Highly Sensitive Detection for Mercury Ions Using Graphene Oxide (GO) Sensors
Abstract
1. Introduction
2. Materials and Methods
Reagents and Experiments
3. Preparation of Sensors
4. Detection of Hg2+
4.1. Amino-Immobilized DNA Sensors for Hg2+ Detection
4.2. Amino-Immobilized DNA Sensors for Hg2+ Detection Combined with SYBR Green I
4.3. The Optimization of SYBR Green l Concentration
4.4. The Optimization of DNA Concentration
5. Results
5.1. Experimental Design
5.2. The Optimization of GO Concentration
5.3. Sensitive Detection
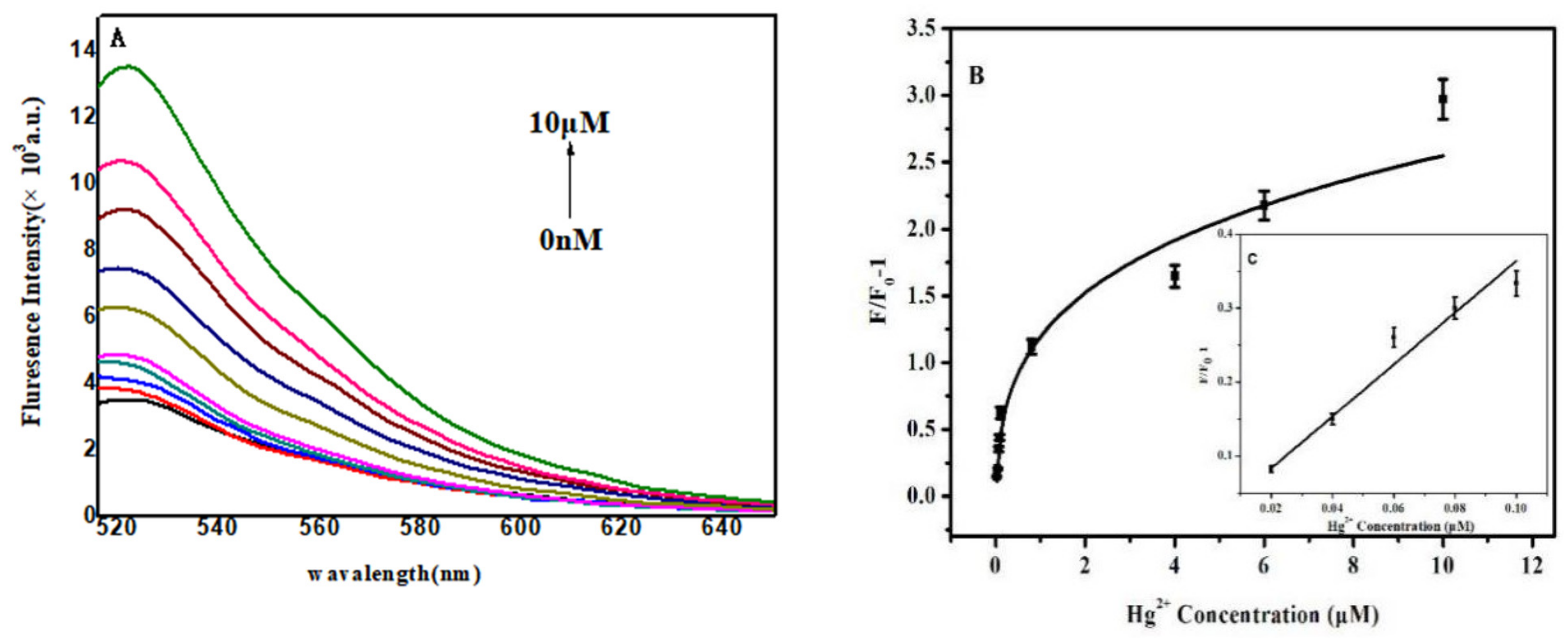
5.4. Construction of FAM-Labeled Single-Stranded DNA Combined with SYBR Green I to Detect Hg2+
5.5. Construction of a FAM-Labeled Double-Stranded Amino-Immobilized DNA Combined with SYBR Green I to Detect Hg2+
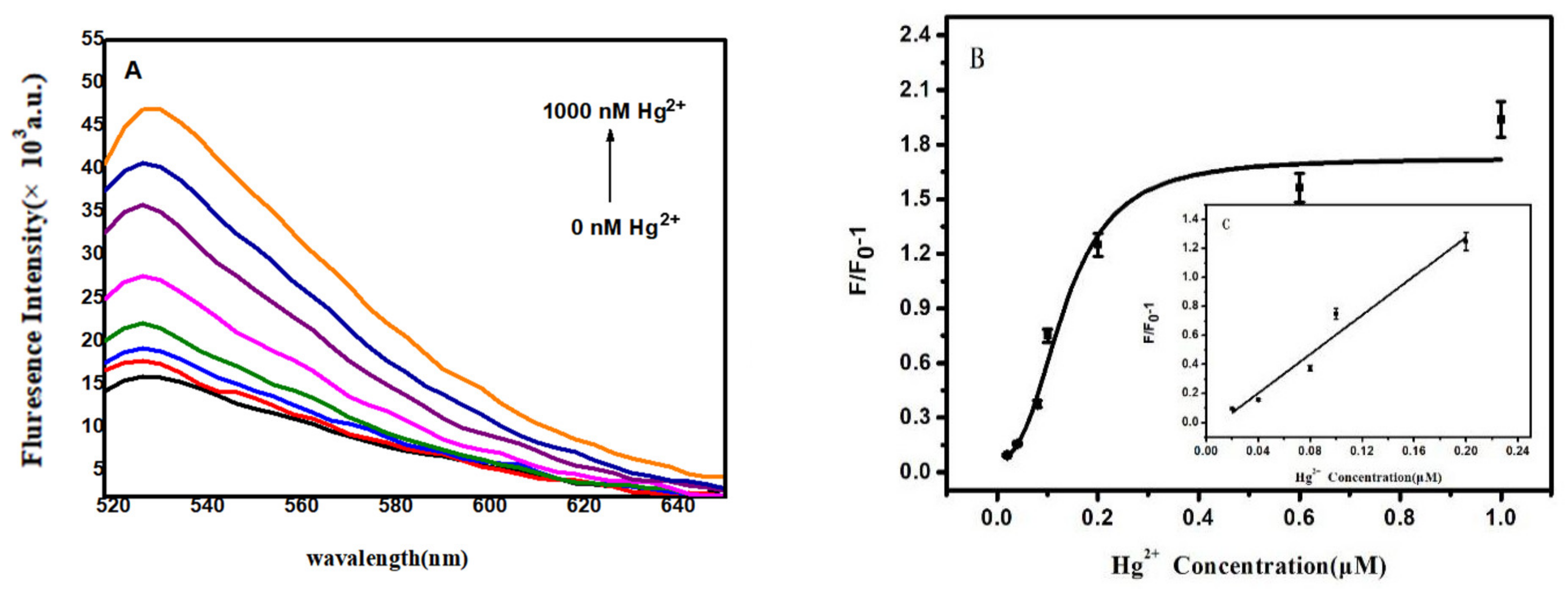
5.5.1. Sensitive Detection
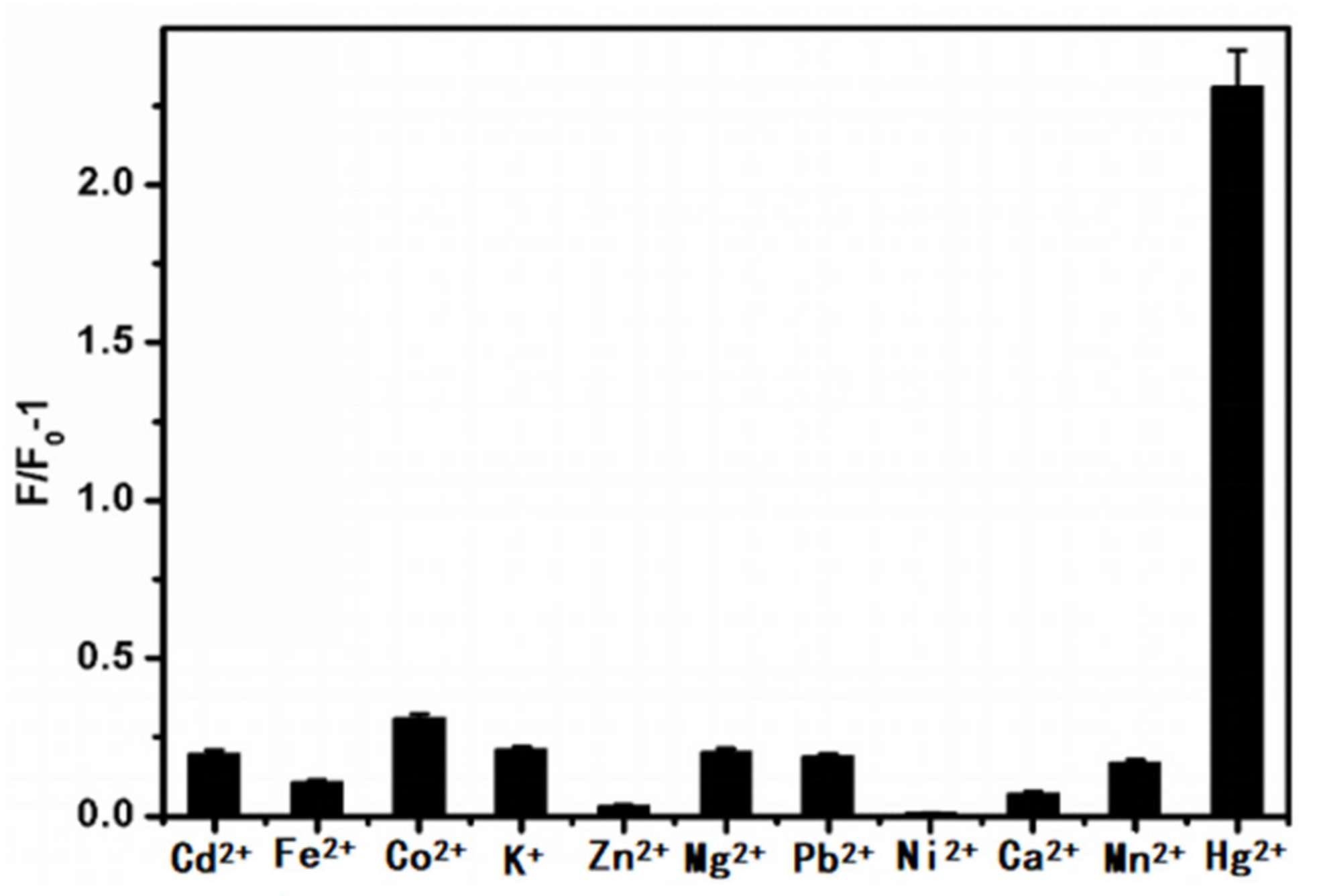
5.5.2. Selective Detection
5.5.3. Determination of Hg2+ in Real Samples
6. Discussions and Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, D.; Wang, S.; Azad, F.; Su, S.C.J.E.; Safety, E. Single-step synthesis of polychromatic carbon quantum dots for macroscopic detection of Hg2+. Ecotoxicol. Environ. Saf. 2020, 190, 110141. [Google Scholar] [CrossRef]
- Hu, J.; Yu, X.; Zhang, X.; Jing, C.; Liu, T.; Hu, X.; Lu, S.; Uvdal, K.; Gao, H.-W.; Hu, Z. Rapid detection of mercury (II) ions and water content by a new rhodamine B-based fluorescent chemosensor. Spectrochim. Acta Part. A Mol. Biomol. Spectrosc. 2020, 241, 118657. [Google Scholar] [CrossRef]
- Yao, L.; Gao, S.; Liu, S.; Bi, Y.; Wang, R.; Qu, H.; Wu, Y.; Mao, Y.; Zheng, L. Single-Atom Enzyme-Functionalized Solution-Gated Graphene Transistor for Real-Time Detection of Mercury Ion. ACS Appl. Mater. Interfaces 2020, 12, 6268–6275. [Google Scholar] [CrossRef]
