Microfluidics-Based POCT for SARS-CoV-2 Diagnostics
Abstract
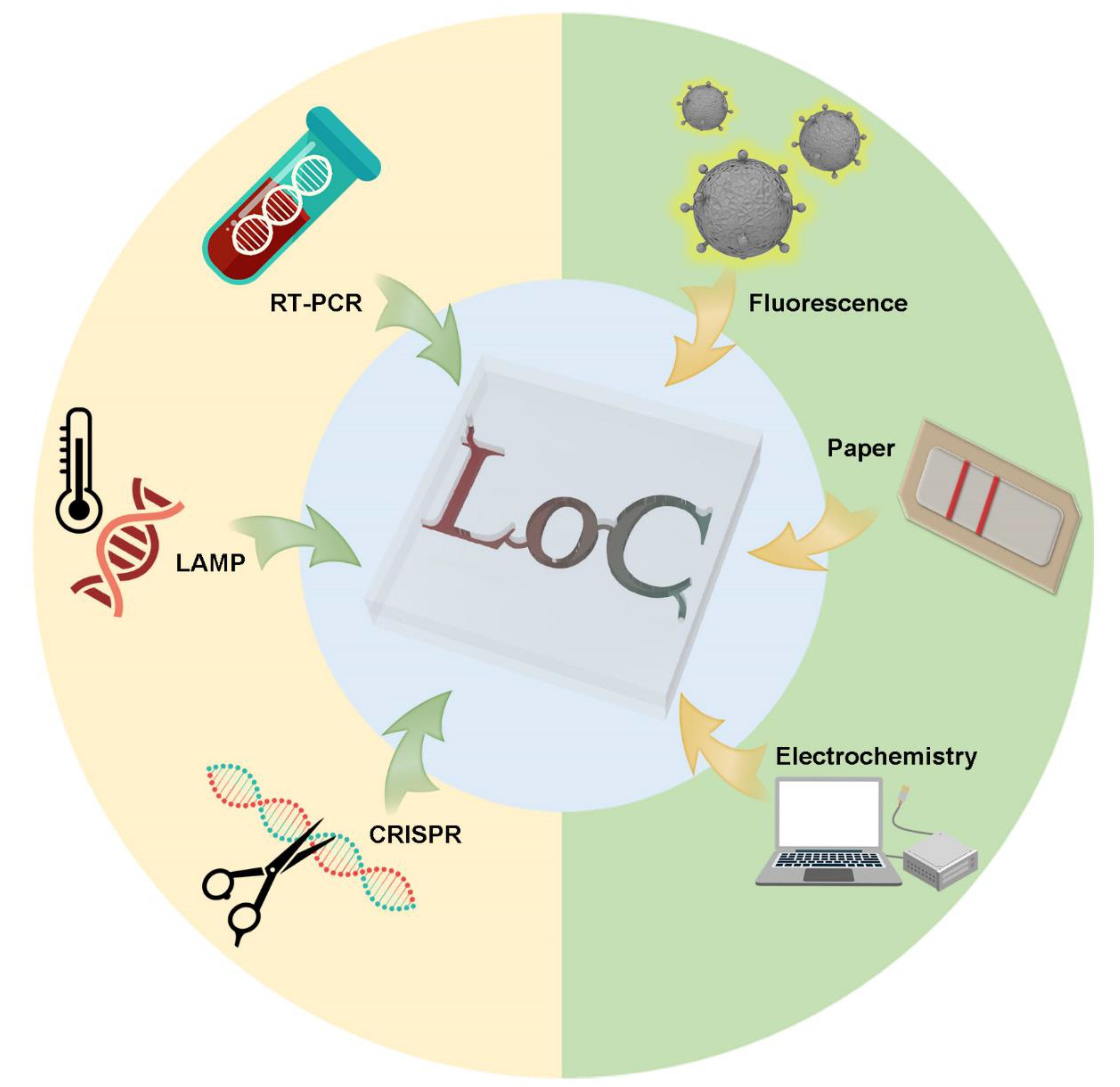
:1. Introduction
2. Nucleic Acid Tests (NAT) Based on Microfluidics
2.1. RT-PCR Tests
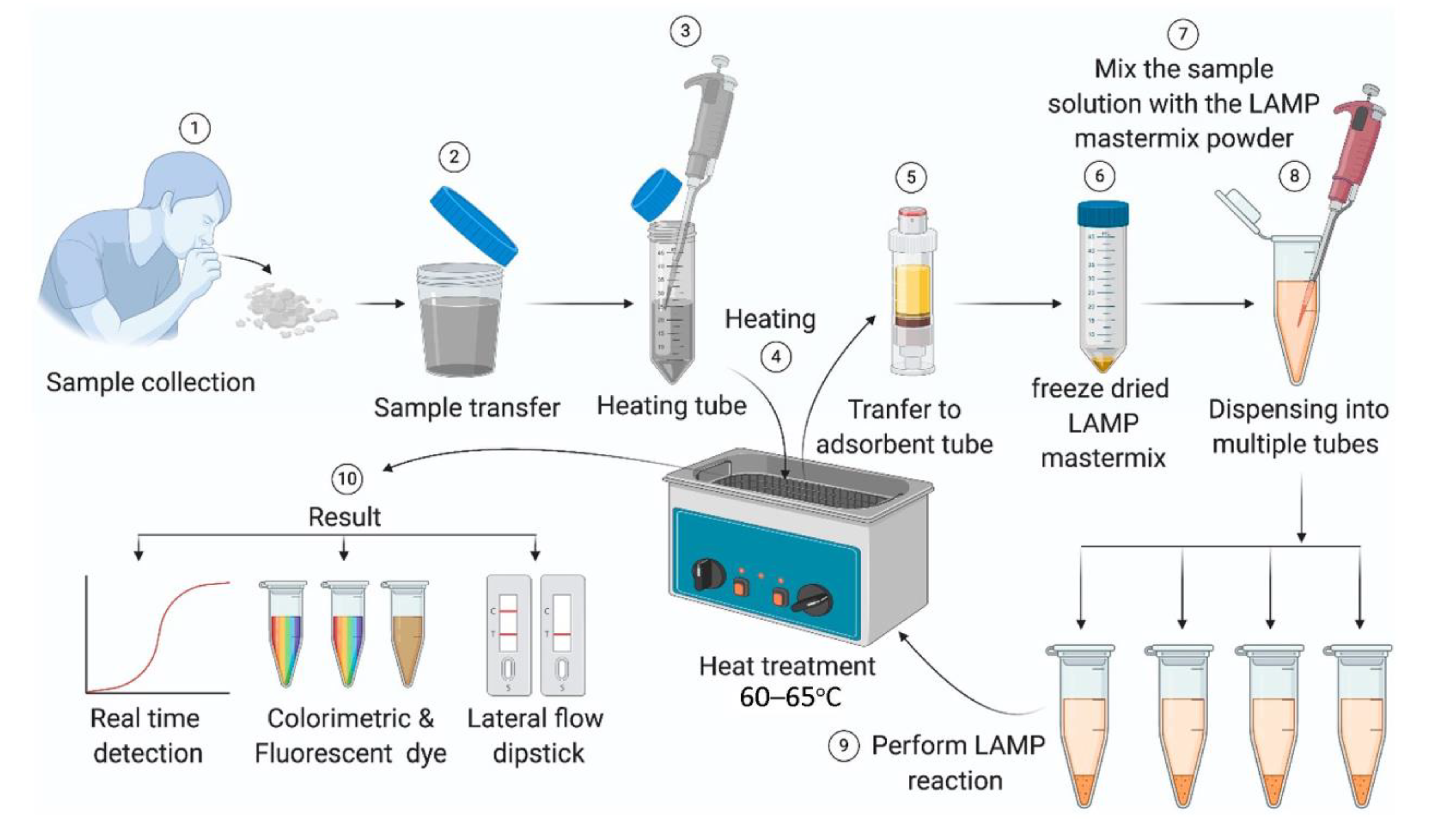
2.2. LAMP Tests
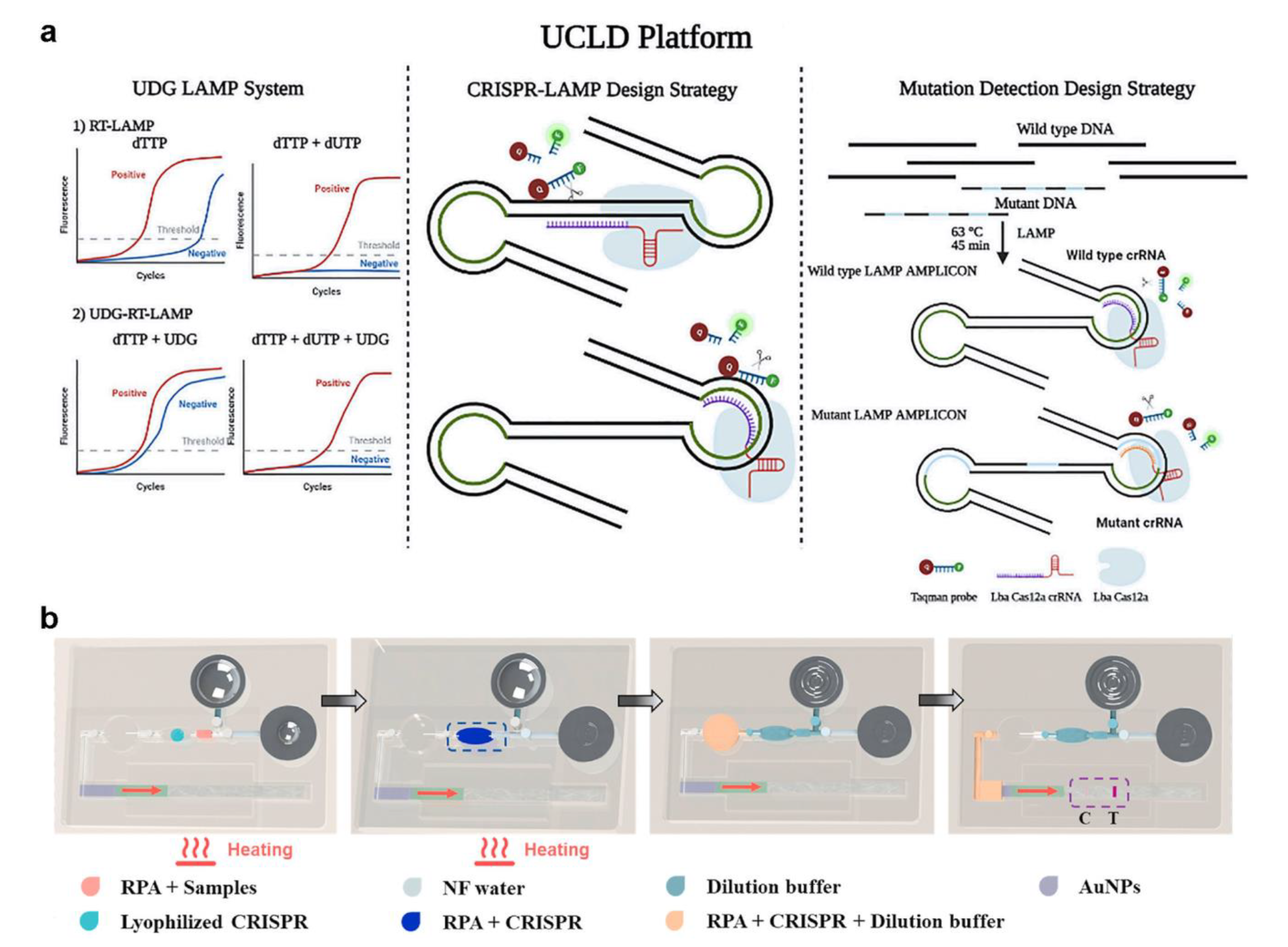
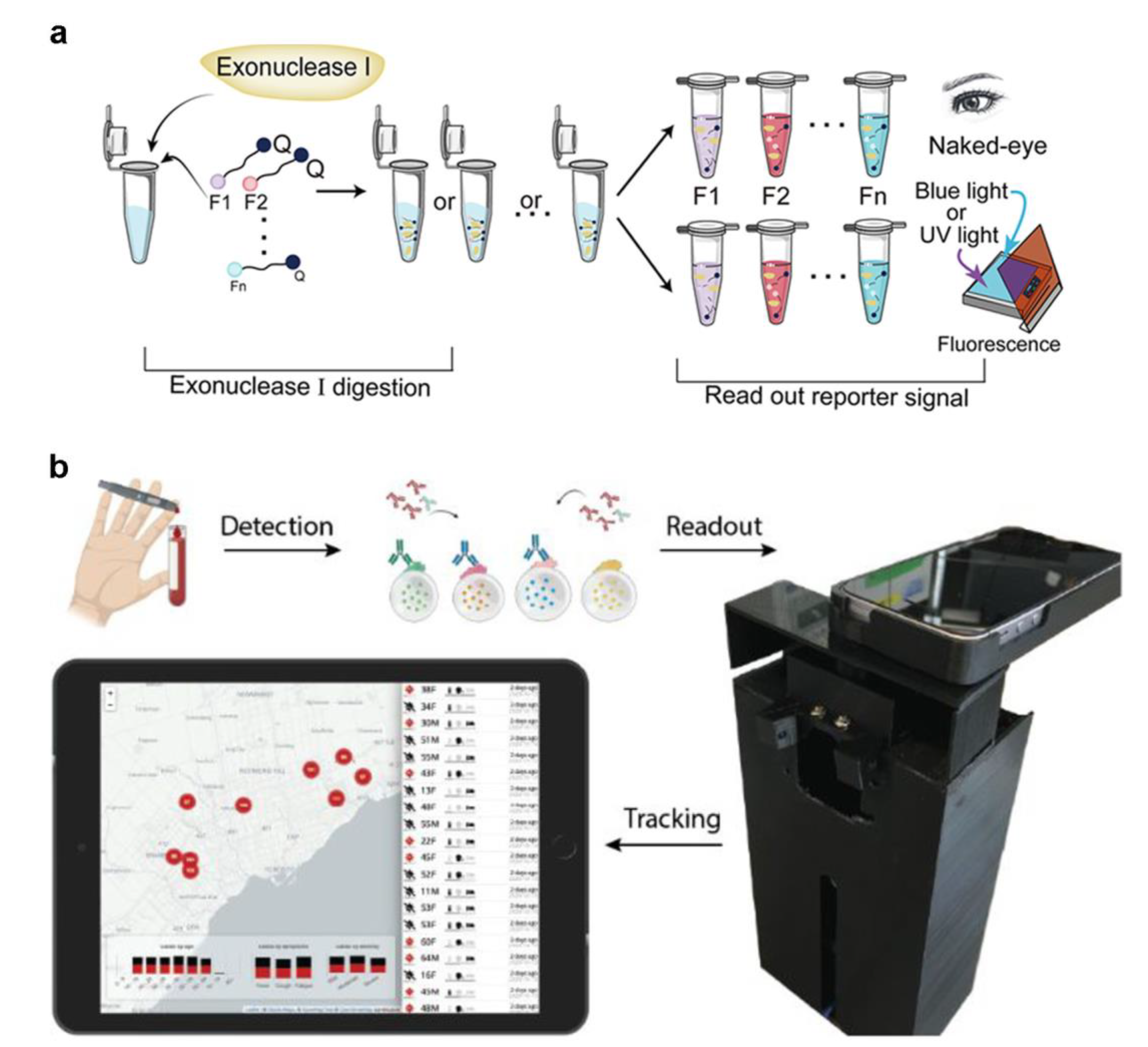
2.3. CRISPR-Associated Proteins System (Cas) Tests
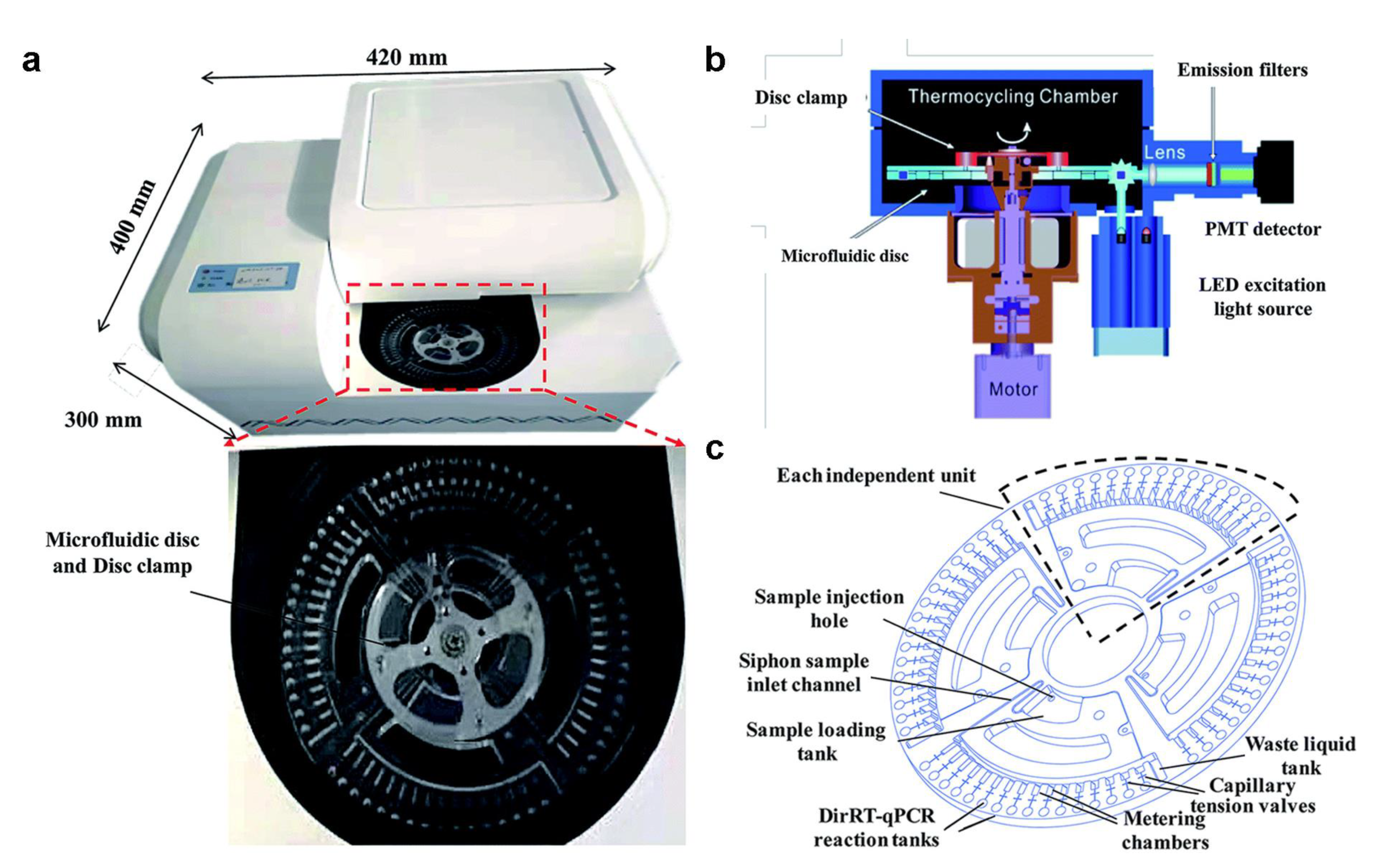
2.4. Commercial NAT Microfluidic Implementation
3. Microfluidics-Based POCT on Immunoassay
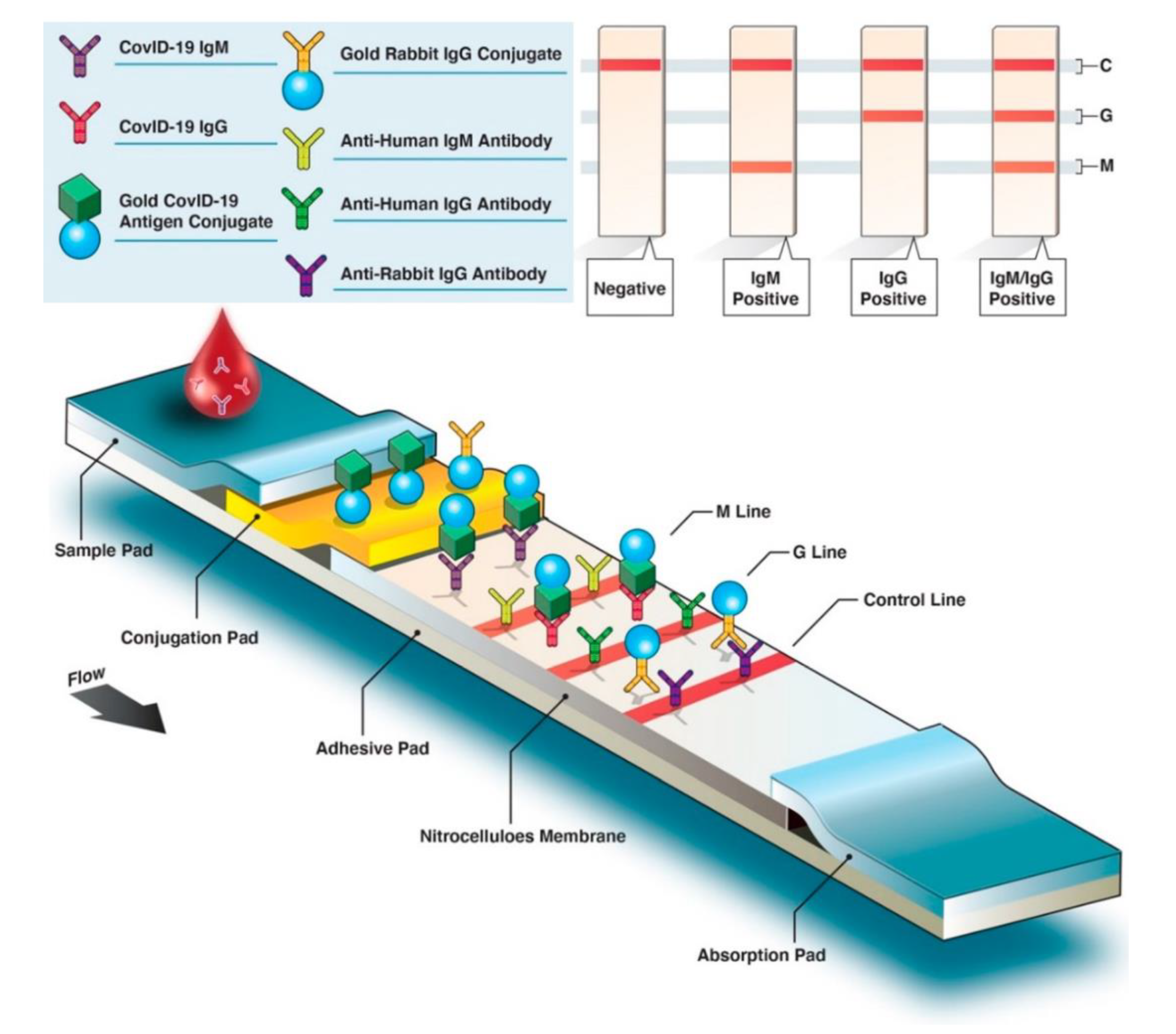
3.1. Fluorescence-Assisted Tests
3.2. Paper-Based Tests
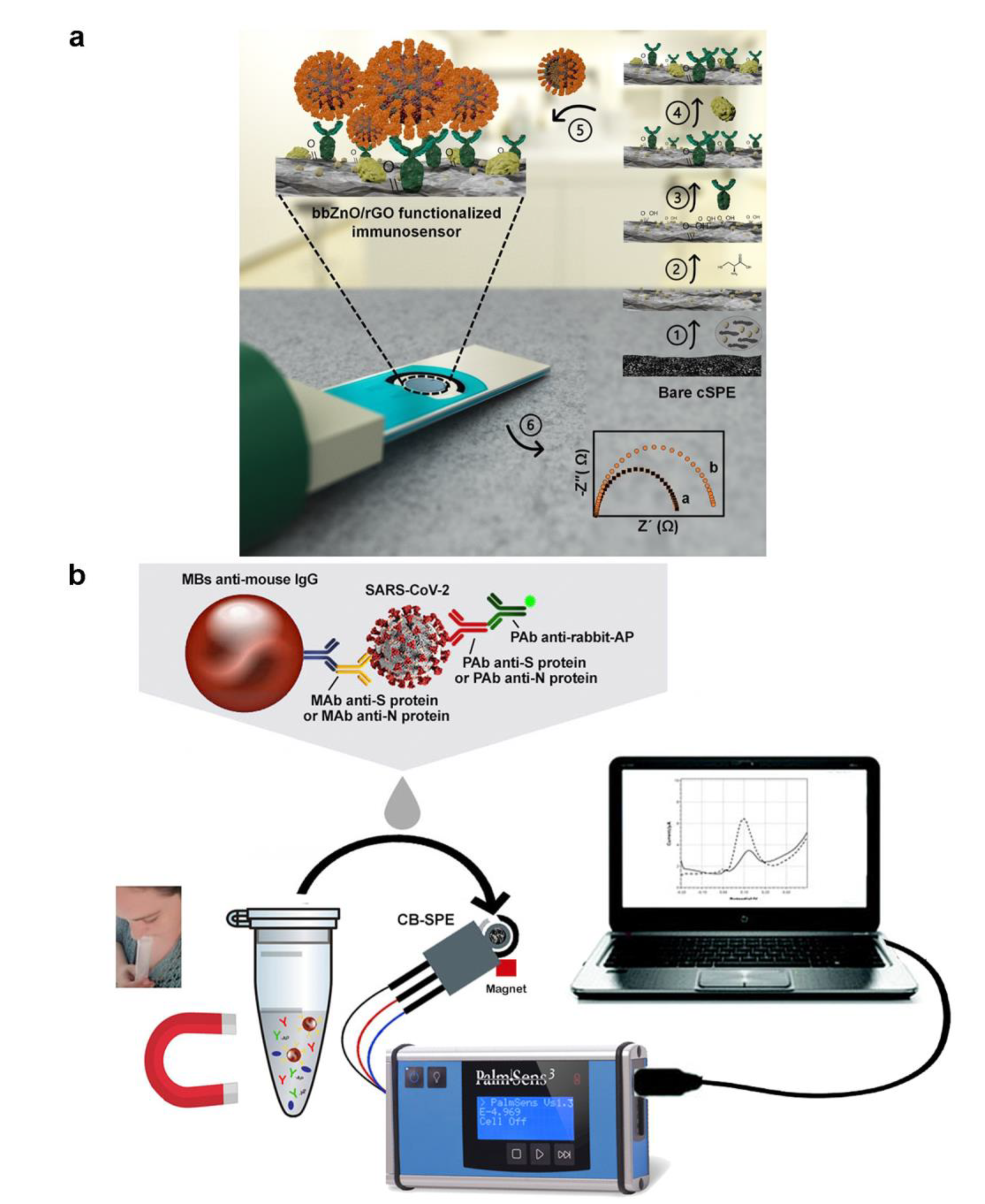
3.3. Electrochemical Biosensors
3.4. Commercial Microfluidics Immunoassay Products
4. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviation
References
- Kim, D.; Lee, J.Y.; Yang, J.S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921. [Google Scholar] [CrossRef]
- Guo, Y.R.; Cao, Q.D.; Hong, Z.S.; Tan, Y.Y.; Chen, S.D.; Jin, H.J.; Tan, K.S.; Wang, D.Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—An update on the status. Military Med. Res. 2020, 7, 11. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.H.; Cai, L.; Cheng, Z.S.; Cheng, H.; Deng, T.; Fan, Y.P.; Fang, C.; Huang, D.; Huang, L.Q.; Huang, Q.; et al. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version). Mil. Med. Res. 2020, 7, 4. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.Y.; Ma, Y.T.; Zhang, J.Y.; Xie, X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020, 17, 259–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Augustin, M.N.; Mbeva, J.B.K. Knowledge, attitudes, and behaviors of healthcare professionals at the start of an Ebola virus epidemic. Infect. Dis. Now 2021, 51, 50–54. [Google Scholar] [CrossRef]
- Sarkar, S. Breaking the chain: Governmental frugal innovation in Kerala to combat the COVID-19 pandemic. Gov. Inf. Q. 2021, 38, 101549. [Google Scholar] [CrossRef]
- Eames, K.T.D.; Keeling, M.J. Contact tracing and disease control. Proc. R. Soc. B-Biol. Sci. 2003, 270, 2565–2571. [Google Scholar] [CrossRef] [Green Version]
- Song, Q.; Sun, X.D.; Dai, Z.Y.; Gao, Y.B.; Gong, X.Q.; Zhou, B.P.; Wu, J.B.; Wen, W.J. Point-of-care testing detection methods for COVID-19. Lab Chip 2021, 21, 1634–1660. [Google Scholar] [CrossRef] [PubMed]
- Mou, L.; Zhang, Y.Y.; Feng, Y.; Hong, H.H.; Xia, Y.; Jiang, X.Y. Multiplexed Lab-on-a-Chip Bioassays for Testing Antibodies against SARS-CoV-2 and Its Variants in Multiple Individuals. Anal. Chem. 2022, 94, 2510–2516. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, Y.; Tang, H.; Zong, N.; Jiang, X.Y. Microfluidics for Biomedical Analysis. Small Methods 2020, 4, 1900451. [Google Scholar] [CrossRef]
- Yang, L.; Yamamoto, T. Quantification of Virus Particles Using Nanopore-Based Resistive-Pulse Sensing Techniques. Front. Microbiol. 2016, 7, 1500. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.L.; Fohlerova, Z.; Pekarek, J.; Basova, E.; Neuzil, P. Recent advances in lab-on-a-chip technologies for viral diagnosis. Biosens. Bioelectron. 2020, 153, 112041. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.F.; Wan, X.H.; Yang, M.Z.; Qian, C.C.; Sohan, A. Wave-shaped microfluidic chip assisted point-of-care testing for accurate and rapid diagnosis of infections. Mil. Med. Res. 2022, 9, 8. [Google Scholar] [CrossRef]
- Yin, B.F.; Qian, C.C.; Wan, X.H.; Sohan, A.; Lin, X.D. Tape integrated self-designed microfluidic chip for point-of-care immunoassays simultaneous detection of disease biomarkers with tunable detection range. Biosens. Bioelectron. 2022, 212, 114429. [Google Scholar] [CrossRef]
- Yin, B.F.; Wan, X.H.; Qian, C.C.; Sohan, A.; Zhou, T.; Yue, W.K. Enzyme Method-Based Microfluidic Chip for the Rapid Detection of Copper Ions. Micromachines 2021, 12, 1380. [Google Scholar] [CrossRef]
- Peeri, N.C.; Shrestha, N.; Rahman, M.S.; Zaki, R.; Tan, Z.Q.; Bibi, S.; Baghbanzadeh, M.; Aghamohammadi, N.; Zhang, W.Y.; Haque, U. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: What lessons have we learned? Int. J. Epidemiol. 2020, 49, 717–726. [Google Scholar] [CrossRef] [Green Version]
