Constriction Channel Based Single-Cell Mechanical Property Characterization †
Abstract
:1. Introduction
| Cell Types | Quantified Parameters | Key Observations | References |
|---|---|---|---|
| Plasmodium falciparum infected RBCs | Channel blockage | Maturation of Plasmodium falciparum decreased the deformability of infected RBCs. | [23] |
| Plasmodium vivax infected RBCs | Channel blockage | No significant decrease in deformability was observed during the maturation of Plasmodium vivax infected RBCs. | [27] |
| Plasmodium falciparum infected RBCs | Transit velocity | The parasite protein Pf155 decreased the transit velocity of ring-stage infected RBCs. | [29] |
| Plasmodium falciparum infected RBCs | Cortical tension | Cortical tensions were quantified as 3.22 ± 0.64 pN/μm (uninfected RBCs), 4.66 ± 1.15 pN/μm (ring-stage infected RBCs), 8.26 ± 2.84 pN/μm (early trophozoite infected RBCs), and 21.38 ± 5.81 pN/μm (late trophozoite infected RBCs). | [30] |
| Plasmodium falciparum infected RBCs | Transit velocity | Artesunate (a drug in malaria) decreased the deformability of infected RBCs while it had no effect on normal RBCs. | [31] |
| Normal and oxidized RBCs | Cortical tension | Cortical tensions were quantified as 20.13 ± 1.47 pN/μm (normal RBCs) and 27.51 ± 3.64 pN/μm (oxidized RBCs). | [32] |
| White blood cells | Transit time | In diseases of sepsis and leukostasis, decreases in the deformability of WBCs were found. | [24] |
| Breast tumor cells | Entry time and transit velocity | Benign breast epithelial cells of MCF-10A had longer entry times than tumor breast cells of MCF-7 with similar sizes. | [25] |
| Lung tumor cells | Instantaneous Young’s modulus | Instantaneous Young’s moduli were quantified as 3.48 ± 0.86 kPa for A549 cells and 2.99 ± 0.38 kPa for 95C cells | [47] |
2. Constriction Channel Based Mechanical Property Characterization of Red Blood Cells

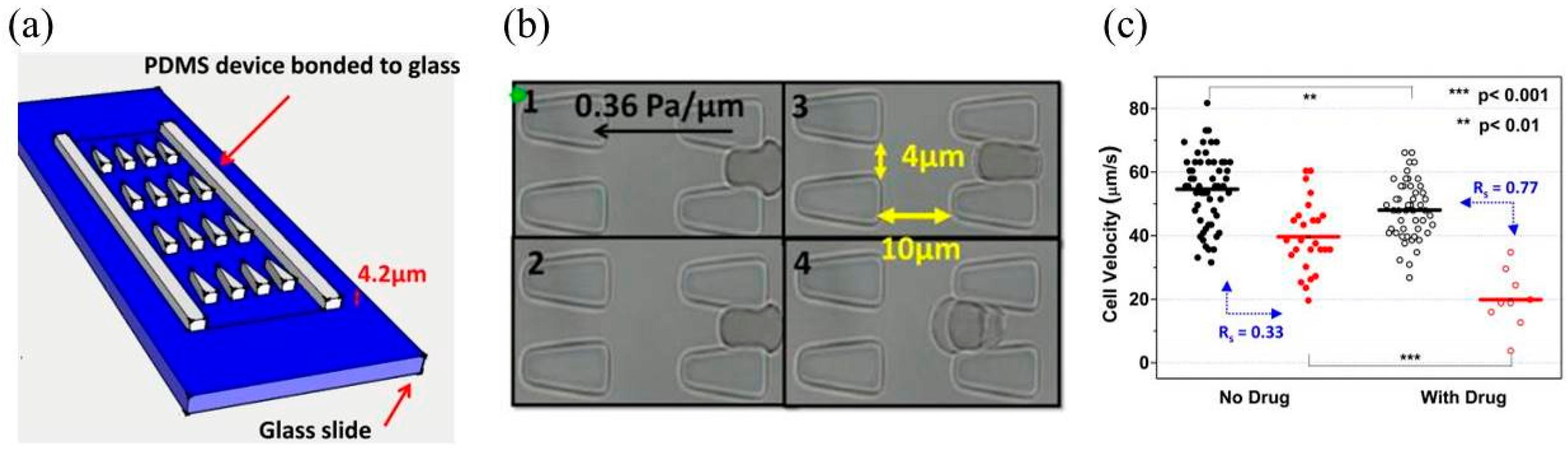
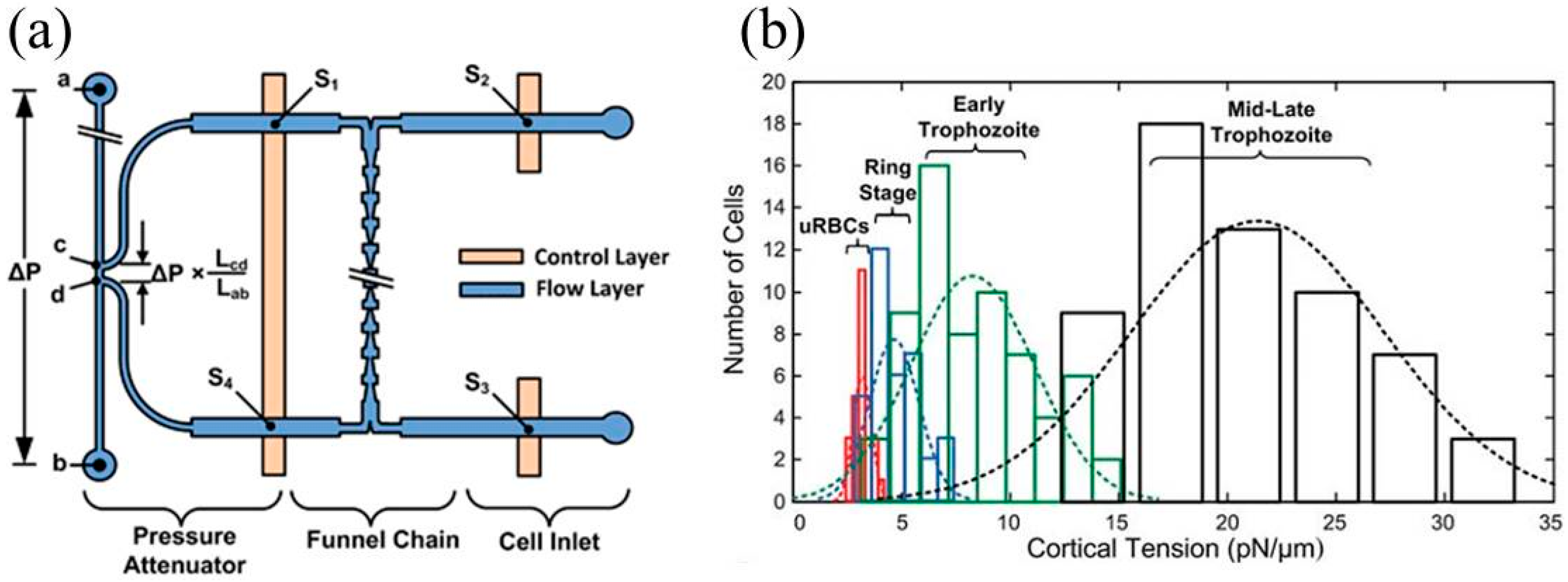
3. Constriction Channel Based Mechanical Property Characterization of White Blood Cells

4. Constriction Channel Based Mechanical Property Characterization of Tumor Cells

5. Mechanical Modeling of the Constriction Channel Design
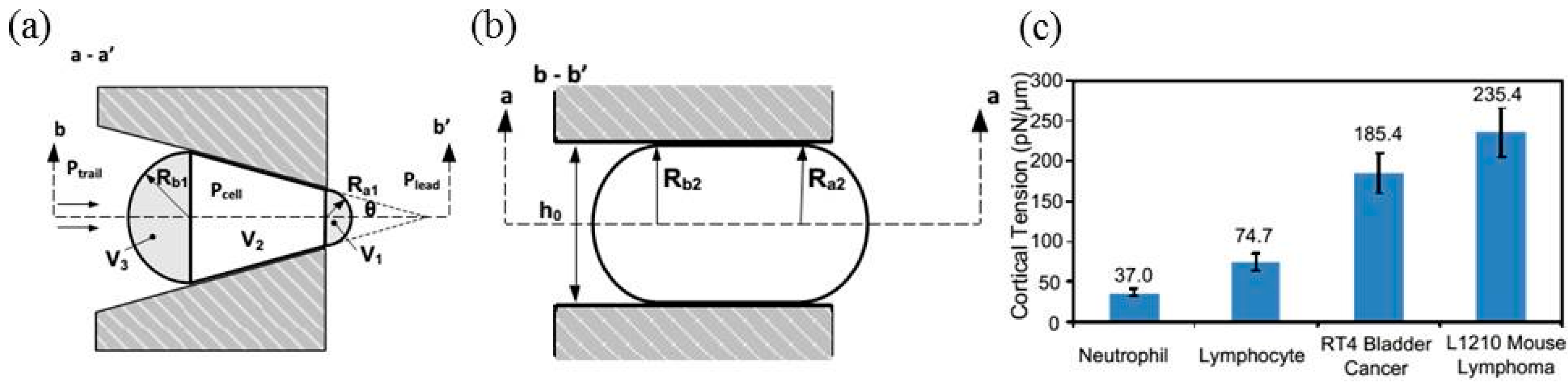
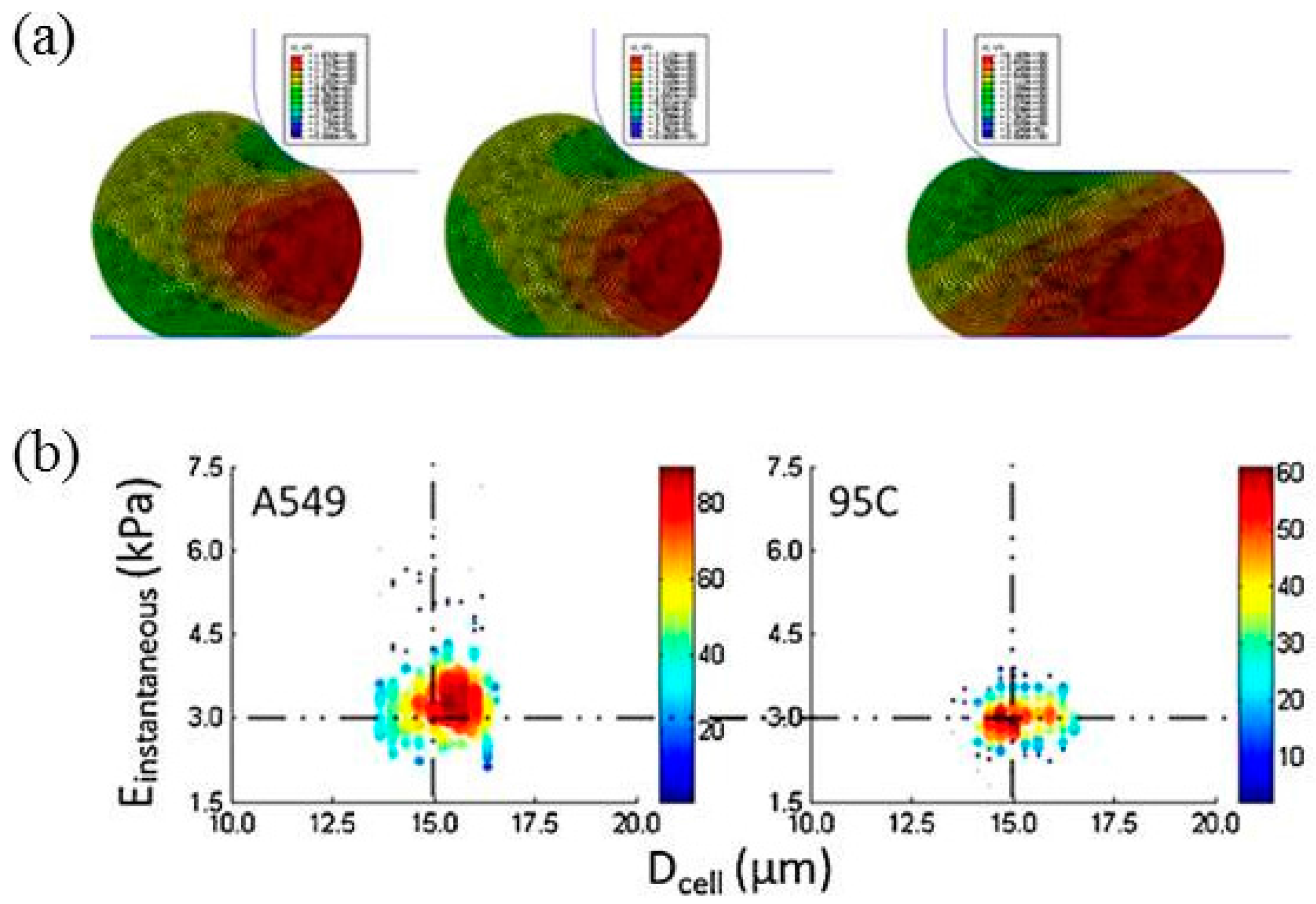