- Frisbie, S.H.; Mitchell, E.J.; Sarkar, B. Urgent need to reevaluate the latest World Health Organization guidelines for toxic inorganic substances in drinking water. Environ. Health Glob. Access Sci. Sources 2015, 14, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Qu, W.; Chen, W.; Zhang, W.; Wang, Z.; Jiang, X. Highly Sensitive, Colorimetric Detection of Mercury(II) in Aqueous Media by Quaternary Ammonium Group-Capped Gold Nanoparticles at Room Temperature. Anal. Chem. 2010, 82, 9606–9610. [Google Scholar] [CrossRef]
- Hu, B.; Hu, L.-L.; Chen, M.-L.; Wang, J.-H. A FRET ratiometric fluorescence sensing system for mercury detection and intracellular colorimetric imaging in live Hela cells. Biosens. Bioelectron. 2013, 49, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Hsu, W.-H.; Jiang, S.-J.; Sahayam, A. Determination of Cu, As, Hg and Pb in vegetable oils by electrothermal vaporization inductively coupled plasma mass spectrometry with palladium nanoparticles as modifier. Talanta 2013, 117, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Xing, A.; Zhao, K. Simultaneous determination of nickel, cobalt and mercury ions in water samples by solid phase extraction using multiwalled carbon nanotubes as adsorbent after chelating with sodium diethyldithiocarbamate prior to high performance liquid chromatography. J. Chromatogr. A 2014, 1360, 76–81. [Google Scholar] [CrossRef]
- Ono, A.; Togashi, H. Highly Selective Oligonucleotide-Based Sensor for Mercury(II) in Aqueous Solutions. Angew. Chem. Int. Ed. 2004, 43, 4300–4302. [Google Scholar] [CrossRef]
- Miyake, Y.; Togashi, H.; Tashiro, M.; Yamaguchi, H.; Oda, S.; Kudo, M.; Tanaka, Y.; Kondo, Y.; Sawa, R.; Fujimoto, T.; et al. MercuryII-mediated formation of thymine-HgII-thymine base pairs in DNA duplexes. J. Am. Chem. Soc. 2006, 128, 2172–2173. [Google Scholar] [CrossRef]