- Prakash, S.; Pinti, M.; Bhushan, B. Theory, fabrication and applications of microfluidic and nanofluidic biosensors. Philos. Trans. R. Soc. A-Math. Phys. Eng. Sci. 2012, 370, 2269–2303. [Google Scholar] [CrossRef] [PubMed]
- Mazzu-Nascimento, T.; Donofrio, F.C.; Bianchi, B.C.; Travensolo, R.D.; Souza, J.B.; Moraes, D.A.; Morbioli, G.G.; Segato, T.P.; Varanda, L.C.; Carrilho, E. Paper-Based Microfluidics Immunoassay for Detection of Canine Distemper Virus. Braz. Arch. Biol. Technol. 2017, 60, e17160317. [Google Scholar] [CrossRef]
- Lu, S.M.; Lin, S.; Zhang, H.R.; Liang, L.G.; Shen, S. Methods of Respiratory Virus Detection: Advances towards Point-of-Care for Early Intervention. Micromachines 2021, 12, 697. [Google Scholar] [CrossRef]
- Zhang, S.Y.; Su, X.S.; Wang, J.; Chen, M.Y.; Li, C.Y.; Li, T.D.; Ge, S.X.; Xia, N.S. Nucleic Acid Testing for Coronavirus Disease 2019: Demand, Research Progression, and Perspective. Crit. Rev. Anal. Chem. 2022, 52, 413–424. [Google Scholar] [CrossRef]
- Su, W.T.; Gao, X.H.; Jiang, L.; Qin, J.H. Microfluidic platform towards point-of-care diagnostics in infectious diseases. J. Chromatogr. A 2015, 1377, 13–26. [Google Scholar] [CrossRef]
- Huang, C.L.; Wang, Y.M.; Li, X.W.; Ren, L.L.; Zhao, J.P.; Hu, Y.; Zhang, L.; Fan, G.H.; Xu, J.Y.; Gu, X.Y.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Gupta, N.; Augustine, S.; Narayan, T.; O’Riordan, A.; Das, A.; Kumar, D.; Luong, J.H.T.; Malhotra, B.D. Point-of-Care PCR Assays for COVID-19 Detection. Biosensors 2021, 11, 141. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.F.; Qian, C.C.; Wang, S.B.; Wan, X.H.; Zhou, T. A Microfluidic Chip-Based MRS Immunosensor for Biomarker Detection via Enzyme-Mediated Nanoparticle Assembly. Front. Chem. 2021, 9, 688442. [Google Scholar] [CrossRef]
- Yin, B.F.; Wan, X.H.; Qian, C.C.; Sohan, A.; Wang, S.B.; Zhou, T. Point-of-Care Testing for Multiple Cardiac Markers Based on a Snail-Shaped Microfluidic Chip. Front. Chem. 2021, 9, 741058. [Google Scholar] [CrossRef]
- Yin, B.F.; Yue, W.K.; Sohan, A.; Zhou, T.; Qian, C.C.; Wan, X.H. Micromixer with Fine-Tuned Mathematical Spiral Structures. ACS Omega 2021, 6, 30779–30789. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.B.; Liu, L.Y.; Tu, Y.P.; Zhang, J.; Miao, G.J.; Zhang, L.L.; Ge, S.X.; Xia, N.S.; Yu, D.L.; Qiu, X.B. Rapid PCR powered by microfluidics: A quick review under the background of COVID-19 pandemic. Trac-Trends Anal. Chem. 2021, 143, 116377. [Google Scholar] [CrossRef]
- Basiri, A.; Heidari, A.; Nadi, M.F.; Fallahy, M.T.P.; Nezamabadi, S.S.; Sedighi, M.; Saghazadeh, A.; Rezaei, N. Microfluidic devices for detection of RNA viruses. Rev. Med. Virol. 2021, 31, e2154. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Chung, H.K.; Pireku, P.K.; Beitzel, B.F.; Sanborn, M.A.; Tang, C.Y.; Hammer, R.D.; Ritter, D.; Wan, X.F.; Berry, I.M.; et al. Rapid High-Throughput Whole-Genome Sequencing of SARS-CoV-2 by Using One-Step Reverse Transcription-PCR Amplification with an Integrated Microfluidic System and Next-Generation Sequencing. J. Clin. Microbiol. 2021, 59, e02784-20. [Google Scholar] [CrossRef]
- Turiello, R.; Dignan, L.M.; Thompson, B.; Poulter, M.; Hickey, J.; Chapman, J.; Landers, J.P. Centrifugal Microfluidic Method for Enrichment and Enzymatic Extraction of Severe Acute Respiratory Syndrome Coronavirus 2 RNA. Anal. Chem. 2022, 94, 3287–3295. [Google Scholar] [CrossRef]
- Malic, L.; Brassard, D.; Da Fonte, D.; Nassif, C.; Mounier, M.; Ponton, A.; Geissler, M.; Shiu, M.; Morton, K.J.; Veres, T. Automated sample-to-answer centrifugal microfluidic system for rapid molecular diagnostics of SARS-CoV-2. Lab Chip 2022, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zong, N.; Ye, F.; Mei, Y.; Qu, J.; Jiang, X. Dual-CRISPR/Cas12a-Assisted RT-RAA for Ultrasensitive SARS-CoV-2 Detection on Automated Centrifugal Microfluidics. Anal. Chem. 2022, 94, 9603–9609. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, C.M.; Carthy, E.; Kinahan, D.J. Biosensing on the Centrifugal Microfluidic Lab-on-a-Disc Platform. Processes 2020, 8, 1360. [Google Scholar] [CrossRef]