6. Discussions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ethier, C.R.; Simmons, C.A. Introductory Biomechanics: From Cells to Organisms; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Fletcher, D.A.; Mullins, R.D. Cell mechanics and the cytoskeleton. Nature 2010, 463, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Di Carlo, D. A mechanical biomarker of cell state in medicine. J. Lab. Autom. 2012, 17, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.Y.H.; Lim, C.T. Biomechanics approaches to studying human diseases. Trends Biotechnol. 2007, 25, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.L.; Goldmann, W.H. Feeling the forces: Atomic force microscopy in cell biology. Life Sci. 2003, 72, 2553–2560. [Google Scholar] [CrossRef]
- Kirmizis, D.; Logothetidis, S. Atomic force microscopy probing in the measurement of cell mechanics. Int. J. Nanomed. 2010, 5, 137–145. [Google Scholar] [CrossRef]
- Lekka, M.; Pogoda, K.; Gostek, J.; Klymenko, O.; Prauzner-Bechcicki, S.; Wiltowska-Zuber, J.; Jaczewska, J.; Lekki, J.; Stachura, Z. Cancer cell recognition--mechanical phenotype. Micron 2012, 43, 1259–1266. [Google Scholar] [CrossRef] [PubMed]
- Cross, S.E.; Jin, Y.S.; Rao, J.; Gimzewski, J.K. Nanomechanical analysis of cells from cancer patients. Nat. Nanotechnol. 2007, 2, 780–783. [Google Scholar] [CrossRef] [PubMed]
- Hochmuth, R.M. Micropipette aspiration of living cells. J. Biomech. 2000, 33, 15–22. [Google Scholar] [CrossRef]
- Shojaei-Baghini, E.; Zheng, Y.; Jewett, M.A.S.; Geddie, W.B.; Sun, Y. Mechanical characterization of benign and malignant urothelial cells from voided urine. Appl. Phys. Lett. 2013, 102, 123704. [Google Scholar] [CrossRef]
- Shojaei-Baghini, E.; Zheng, Y.; Sun, Y. Automated micropipette aspiration of single cells. Ann. Biomed. Eng. 2013, 41, 1208–1216. [Google Scholar] [CrossRef] [PubMed]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Wootton, R.C.; Demello, A.J. Microfluidics: Exploiting elephants in the room. Nature 2010, 464, 839–840. [Google Scholar] [CrossRef] [PubMed]
- Weaver, W.M.; Tseng, P.; Kunze, A.; Masaeli, M.; Chung, A.J.; Dudani, J.S.; Kittur, H.; Kulkarni, R.P.; Di Carlo, D. Advances in high-throughput single-cell microtechnologies. Curr. Opin. Biotechnol. 2014, 25, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Sims, C.E.; Allbritton, N.L. Analysis of single mammalian cells on-chip. Lab Chip 2007, 7, 423–440. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Wong, P.K.; Park, J.; Levchenko, A.; Sun, Y. Microengineered platforms for cell mechanobiology. Annu. Rev. Biomed. Eng. 2009, 11, 203–233. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Nguyen, J.; Wei, Y.; Sun, Y. Recent advances in microfluidic techniques for single-cell biophysical characterization. Lab Chip 2013, 13, 2464–2483. [Google Scholar] [CrossRef] [PubMed]
