 |
| Structure | Line (L)/Trench (T) Width (μm) | Inspected Comb Structure Area (mm2) | Number of Cells Sampled (n) | Coverage (cell/mm2) | L/S | % of Population Aligned ±10° from Lines |
|---|---|---|---|---|---|---|
| 1 | 0.18 | 1.8 | 281 | 156 | 2.2 + 0.7 | 63.0 + 1.4 |
| 2 | 0.25 | 1.8 | 171 | 95 | 2.3 + 0.8 | 55.6 + 4.1 |
| 3 | 0.5 | 1.8 | 235 | 131 | 2.1 + 0.6 | 53.2 + 7.4 |
| 4 | 1 | 1.8 | 197 | 109 | 2.7 + 0.9 | 77.7 + 2.0 |
| 5 | 2 | 1.8 | 179 | 99 | 2.8 + 1.4 | 68.7 + 4.9 |
| 6 | 5 | 1.8 | 238 | 132 | 2.4 + 1.2 | 68.0 + 6.2 |
| 7 | 10 | 1.8 | 159 | 88 | 2.3 + 0.8 | 71.7 + 8.6 |
| 8 | 50 | 1.8 | 337 | 187 | 1.6 + 0.3 | 18.7 + 7.7 |
| 9 | 100 | 6.6 | 947 | 143 | 1.5 + 0.4 | 17.4 + 0.3 |
| 10 | blanket Ta | 1.8 | 303 | 168 | 1.5 + 0.4 | 8.6 + 5.5 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moussa, H.I.; Logan, M.; Wong, K.; Rao, Z.; Aucoin, M.G.; Tsui, T.Y. Nanoscale-Textured Tantalum Surfaces for Mammalian Cell Alignment. Micromachines 2018, 9, 464. https://doi.org/10.3390/mi9090464
Moussa HI, Logan M, Wong K, Rao Z, Aucoin MG, Tsui TY. Nanoscale-Textured Tantalum Surfaces for Mammalian Cell Alignment. Micromachines. 2018; 9(9):464. https://doi.org/10.3390/mi9090464
Chicago/Turabian StyleMoussa, Hassan I., Megan Logan, Kingsley Wong, Zheng Rao, Marc G. Aucoin, and Ting Y. Tsui. 2018. "Nanoscale-Textured Tantalum Surfaces for Mammalian Cell Alignment" Micromachines 9, no. 9: 464. https://doi.org/10.3390/mi9090464