- Ji, M.H.; Xia, Y.; Loo, J.F.C.; Li, L.; Ho, H.P.; He, J.N.; Gu, D.Y. Automated multiplex nucleic acid tests for rapid detection of SARS-CoV-2, influenza A and B infection with direct reverse-transcription quantitative PCR (dirRT-qPCR) assay in a centrifugal microfluidic platform. RSC Adv. 2020, 10, 34088–34098. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Chu, C.H.; Sarioglu, A.F. Point-of-Care Toolkit for Multiplex Molecular Diagnosis of SARS-CoV-2 and Influenza A and B Viruses. ACS Sens. 2021, 6, 3204–3213. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Yao, X.G.; Zhao, Y.L.; Wu, J.M.; Huang, P.; Pan, C.H.; Liu, S.W.; Pan, C.G. Comparative review of respiratory diseases caused by coronaviruses and influenza A viruses during epidemic season. Microbes Infect. 2020, 22, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Solomon, D.A.; Sherman, A.C.; Kanjilal, S. Influenza in the COVID-19 Era. JAMA-J. Am. Med. Assoc. 2020, 324, 1342–1343. [Google Scholar] [CrossRef]
- Choreno-Parra, J.A.; Jimenez-Alvarez, L.A.; Cruz-Lagunas, A.; Rodriguez-Reyna, T.S.; Ramirez-Martinez, G.; Sandoval-Vega, M.; Hernandez-Garcia, D.L.; Choreno-Parra, E.M.; Balderas-Martinez, Y.I.; Martinez-Sanchez, M.E.; et al. Clinical and Immunological Factors That Distinguish COVID-19 From Pandemic Influenza A(H1N1). Front. Immunol. 2021, 12, 593595. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Long, L.Y.; Zhang, D.T.; Yan, T.T.; Cui, S.J.; Yang, P.; Wang, Q.Y.; Ren, S.M. Potential False-Negative Nucleic Acid Testing Results for Severe Acute Respiratory Syndrome Coronavirus 2 from Thermal Inactivation of Samples with Low Viral Loads. Clin. Chem. 2020, 66, 794–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.Y.; Pei, F.Y.; Ji, M.Y.; Wang, L.; Zhao, H.L.; Li, H.J.; Yang, W.H.; Wang, Q.X.; Zhao, Q.Q.; Wang, Y.S. Sensitivity evaluation of 2019 novel coronavirus (SARS-CoV-2) RT-PCR detection kits and strategy to reduce false negative. PLoS ONE 2020, 15, e0241469. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.J.; Lu, J.M.; Yu, T.; Long, Y.; Liu, G.Z. Advances in nucleic acid amplification techniques (NAATs): COVID-19 point-of-care diagnostics as an example. Biosens. Bioelectron. 2022, 206, 114109. [Google Scholar] [CrossRef] [PubMed]
- Panpradist, N.; Kline, E.C.; Atkinson, R.G.; Roller, M.; Wang, Q.; Hull, I.T.; Kotnik, J.H.; Oreskovic, A.K.; Bennett, C.; Leon, D.; et al. Harmony COVID-19: A ready-to-use kit, low-cost detector, and smartphone app for point-of-care SARS-CoV-2 RNA detection. Sci. Adv. 2021, 7, eabj1281. [Google Scholar] [CrossRef] [PubMed]
- Park, G.S.; Ku, K.; Baek, S.H.; Kim, S.J.; Kim, S.I.; Kim, B.T.; Maeng, J.S. Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays Targeting Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J. Mol. Diagn. 2020, 22, 729–735. [Google Scholar] [CrossRef]
- Jiang, M.H.; Pan, W.H.; Arasthfer, A.; Fang, W.J.; Ling, L.Y.; Fang, H.; Daneshnia, F.; Yu, J.; Liao, W.Q.; Pei, H.; et al. Development and Validation of a Rapid, Single-Step Reverse Transcriptase Loop-Mediated Isothermal Amplification (RT-LAMP) System Potentially to Be Used for Reliable and High-Throughput Screening of COVID-19. Front. Cell. Infect. Microbiol. 2020, 10, 331. [Google Scholar] [CrossRef] [PubMed]
- Augustine, R.; Hasan, A.; Das, S.; Ahmed, R.; Mori, Y.; Notomi, T.; Kevadiya, B.D.; Thakor, A.S. Loop-Mediated Isothermal Amplification (LAMP): A Rapid, Sensitive, Specific, and Cost-Effective Point-of-Care Test for Coronaviruses in the Context of COVID-19 Pandemic. Biology 2020, 9, 182. [Google Scholar] [CrossRef] [PubMed]
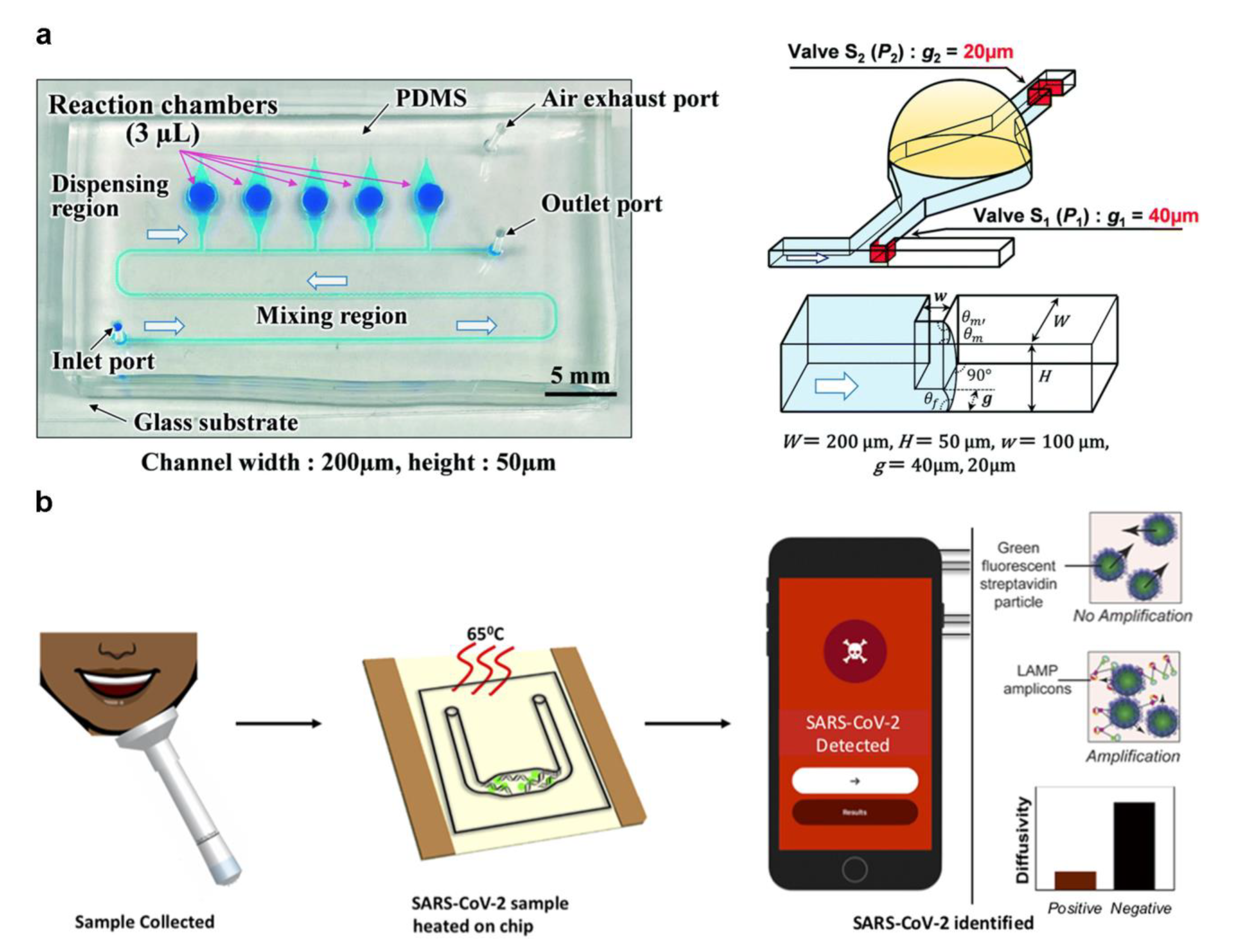
- Natsuhara, D.; Saito, R.; Aonuma, H.; Sakurai, T.; Okamoto, S.; Nagai, M.; Kanuka, H.; Shibata, T. A method of sequential liquid dispensing for the multiplexed genetic diagnosis of viral infections in a microfluidic device. Lab Chip 2021, 21, 4779–4790. [Google Scholar] [CrossRef]
- Lyu, W.Y.; Zhang, J.J.; Yu, Y.; Xu, L.; Shen, F. Slip formation of a high-density droplet array for nucleic acid quantification by digital LAMP with a random-access system. Lab Chip 2021, 21, 3086–3093. [Google Scholar] [CrossRef]
- De Oliveira, K.G.; Estrela, P.F.N.; Mendes, G.D.; dos Santos, C.A.; Silveira-Lacerda, E.D.; Duarte, G.R.M. Rapid molecular diagnostics of COVID-19 by RT-LAMP in a centrifugal polystyrene-toner based microdevice with end-point visual detection. Analyst 2021, 146, 11. [Google Scholar] [CrossRef]