- Lincoln, B.; Erickson, H.M.; Schinkinger, S.; Wottawah, F.; Mitchell, D.; Ulvick, S.; Bilby, C.; Guck, J. Deformability-based flow cytometry. Cytom. Part A 2004, 59, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Guck, J.; Schinkinger, S.; Lincoln, B.; Wottawah, F.; Ebert, S.; Romeyke, M.; Lenz, D.; Erickson, H.M.; Ananthakrishnan, R.; Mitchell, D.; et al. Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys. J. 2005, 88, 3689–3698. [Google Scholar] [CrossRef] [PubMed]
- Gossett, D.R.; Tse, H.T.; Lee, S.A.; Ying, Y.; Lindgren, A.G.; Yang, O.O.; Rao, J.; Clark, A.T.; Di Carlo, D. Hydrodynamic stretching of single cells for large population mechanical phenotyping. Proc. Natl. Acad. Sci. USA 2012, 109, 7630–7635. [Google Scholar] [CrossRef] [PubMed]
- Yaginuma, T.; Oliveira, M.S.; Lima, R.; Ishikawa, T.; Yamaguchi, T. Human red blood cell behavior under homogeneous extensional flow in a hyperbolic-shaped microchannel. Biomicrofluidics 2013, 7, 054110. [Google Scholar] [CrossRef] [PubMed]
- Otto, O.; Rosendahl, P.; Mietke, A.; Golfier, S.; Herold, C.; Klaue, D.; Girardo, S.; Pagliara, S.; Ekpenyong, A.; Jacobi, A.; et al. Real-time deformability cytometry: On-the-fly cell mechanical phenotyping. Nat. Meth. 2015, 12, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Shelby, J.P.; White, J.; Ganesan, K.; Rathod, P.K.; Chiu, D.T. A microfluidic model for single-cell capillary obstruction by Plasmodium falciparum infected erythrocytes. Proc. Natl. Acad. Sci. USA 2003, 100, 14618–14622. [Google Scholar] [CrossRef] [PubMed]
- Rosenbluth, M.J.; Lam, W.A.; Fletcher, D.A. Analyzing cell mechanics in hematologic diseases with microfluidic biophysical flow cytometry. Lab Chip 2008, 8, 1062–1070. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.W.; Li, Q.S.; Lee, G.Y.H.; Kumar, A.P.; Ong, C.N.; Lim, C.T. Deformability study of breast cancer cells using microfluidics. Biomed. Microdevices 2009, 11, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Abkarian, M.; Faivre, M.; Stone, H.A. High-speed microfluidic differential manometer for cellular-scale hydrodynamics. Proc. Natl. Acad. Sci. USA 2006, 103, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Handayani, S.; Chiu, D.T.; Tjitra, E.; Kuo, J.S.; Lampah, D.; Kenangalem, E.; Renia, L.; Snounou, G.; Price, R.N.; Anstey, N.M.; et