- Sathishkumar, K.; Narenkumar, J.; Selvi, A.; Murugan, K.; Babujanarthanam, R.; Rajasekar, A. Treatment of soak liquor and bioelectricity generation in dual chamber microbial fuel cell. Environ. Sci. Pollut. Res. 2018, 25, 11424–11430. [Google Scholar] [CrossRef]
- Wang, Y.-W.; Wang, L.; An, F.; Xu, H.; Yin, Z.; Tang, S.; Yang, H.-H.; Song, H. Graphitic carbon nitride supported platinum nanocomposites for rapid and sensitive colorimetric detection of mercury ions. Anal. Chim. Acta 2017, 980, 72–78. [Google Scholar] [CrossRef]
- Xue, X.; Wang, F.; Liu, X. One-Step, Room Temperature, Colorimetric Detection of Mercury (Hg2+) Using DNA/Nanoparticle Conjugates. J. Am. Chem. Soc. 2008, 130, 3244–3245. [Google Scholar] [CrossRef]
- Chen, G.-H.; Chen, W.-Y.; Yen, Y.-C.; Wang, C.-W.; Chang, H.-T.; Chen, C.-F. Detection of Mercury(II) Ions Using Colorimetric Gold Nanoparticles on Paper-Based Analytical Devices. Anal. Chem. 2014, 86, 6843–6849. [Google Scholar] [CrossRef]
- Xiong, E.; Wu, L.; Zhou, J.; Yu, P.; Zhang, X.; Chen, J. A ratiometric electrochemical biosensor for sensitive detection of Hg2+ based on thymine–Hg2+ –thymine structure. Anal. Chim. Acta 2015, 853, 242–248. [Google Scholar] [CrossRef]
- Yang, F.; Zuo, X.; Li, Z.; Deng, W.; Shi, J.; Zhang, G.; Huang, Q.; Song, S.; Fan, C. A Bubble-Mediated Intelligent Microscale Electrochemical Device for Single-Step Quantitative Bioassays. Adv. Mater. 2014, 26, 4671–4676. [Google Scholar] [CrossRef]
- Zhao, Y.; Yamaguchi, Y.; Ni, Y.; Li, M.; Dou, X. A SERS-based capillary sensor for the detection of mercury ions in environmental water. Spectrochim. Acta Part. A Mol. Biomol. Spectrosc. 2020, 233, 118193. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Zhang, H.; Fu, H.; Wei, Y.; Cai, W. Raman reporter-assisted Au nanorod arrays SERS nanoprobe for ultrasensitive detection of mercuric ion (Hg2+) with superior anti-interference performances. J. Hazard. Mater. 2020, 398, 122890. [Google Scholar] [CrossRef] [PubMed]