- Colbert, A.J.J.; Lee, D.H.; Clayton, K.N.; Wereley, S.T.; Linnes, J.C.; Kinzer-Ursem, T.L. PD-LAMP smartphone detection of SARS-CoV-2 on chip. Anal. Chim. Acta 2022, 1203, 339702. [Google Scholar] [CrossRef]
- Lim, J.; Stavins, R.; Kindratenko, V.; Baek, J.; Wang, L.Y.; White, K.; Kumar, J.; Valera, E.; King, W.P.; Bashir, R. Microfluidic point-of-care device for detection of early strains and B.1.1.7 variant of SARS-CoV-2 virus. Lab Chip 2022, 22, 1297–1309. [Google Scholar] [CrossRef]
- Nzelu, C.O.; Kato, H.; Peters, N.C. Loop-mediated isothermal amplification (LAMP): An advanced molecular point-of-care technique for the detection of Leishmania infection. PLoS Neglect. Trop. Dis. 2019, 13, e0007698. [Google Scholar] [CrossRef] [Green Version]
- Chaouch, M. Loop-mediated isothermal amplification (LAMP): An effective molecular point-of-care technique for the rapid diagnosis of coronavirus SARS-CoV-2. Rev. Med. Virol. 2021, 31, e2215. [Google Scholar] [CrossRef]
- Anahtar, M.N.; McGrath, G.E.G.; Rabe, B.A.; Tanner, N.A.; White, B.A.; Lennerz, J.K.M.; Branda, J.A.; Cepko, C.L.; Rosenberg, E.S. Clinical Assessment and Validation of a Rapid and Sensitive SARS-CoV-2 Test Using Reverse Transcription Loop-Mediated Isothermal Amplification Without the Need for RNA Extraction. Open Forum Infect. Dis. 2021, 8, ofaa631. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.M.; Mali, P. Therapeutic genome engineering via CRISPR-Cas systems. Wiley Interdiscip. Rev.-Syst. Biol. 2017, 9, e1380. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M.R. Molecular Mechanisms of RNA Targeting by Cas13-containing Type VI CRISPR-Cas Systems. J. Mol. Biol. 2019, 431, 66–87. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Zhao, W.; Zhao, W.; Si, Y.Y.; Chen, N.Z.; Chen, X.; Zhang, X.L.; Fan, L.Y.; Sui, G.D. Universally Stable and Precise CRISPR-LAMP Detection Platform for Precise Multiple Respiratory Tract Virus Diagnosis Including Mutant SARS-CoV-2 Spike N501Y. Anal. Chem. 2021, 93, 16184–16193. [Google Scholar] [CrossRef]
- Ramachandran, A.; Huyke, D.A.; Sharma, E.; Sahoo, M.K.; Huang, C.H.; Banaei, N.; Pinsky, B.A.; Santiago, J.G. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 29518–29525. [Google Scholar] [CrossRef]
- Silva, F.S.R.; Erdogmus, E.; Shokr, A.; Kandula, H.; Thirumalaraju, P.; Kanakasabapathy, M.K.; Hardie, J.M.; Pacheco, L.G.C.; Li, J.Z.; Kuritzkes, D.R.; et al. SARS-CoV-2 RNA Detection by a Cellphone-Based Amplification-Free System with CRISPR/CAS-Dependent Enzymatic (CASCADE) Assay. Adv. Mater. Technol. 2021, 6, 2100602. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ding, X.; Yin, K.; Avery, L.; Ballesteros, E.; Liu, C. Instrument-free, CRISPR-based diagnostics of SARS-CoV-2 using self-contained microfluidic system. Biosens. Bioelectron. 2022, 199, 113865. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, H.; Salehiabar, M.; Barsbay, M.; Ghaffarlou, M.; Kavetskyy, T.; Sharafi, A.; Davaran, S.; Chauhan, S.C.; Danafar, H.; Kaboli, S.; et al. CRISPR Systems for COVID-19 Diagnosis. ACS Sens. 2021, 6, 1430–1445. [Google Scholar] [CrossRef] [PubMed]
- Suea-Ngam, A.; Bezinge, L.; Mateescu, B.; Howes, P.D.; deMello, A.J.; Richards, D.A. Enzyme-Assisted Nucleic Acid Detection for Infectious Disease Diagnostics: Moving toward the Point-of-Care. ACS Sens. 2020, 5, 2701–2723. [Google Scholar] [CrossRef] [PubMed]
- Habimana, J.D.; Huang, R.Q.; Muhoza, B.; Kalisa, Y.N.; Han, X.B.; Deng, W.Y.; Li, Z.Y. Mechanistic insights of CRISPR/Cas nucleases for programmable targeting and early-stage diagnosis: A review. Biosens. Bioelectron. 2022, 203, 114033. [Google Scholar] [CrossRef]
- Spindiag Declares CE-Conformity for Two New Rhonda PCR Rapid Tests for Use at the Point of Care (POC)—Spindiag. Available online: https://www.spindiag.de/ce-conformity-for-two-new-rhonda-pcr-rapid-tests/ (accessed on 15 July 2022).
- ID NOW COVID-19 Testing|Abbott U.S. Available online: https://www.abbott.com/IDNOW.html (accessed on 5 June 2022).
- Credo Diagnostics Biomedical VitaPCR SARS-CoV-2 Assay Garners CE Mark. Credo Diagnostics Biomedical Pte. Ltd. Available online: https://www.credodxbiomed.com/en/news/100-credo-diagnostics-biomedical-vitapcr-sars-cov-2-assay-garners-ce-mark (accessed on 5 June 2022).