al. High deformability of Plasmodium vivax-infected red blood cells under microfluidic conditions. J. Infect. Dis. 2009, 199, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Quinn, D.J.; Pivkin, I.; Wong, S.Y.; Chiam, K.H.; Dao, M.; Karniadakis, G.E.; Suresh, S. Combined simulation and experimental study of large deformation of red blood cells in microfluidic systems. Ann. Biomed. Eng. 2011, 39, 1041–1050. [Google Scholar] [CrossRef] [PubMed]
- Diez-Silva, M.; Park, Y.; Huang, S.; Bow, H.; Mercereau-Puijalon, O.; Deplaine, G.; Lavazec, C.; Perrot, S.; Bonnefoy, S.; Feld, M.S.; et al. Pf155/RESA protein influences the dynamic microcirculatory behavior of ring-stage Plasmodium falciparum infected red blood cells. Sci. Rep. 2012, 2, 614. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Reiling, S.J.; Rohrbach, P.; Ma, H. Microfluidic biomechanical assay for red blood cells parasitized by Plasmodium falciparum. Lab Chip 2012, 12, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Undisz, A.; Diez-Silva, M.; Bow, H.; Dao, M.; Han, J. Dynamic deformability of Plasmodium falciparum-infected erythrocytes exposed to artesunate in vitro. Integr. Biol. 2013, 5, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Kwan, J.M.; Guo, Q.; Kyluik-Price, D.L.; Ma, H.; Scott, M.D. Microfluidic analysis of cellular deformability of normal and oxidatively damaged red blood cells. Am. J. Hematol. 2013, 88, 682–689. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Feng, J.J. Simulation of malaria-infected red blood cells in microfluidic channels: Passage and blockage. Biomicrofluidics 2013, 7, 44115. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, S.; Kuroda, K.; Tsai, C.D.; Fukui, W.; Arai, F.; Kaneko, M. Red blood cell fatigue evaluation based on the close-encountering point between extensibility and recoverability. Lab Chip 2014, 14, 1135–1141. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Duffy, S.P.; Matthews, K.; Santoso, A.T.; Scott, M.D.; Ma, H. Microfluidic analysis of red blood cell deformability. J. Biomech. 2014, 47, 1767–1776. [Google Scholar] [CrossRef] [PubMed]
- Myrand-Lapierre, M.-E.; Deng, X.; Ang, R.R.; Matthews, K.; Santoso, A.T.; Ma, H. Multiplexed fluidic plunger mechanism for the measurement of red blood cell deformability. Lab Chip 2015, 15, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Herricks, T.; Antia, M.; Rathod, P.K. Deformability limits of Plasmodium falciparum-infected red blood cells. Cell. Microbiol. 2009, 11, 1340–1353. [Google Scholar] [CrossRef] [PubMed]