- Sadrolhosseini, A.R.; Noor, A.S.M.; Bahrami, A.; Lim, H.N.; Talib, Z.A.; Mahdi, M.A. Application of Polypyrrole Multi-Walled Carbon Nanotube Composite Layer for Detection of Mercury, Lead and Iron Ions Using Surface Plasmon Resonance Technique. PLoS ONE 2014, 9, e93962. [Google Scholar] [CrossRef]
- Liu, T.; Dong, J.X.; Liu, S.G.; Li, N.; Lin, S.M.; Fan, Y.Z.; Lei, J.L.; Luo, H.Q.; Li, N.B. Carbon quantum dots prepared with polyethyleneimine as both reducing agent and stabilizer for synthesis of Ag/CQDs composite for Hg2+ ions detection. J. Hazard. Mater. 2016, 322, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Li, X.-P.; Jiang, J.-H.; Yu, R.-Q. A highly sensitive label-free sensor for Mercury ion (Hg2+) by inhibiting thioflavin T as DNA G-quadruplexes fluorescent inducer. Talanta 2014, 122, 85–90. [Google Scholar] [CrossRef]
- Chiang, C.-K.; Huang, C.-C.; Liu, C.-W.; Chang, H.-T. Oligonucleotide-Based Fluorescence Probe for Sensitive and Selective Detection of Mercury(II) in Aqueous Solution. Anal. Chem. 2008, 80, 3716–3721. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Ding, J.; Liu, J. 2-Aminopurine-modified DNA homopolymers for robust and sensitive detection of mercury and silver. Biosens. Bioelectron. 2017, 87, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Xu, L.; Zhou, X.; Qin, J.; Yang, C. Designing label-free DNA sequences to achieve controllable turn-off/on fluorescence response for Hg2+ detection. Chem. Commun. 2011, 47, 8010–8012. [Google Scholar] [CrossRef] [PubMed]
- Zareh Jonaghani, M.; Zali-Boeini, H. Highly selective fluorescent and colorimetric chemosensor for detection of Hg2+ ion in aqueous media. Spectrochim. Acta Part. A Mol. Biomol. Spectrosc. 2017, 178, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Zhang, T.-T.; Zhao, W.; Xu, J.-J.; Chen, H.-Y. A colorimetric/fluorescent dual-mode sensor for ultra-sensitive detection of Hg 2+. Talanta 2017, 165, 570–576. [Google Scholar] [CrossRef]