- POC Respiratory Panel Test|Respiratory EZ Panel|BioFire Diagnostics. Available online: https://www.biofiredx.com/products/the-filmarray-panels/filmarray-respiratory-panel-ez (accessed on 5 June 2022).
- 1copy COVID-19 qPCR Multi Kit—Instructions for Use. Available online: https://www.fda.gov/media/137935/download (accessed on 5 June 2022).
- TEST COVID-19—Biosynex. Available online: https://www.biosynex.com/en/pharmacie-para-test-covid-19/ (accessed on 5 June 2022).
- Foaming Test—Covid-19 Screening Test Kit|PharmaNona. Available online: https://foamingtest.com/en (accessed on 5 June 2022).
- AQ-TOP COVID-19 Rapid Detection Kit PLUS—Instructions for Use. Available online: https://www.fda.gov/media/142800/download (accessed on 5 June 2022).
- COVID-19 Mutation Multiplex RT-PCR Detection Kit (Lyophilized). Available online: https://www.chkbiotech.com/covid-19-mutation-multiplex-rt-pcr-detection-kit-lyophilized-product/ (accessed on 5 June 2022).
- Zyobio_INSERTO.pdf. Available online: www.bvs.hn/COVID-19/Plataforma/Zyobio_INSERTO.pdf (accessed on 5 June 2022).
- Montesinos, I.; Gruson, D.; Kabamba, B.; Dahma, H.; Van den Wijngaert, S.; Reza, S.; Carbone, V.; Vandenberg, O.; Gulbis, B.; Wolff, F.; et al. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies. J. Clin. Virol. 2020, 128, 104413. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.H.; Kim, M.W.; Park, C.Y.; Choi, C.-S.; Kailasa, S.K.; Park, J.P.; Park, T.J. Development of a rapid and sensitive electrochemical biosensor for detection of human norovirus via novel specific binding peptides. Biosens. Bioelectron. 2019, 123, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.H.; Ma, Y.D.; Fu, C.Y.; Lee, G.B. A structure-free digital microfluidic platform for detection of influenza a virus by using magnetic beads and electromagnetic forces. Lab Chip 2020, 20, 789–797. [Google Scholar] [CrossRef]
- Shi, B.; Li, Y.M.; Wu, D.; Wu, W.M. A handheld continuous-flow real-time fluorescence qPCR system with a PVC microreactor. Analyst 2020, 145, 2767–2773. [Google Scholar] [CrossRef] [PubMed]
- Richards, G.A.; Baker, T.; Amin, P.; Council World Federation, Societies. Ebola virus disease: Report from the task force on tropical diseases by the World Federation of Societies of Intensive and Critical Care Medicine. J. Crit. Care 2018, 43, 352–355. [Google Scholar] [CrossRef]
- Shen, K.M.; Sabbavarapu, N.M.; Fu, C.Y.; Jan, J.T.; Wang, J.R.; Hung, S.C.; Lee, G.B. An integrated microfluidic system for rapid detection and multiple subtyping of influenza A viruses by using glycan-coated magnetic beads and RT-PCR. Lab Chip 2019, 19, 1277–1286. [Google Scholar] [CrossRef] [PubMed]
- Black, J.A.; Hamilton, E.; Hueros, R.A.R.; Parks, J.W.; Hawkins, A.R.; Schmidt, H. Enhanced Detection of Single Viruses On-Chip via Hydrodynamic Focusing. IEEE J. Sel. Top. Quantum Electron. 2019, 25, 7201206. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kang, M.; Park, E.; Chung, D.R.; Kim, J.; Hwang, E.S. A Simple and Multiplex Loop-Mediated Isothermal Amplification (LAMP) Assay for Rapid Detection of SARS-CoV. BioChip J. 2019, 13, 341–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tourlousse, D.M.; Ahmad, F.; Stedtfeld, R.D.; Seyrig, G.; Tiedje, J.M.; Hashsham, S.A. A polymer microfluidic chip for quantitative detection of multiple water- and foodborne pathogens using real-time fluorogenic loop-mediated isothermal amplification. Biomed. Microdevices 2012, 14, 769–778. [Google Scholar] [CrossRef]
- Iswardy, E.; Tsai, T.C.; Cheng, I.F.; Ho, T.C.; Perng, G.C.; Chang, H.C. A bead-based immunofluorescence-assay on a microfluidic dielectrophoresis platform for rapid dengue virus detection. Biosens. Bioelectron. 2017, 95, 174–180. [Google Scholar] [CrossRef]