- Bow, H.; Pivkin, I.V.; Diez-Silva, M.; Goldfless, S.J.; Dao, M.; Niles, J.C.; Suresh, S.; Han, J. A microfabricated deformability-based flow cytometer with application to malaria. Lab Chip 2011, 11, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.K.; Soriano, A.; Mahadevan, L.; Higgins, J.M.; Bhatia, S.N. A biophysical indicator of vaso-occlusive risk in sickle cell disease. Sci. Transl. Med. 2012, 4, 123ra26. [Google Scholar] [CrossRef] [PubMed]
- Zeng, N.F.; Ristenpart, W.D. Mechanical response of red blood cells entering a constriction. Biomicrofluidics 2014, 8, 064123. [Google Scholar] [CrossRef] [PubMed]
- Gabriele, S.; Benoliel, A.M.; Bongrand, P.; Theodoly, O. Microfluidic investigation reveals distinct roles for actin cytoskeleton and myosin II activity in capillary leukocyte trafficking. Biophys. J. 2009, 96, 4308–4318. [Google Scholar] [CrossRef] [PubMed]
- Sharei, A.; Zoldan, J.; Adamo, A.; Sim, W.Y.; Cho, N.; Jackson, E.; Mao, S.; Schneider, S.; Han, M.-J.; Lytton-Jean, A.; et al. A vector-free microfluidic platform for intracellular delivery. Proc. Natl. Acad. Sci. USA 2013, 110, 2082–2087. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.S.; Vanapalli, S.A. Probing the mechanical properties of brain cancer cells using a microfluidic cell squeezer device. Biomicrofluidics 2013, 7, 11806. [Google Scholar] [CrossRef] [PubMed]
- Mak, M.; Erickson, D. A serial micropipette microfluidic device with applications to cancer cell repeated deformation studies. Integr. Biol. 2013, 5, 1374–1384. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.M.; Liu, A.P. A microfluidic pipette array for mechanophenotyping of cancer cells and mechanical gating of mechanosensitive channels. Lab Chip 2015, 15, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Park, S.; Ma, H. Microfluidic micropipette aspiration for measuring the deformability of single cells. Lab Chip 2012, 12, 2687–2695. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.N.; Chen, D.Y.; Zhao, Y.; Wei, C.; Zhao, X.T.; Yue, W.T.; Long, R.; Wang, J.B.; Chen, J. A constriction channel based microfluidic system enabling continuous characterization of cellular instantaneous Young’s modulus. Sens. Actuat. B Chem. 2014, 202, 1183–1189. [Google Scholar] [CrossRef]