- Lee, S.H.; Baek, Y.H.; Kim, Y.H.; Choi, Y.K.; Song, M.S.; Ahn, J.Y. One-Pot Reverse Transcriptional Loop-Mediated Isothermal Amplification (RT-LAMP) for Detecting MERS-CoV. Front. Microbiol. 2017, 7, 2166. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.D.; Li, K.H.; Chen, Y.H.; Lee, Y.M.; Chou, S.T.; Lai, Y.Y.; Huang, P.C.; Ma, H.P.; Lee, G.B. A sample-to-answer, portable platform for rapid detection of pathogens with a smartphone interface. Lab Chip 2019, 19, 3804–3814. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Aggarwal, K.; Vinitha, T.U.; Nguyen, T.; Han, J.; Ahn, C.H. A new microchannel capillary flow assay (MCFA) platform with lyophilized chemiluminescence reagents for a smartphone-based POCT detecting malaria. Microsyst. Nanoeng. 2020, 6, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mauk, M.; Song, J.Z.; Bau, H.H.; Gross, R.; Bushman, F.D.; Collman, R.G.; Liu, C.C. Miniaturized devices for point of care molecular detection of HIV. Lab Chip 2017, 17, 382–394. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.C.; Wang, K.; Xu, H.; Yan, W.Q.; Jin, Q.H.; Cui, D.X. Detection platforms for point-of-care testing based on colorimetric, luminescent and magnetic assays: A review. Talanta 2019, 202, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.S.; Tao, D.G.; Fu, Y.H.; Xu, B.R.; Tang, Y.; Steinaa, L.; Hemmink, J.D.; Pan, W.Y.; Huang, X.; Nie, X.W.; et al. Rapid Visual CRISPR Assay: A Naked-Eye Colorimetric Detection Method for Nucleic Acids Based on CRISPR/Cas12a and a Convolutional Neural Network. ACS Synth. Biol. 2022, 11, 383–396. [Google Scholar] [CrossRef]
- Zhang, Y.W.; Malekjahani, A.; Udugama, B.N.; Kadhiresan, P.; Chen, H.M.; Osborne, M.; Franz, M.; Kucera, M.; Plenderleith, S.; Yip, L.; et al. Surveilling and Tracking COVID-19 Patients Using a Portable Quantum Dot Smartphone Device. Nano Lett. 2021, 21, 5209–5216. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.L.; Jiang, H.; Li, X.Q.; Lv, X.F. Recent advances in sensitivity enhancement for lateral flow assay. Microchim. Acta 2021, 188, 379. [Google Scholar] [CrossRef]
- Hristov, D.; Rijal, H.; Gomez-Marquez, J.; Hamad-Schifferli, K. Developing a Paper-Based Antigen Assay to Differentiate between Coronaviruses and SARS-CoV-2 Spike Variants. Anal. Chem. 2021, 93, 7825–7832. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.F.; Kacherovsky, N.; Panpradist, N.; Wan, R.; Liang, J.; Zhang, B.; Salipante, S.J.; Lutz, B.R.; Pun, S.H. Aptamer Sandwich Lateral Flow Assay (AptaFlow) for Antibody-Free SARS-CoV-2 Detection. Anal. Chem. 2022, 94, 7278–7285. [Google Scholar] [CrossRef]
- Boehringer, H.R.; O’Farrell, B.J. Lateral Flow Assays in Infectious Disease Diagnosis. Clin. Chem. 2021, 68, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wen, T.; Shi, F.J.; Zeng, X.Y.; Jiao, Y.J. Rapid Detection of IgM Antibodies against the SARS-CoV-2 Virus via Colloidal Gold Nanoparticle-Based Lateral-Flow Assay. ACS Omega 2020, 5, 12550–12556. [Google Scholar] [CrossRef]
- Chen, R.; Ren, C.P.; Liu, M.; Ge, X.P.; Qu, M.S.; Zhou, X.B.; Liang, M.F.; Liu, Y.; Li, F.Y. Early Detection of SARS-CoV-2 Seroconversion in Humans with Aggregation-Induced Near-Infrared Emission Nanoparticle-Labeled Lateral Flow Immunoassay. ACS Nano 2021, 15, 8996–9004. [Google Scholar] [CrossRef]
- Grant, B.D.; Anderson, C.E.; Williford, J.R.; Alonzo, L.F.; Glukhova, V.A.; Boyle, D.S.; Weigl, B.H.; Nichols, K.P. SARS-CoV-2 Coronavirus Nucleocapsid Antigen-Detecting Half-Strip Lateral Flow Assay Toward the Development of Point of Care Tests Using Commercially Available Reagents. Anal. Chem. 2020, 92, 11305–11309. [Google Scholar] [CrossRef]
- Wen, T.; Huang, C.; Shi, F.J.; Zeng, X.Y.; Lu, T.; Ding, S.N.; Jiao, Y.J. Development of a lateral flow immunoassay strip for rapid detection of IgG antibody against SARS-CoV-2 virus. Analyst 2020, 145, 5345–5352. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.H.; Zhang, Z.G.; Zhai, X.M.; Li, Y.Y.; Lin, L.; Zhao, H.; Bian, L.; Li, P.; Yu, L.; Wu, Y.S.; et al. Rapid and Sensitive Detection of anti-SARS-CoV-2 IgG, Using Lanthanide-Doped Nanoparticles-Based Lateral Flow Immunoassay. Anal. Chem. 2020, 92, 7226–7231. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, A.; Meurant, R.; Ardakani, A. COVID-19 Serological Tests: How Well Do They Actually Perform? Diagnostics 2020, 10, 453. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Nimse, S.B.; Kim, J.; Song, K.S.; Kim, T. Development of a Lateral Flow Strip Membrane Assay for Rapid and Sensitive Detection of the SARS-CoV-2. Anal. Chem. 2020, 92, 14139–14144. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, X.X.; Han, L.M.; Chen, T.; Wang, L.C.; Li, H.; Li, S.; He, L.F.; Fu, X.Y.; Chen, S.J.; et al. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. 2020, 166, 112437. [Google Scholar] [CrossRef]