- Lange, J.R.; Steinwachs, J.; Kolb, T.; Lautscham, L.A.; Harder, I.; Whyte, G.; Fabry, B. Microconstriction arrays for high-throughput quantitative measurements of cell mechanical properties. Biophys. J. 2015, 109, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Preira, P.; Grandne, V.; Forel, J.M.; Gabriele, S.; Camara, M.; Theodoly, O. Passive circulating cell sorting by deformability using a microfluidic gradual filter. Lab Chip 2013, 13, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Mak, M.; Reinhart-King, C.A.; Erickson, D. Elucidating mechanical transition effects of invading cancer cells with a subnucleus-scaled microfluidic serial dimensional modulation device. Lab Chip 2013, 13, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Preira, P.; Valignat, M.-P.; Bico, J.; Theodoly, O. Single cell rheometry with a microfluidic constriction: Quantitative control of friction and fluid leaks between cell and channel walls. Biomicrofluidics 2013, 7, 024111. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.H.; Sakuma, S.; Arai, F.; Kaneko, M. A new dimensionless index for evaluating cell stiffness-based deformability in microchannel. IEEE Trans. Bio Med. Eng. 2014, 61, 1187–1195. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, X.T.; Chen, D.Y.; Luo, Y.N.; Jiang, M.; Wei, C.; Long, R.; Yue, W.T.; Wang, J.B.; Chen, J. Tumor cell characterization and classification based on cellular specific membrane capacitance and cytoplasm conductivity. Biosens. Bioelectron. 2014, 57, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, D.; Luo, Y.; Li, H.; Deng, B.; Huang, S.-B.; Chiu, T.-K.; Wu, M.-H.; Long, R.; Hu, H.; et al. A microfluidic system for cell type classification based on cellular size-independent electrical properties. Lab Chip 2013, 13, 2272–2277. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, D.; Li, H.; Luo, Y.; Deng, B.; Huang, S.B.; Chiu, T.K.; Wu, M.H.; Long, R.; Hu, H.; et al. A microfluidic system enabling continuous characterization of specific membrane capacitance and cytoplasm conductivity of single cells in suspension. Biosens. Bioelectron. 2013, 43, 304–307. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xue, C.; Zhao, Y.; Chen, D.; Wu, M.H.; Wang, J. Microfluidic Impedance Flow Cytometry Enabling High-Throughput Single-Cell Electrical Property Characterization. Int. J. Mol. Sci. 2015, 16, 9804–9830. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Ristenpart, W.D.; Stone, H.A. Dynamics of shear-induced ATP release from red blood cells. Proc. Natl. Acad. Sci. USA 2008, 105, 16432–16437. [Google Scholar] [CrossRef] [PubMed]