- Wang, D.M.; He, S.G.; Wang, X.H.; Yan, Y.Q.; Liu, J.Z.; Wu, S.M.; Liu, S.G.; Lei, Y.; Chen, M.; Li, L.; et al. Rapid lateral flow immunoassay for the fluorescence detection of SARS-CoV-2 RNA. Nat. Biomed. Eng 2020, 4, 1150–1158. [Google Scholar] [CrossRef] [PubMed]
- Shaffaf, T.; Forouhi, S.; Ghafar-Zadeh, E. Towards Fully Integrated Portable Sensing Devices for COVID-19 and Future Global Hazards: Recent Advances, Challenges, and Prospects. Micromachines 2021, 12, 915. [Google Scholar] [CrossRef]
- Drobysh, M.; Ramanaviciene, A.; Viter, R.; Ramanavicius, A. Affinity Sensors for the Diagnosis of COVID-19. Micromachines 2021, 12, 390. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, A.K.; Dhau, J.S.; Gohel, H.; Mishra, Y.K.; Kateb, B.; Kim, N.Y.; Goswami, D.Y. Electrochemical SARS-CoV-2 Sensing at Point-of-Care and Artificial Intelligence for Intelligent COVID-19 Management. ACS Appl. Bio Mater. 2020, 3, 7306–7325. [Google Scholar] [CrossRef]
- De Eguilaz, M.R.; Cumba, L.R.; Forster, R.J. Electrochemical detection of viruses and antibodies: A mini review. Electrochem. Commun. 2020, 116, 106762. [Google Scholar] [CrossRef]
- Thiyagarajan, N.; Chang, J.L.; Senthilkumar, K.; Zen, J.M. Disposable electrochemical sensors: A mini review. Electrochem. Commun. 2014, 38, 86–90. [Google Scholar] [CrossRef]
- Beduk, T.; Beduk, D.; de Oliveira, J.I.; Zihnioglu, F.; Cicek, C.; Sertoz, R.; Arda, B.; Goksel, T.; Turhan, K.; Salama, K.N.; et al. Rapid Point-of-Care COVID-19 Diagnosis with a Gold-Nanoarchitecture-Assisted Laser-Scribed Graphene Biosensor. Anal. Chem. 2021, 93, 8585–8594. [Google Scholar] [CrossRef] [PubMed]
- Li, J.R.; Lillehoj, P.B. Microfluidic Magneto Immunosensor for Rapid, High Sensitivity Measurements of SARS-CoV-2 Nucleocapsid Protein in Serum. ACS Sens. 2021, 6, 1270–1278. [Google Scholar] [CrossRef] [PubMed]
- Haghayegh, F.; Salahandish, R.; Hassani, M.; Sanati-Nezhad, A. Highly Stable Buffer-Based Zinc Oxide/Reduced Graphene Oxide Nanosurface Chemistry for Rapid Immunosensing of SARS-CoV-2 Antigens. ACS Appl. Mater. Interfaces 2022, 14, 10844–10855. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Hu, C.S.; Jahan, S.; Yuan, B.; Saleh, M.S.; Ju, E.G.; Gao, S.J.; Panat, R. Sensing of COVID-19 Antibodies in Seconds via Aerosol Jet Nanoprinted Reduced-Graphene-Oxide-Coated 3D Electrodes. Adv. Mater. 2021, 33, 2006647. [Google Scholar] [CrossRef] [PubMed]
- Fabiani, L.; Saroglia, M.; Galata, G.; De Santis, R.; Fillo, S.; Luca, V.; Faggioni, G.; D’Amore, N.; Regalbuto, E.; Salvatori, P.; et al. Magnetic beads combined with carbon black -based screen-printed electrodes for COVID-19: A reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens. Bioelectron. 2021, 171, 112686. [Google Scholar] [CrossRef]
- Yakoh, A.; Pimpitak, U.; Rengpipat, S.; Hirankarn, N.; Chailapakul, O.; Chaiyo, S. Paper-based electrochemical biosensor for diagnosing COVID-19: Detection of SARS-CoV-2 antibodies and antigen. Biosens. Bioelectron. 2021, 176, 112912. [Google Scholar] [CrossRef] [PubMed]
- Alafeef, M.; Dighe, K.; Moitra, P.; Pan, D. Rapid, Ultrasensitive, and Quantitative Detection of SARS-CoV-2 Using Antisense Oligonucleotides Directed Electrochemical Biosensor Chip. ACS Nano 2020, 14, 17028–17045. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Liu, F.; Xie, W.; Zhou, T.C.; OuYang, J.; Jin, L.; Li, H.; Zhao, C.Y.; Zhang, L.; Wei, J.; et al. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sens. Actuator B-Chem. 2021, 327, 128899. [Google Scholar] [CrossRef]
- Torrente-Rodriguez, R.M.; Lukas, H.; Tu, J.B.; Min, J.H.; Yang, Y.R.; Xu, C.H.; Rossiter, H.B.; Gao, W. SARS-CoV-2 RapidPlex: A Graphene-Based Multiplexed Telemedicine Platform for Rapid and Low-Cost COVID-19 Diagnosis and Monitoring. Matter 2020, 3, 19. [Google Scholar] [CrossRef]
- Products Catalog—Biosurfit. Available online: https://www.biosurfit.com/en/products/spinit-covid-antibody/ (accessed on 15 July 2022).
- Schnurra, C.; Reiners, N.; Biemann, R.; Kaiser, T.; Trawinski, H.; Jassoy, C. Comparison of the diagnostic sensitivity of SARS-CoV-2 nucleoprotein and glycoprotein-based antibody tests. J. Clin. Virol. 2020, 129, 104544. [Google Scholar] [CrossRef]
- Antigen test ELISA—AAZ|COVID-19 AAZ. Available online: https://www.covid19aaz.com/en/antigen-test-elisa (accessed on 8 June 2022).
- Gremmels, H.; Winkel, B.M.F.; Schuurman, R.; Rosingh, A.; Rigter, N.A.M.; Rodriguez, O.; Ubijaan, J.; Wensing, A.M.J.; Bonten, M.J.M.; Hofstra, L.M. Real-life validation of the Panbio (TM) COVID-19 antigen rapid test (Abbott) in community-dwelling subjects with symptoms of potential SARS-CoV-2 infection. EClinicalMedicine 2021, 31, 100677. [Google Scholar] [CrossRef] [PubMed]
- SARS-CoV-2 Neutralizing Antibody, Coronavirus, COVID-19, Vaccine, SARS, Rapid Test. Available online: https://www.accubiotech.com/product-accu-tell-lt;sup-gt;-lt;-sup-gt;-sars-cov-2-neutralizing-antibody-rapid-test-cassette.html (accessed on 8 June 2022).
- Actim® SARS-CoV-2—Actim. Available online: https://www.actimtest.com/actim-sars-cov-2/ (accessed on 8 June 2022).
- TestNOW-COVID-19-e-Brochure-NEW. Available online: https://affimedix.com/wp-content/uploads/2021/10/TestNOW-COVID-19-e-Brochure-NEW.pdf (accessed on 30 May 2022).
- 2019-nCoV Antigen Device (Anterior Nasal Swab). Available online: www.amslabs.co.uk (accessed on 17 March 2022).
| Product Name | Manufacturer | Method | Time (min/test) | Target | Sample Type | Sensitivity 1/Specificity 2 | Storage and Stability | Limit of Detection | Reference |
|---|---|---|---|---|---|---|---|---|---|
| ID NOW™ COVID-19 | Abbott Diagnostics Scarborough, Inc. | NEAR | 15 | COVID-19 RdRp gene | Nasal, Throat, and Nasopharyngeal swabs | 93.3%/98.4% | 2–30 °C | 125 genome equivalents per mL | [67] |
| VitaPCR™ SARS-CoV-2 Gen2 Assay | Credo Diagnostics Biomedical Pte. Ltd. | Real-Time RT-PCR | 20 | N1, N2-Nucleocapsid Gene | Nasopharyngeal and Oropharyngeal swabs | 100%/100% | +2 °C/+8 °C | 30 copies/reaction | [68] |
| The BioFire® Respiratory 2.1-EZ (RP2.1- EZ) Panel (EUA) | BioFire Diagnostics, LLC | Nested multiplex PCR | 45 | Spike protein gene, Membrane protein gene | Nasopharyngeal swabs | 97.1%/99.3% | 15–25 °C | 500 copies/L | [69] |
| 1copy COVID-19 qPCR Kit | 1drop Inc | RT-PCR | 22 | E gene for beta coronavirus and the RdRp gene for SARS-CoV-2 | Nasopharyngeal and Oropharyngeal swabs | -/- | >20 °C | 200 copies/mL | [70] |
| Biosynex COVID-19 Ag+ BSS Rapid Test | BIOSYNEX S.A., Switzerland | RT-PCR | 10 | N-protein detection | Nasopharyngeal swab | 97.5%/100% | 2–8 °C | 750 TCID50/mL | [71] |
| Foaming Test | PharmaNona | POC/Near POC | 1 | Nucleic acid | Urine | 92%/89% | - | - | [72] |
| AQ-TOP COVID-9 Rapid Detection Kit PLUS | SEASUN BIOMATERIALS | real-time Hyper RT-PCR | 30 | SARS-CoV-2 Orf1ab and the human RNase P gene | Bronchoalveolar lavage, Mid-turbinate swab, Nasal swab, Nasopharyngeal swab, Oropharyngeal swab, Sputum | -/100% | −20 °C | 1 copy/uL in single reaction | [73] |
| Novel Coronavirus (2019-nCoV) RT-PCR Detection Kit (Lyophilized) | Shanghai Chuangkun Bitech Inc. | RT-PCR | 70 | COVID-19 S gene | Nasopharyngeal swab | 95%/100% | - | 500 copies/uL | [74] |
| SARS-CoV-2 IgM/IgG Antibody Assay Kit | Zybio Inc | Colloidal Gold method | 15 | Serum, Plasma, Whole Blood | IgM 98.67%, IgG 95%/IgM 91.11%, IgG 95% | 4–30 °C | Not Applicable | [75] |
| Product Name | Manufacturer | Method | Time (min) | Sampling | Accuracy 1 | Sensitivity 2 | Specificity 3 | Detection Principle | Reference |
|---|---|---|---|---|---|---|---|---|---|
| ANTIGEN RAPID TEST CASSETTE SARS-CoV-2 (SWAB) | A. Menarini Diagnostics, Italy | Immunoassay | 15 | Nasopharyngeal swab | 98.74% (95%CI: 96.80–99.66%) | 96.72% (95%CI: 88.65–99.60%) | 99.22% (95%CI: 97.21–99.91%) | Nucleocapside protein | [121] |
| COV-QUANTO | AAZ-LMB, France | ELISA | 210 | Plasma, serum | 85% (CI: range: 85–115 (Antigen)) | 93% | 98.4% | Immuno-Antigen | [122] |
| Panbio COVID-19 Ag Rapid Test | Abbott Rapid Diagnostic, Switzerland | Near-POC | 15 | Nasal swab, nasopharyngeal swab | 100% | 98% | 99% | ImmunoAssay-Antigen | [123] |
| ACCU-TELL® SARS-CoV-2 Neutralizing Antibody Cassette | AccuBioTech Co., Ltd., China | Near-POC | 10 | Serum, whole blood | 100% (Neutralizing antibody) | 100% (Neutralizing antibody) | 100% (Neutralizing antibody) | Immuno-Antibody | [124] |
| Flowflex SARS-CoV-2 Antigen Rapid Test | Acon Biotech (Hangzhou) Co., Ltd., China | Lab-based, near-POC/POC | 15 | Nasal swab, nasopharyngeal swab | 98.7% (Nasopharyngeal swab) 98.8% (nasal swab) | 97.1% (Nasal swab) 97.6% (nasopharyngeal Swab) | 99.5% (Nasal swab) 99.4% (nasopharyngeal Swab) | Nucleocapsid protein | [124] |
| Actim SARS-CoV-2 | Actim Oy, Finland | Immunochromatography | 15 | Nasopharyngeal swab | 98% (Ct values < 33) | 100 (Ct values < 25), 98% (Ct values < 30). 96% (Ct values < 33), 88% (Ct values 9.3–39.8) | 100% (Antigen) | Immuno-Antigen | [125] |
| InfectCheck®-COVID-19 IgG/IgM Test | Affimedix, Inc., United States | Immunochromatography | 15 | Plasma, serum, whole blood | 99.8% | 98.3% | 99.7% | Immuno-Antibody | [126] |
| 2019-nCoV Antigen Device (Anterior Nasal Swab) | AMS UK (NI) Ltd., United Kingdom | Immunochromatography | Anterior nasal swab | 98.1% | 96.3% | 99.5% | Nucleocapsid protein | [127] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, B.; Wan, X.; Sohan, A.S.M.M.F.; Lin, X. Microfluidics-Based POCT for SARS-CoV-2 Diagnostics. Micromachines 2022, 13, 1238. https://doi.org/10.3390/mi13081238
Yin B, Wan X, Sohan ASMMF, Lin X. Microfluidics-Based POCT for SARS-CoV-2 Diagnostics. Micromachines. 2022; 13(8):1238. https://doi.org/10.3390/mi13081238
Chicago/Turabian StyleYin, Binfeng, Xinhua Wan, A. S. M. Muhtasim Fuad Sohan, and Xiaodong Lin. 2022. "Microfluidics-Based POCT for SARS-CoV-2 Diagnostics" Micromachines 13, no. 8: 1238. https://doi.org/10.3390/mi13081238
APA StyleYin, B., Wan, X., Sohan, A. S. M. M. F., & Lin, X. (2022). Microfluidics-Based POCT for SARS-CoV-2 Diagnostics. Micromachines, 13(8), 1238. https://doi.org/10.3390/mi13081238