- Szeto, G.L.; Van Egeren, D.; Worku, H.; Sharei, A.; Alejandro, B.; Park, C.; Frew, K.; Brefo, M.; Mao, S.; Heimann, M.; et al. Microfluidic squeezing for intracellular antigen loading in polyclonal B-cells as cellular vaccines. Sci. Rep. 2015, 5, 10276. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.B.; Zhao, Z.; Chen, D.Y.; Lee, H.C.; Luo, Y.N.; Chiu, T.K.; Wang, J.B.; Chen, J.; Wu, M.H. A clogging-free microfluidic platform with an incorporated pneumatically-driven membrane-based active valve enabling specific membrane capacitance and cytoplasm conductivity characterization of single cells. Sens. Actuat. B Chem. 2014, 190, 928–936. [Google Scholar] [CrossRef]
- Beattie, W.; Qin, X.; Wang, L.; Ma, H. Clog-free cell filtration using resettable cell traps. Lab Chip 2014, 14, 2657–2665. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, D.; Luo, Y.; Chen, F.; Zhao, X.; Jiang, M.; Yue, W.; Long, R.; Wang, J.; Chen, J. Simultaneous characterization of instantaneous Young’s modulus and specific membrane capacitance of single cells using a microfluidic system. Sensors 2015, 15, 2763–2773. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zheng, Y.; Tan, Q.; Shojaei-Baghini, E.; Zhang, Y.L.; Li, J.; Prasad, P.; You, L.; Wu, X.Y.; Sun, Y. Classification of cell types using a microfluidic device for mechanical and electrical measurement on single cells. Lab Chip 2011, 11, 3174–3181. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Shojaei-Baghini, E.; Azad, A.; Wang, C.; Sun, Y. High-throughput biophysical measurement of human red blood cells. Lab Chip 2012, 12, 2560–2567. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, C.; Wang, J.; Zhao, Y.; Chen, D.; Yue, W.; Chen, J. Constriction Channel Based Single-Cell Mechanical Property Characterization. Micromachines 2015, 6, 1794-1804. https://doi.org/10.3390/mi6111457
Xue C, Wang J, Zhao Y, Chen D, Yue W, Chen J. Constriction Channel Based Single-Cell Mechanical Property Characterization. Micromachines. 2015; 6(11):1794-1804. https://doi.org/10.3390/mi6111457
Chicago/Turabian StyleXue, Chengcheng, Junbo Wang, Yang Zhao, Deyong Chen, Wentao Yue, and Jian Chen. 2015. "Constriction Channel Based Single-Cell Mechanical Property Characterization" Micromachines 6, no. 11: 1794-1804. https://doi.org/10.3390/mi6111457
APA StyleXue, C., Wang, J., Zhao, Y., Chen, D., Yue, W., & Chen, J. (2015). Constriction Channel Based Single-Cell Mechanical Property Characterization. Micromachines, 6(11), 1794-1804. https://doi.org/10.3390/mi6111457