- Tan, L.; Chen, Z.; Zhang, C.; Wei, X.; Lou, T.; Zhao, Y. Colorimetric Detection of Hg2+ Based on the Growth of Aptamer-Coated AuNPs: The Effect of Prolonging Aptamer Strands. Small 2017, 13. [Google Scholar] [CrossRef]
- Jarzyńska, G.; Falandysz, J. The determination of mercury in mushrooms by CV-AAS and ICP-AES techniques. J. Environ. Sci. Health Part. A 2011, 46, 569–573. [Google Scholar] [CrossRef]
- Huang, J.; Wang, Z.; Kim, J.-K.; Su, X.; Li, Z. Detecting Arbitrary DNA Mutations Using Graphene Oxide and Ethidium Bromide. Anal. Chem. 2015, 87, 12254–12261. [Google Scholar] [CrossRef]
- Wang, J.; Liu, B. Highly sensitive and selective detection of Hg2+ in aqueous solution with mercury-specific DNA and Sybr Green, I. Chem. Commun. 2008, 4759–4761. [Google Scholar] [CrossRef]
- Li, H.; Zhai, J.; Tian, J.; Luo, Y.; Sun, X. Carbon nanoparticle for highly sensitive and selective fluorescent detection of mercury(II) ion in aqueous solution. Biosens. Bioelectron. 2011, 26, 4656–4660. [Google Scholar] [CrossRef] [PubMed]
- Weng, S.; Wu, N.; Qiu, H.; Zheng, Y.; Chen, M.; Weng, S.; Chen, Y.; Lin, X. A robust and versatile signal-on fluorescence sensing strategy based on SYBR Green I dye and graphene oxide. Int. J. Nanomed. 2014, 10, 147–156. [Google Scholar] [CrossRef][Green Version]
- Gao, L.; Liu, C.; Li, R.; Xia, N.; Xiong, Y. Highly sensitive detection of 2+ using covalent linking single-strand DNA to the surface of graphene oxide with co-anchor strand. Anal. Methods 2019, 11, 4416–4420. [Google Scholar] [CrossRef]
- Hummers, W.S., Jr.; Offeman, R.E. Preparation of Graphitic Oxide. ACS Publ. 1958, 208, 1334–1339. [Google Scholar] [CrossRef]
- Gao, L.; Li, Q.; Li, R.; Deng, Z.; Brady, B.; Xia, N.; Chen, G.; Zhou, Y.; Xia, H.; Chen, K.; et al. Protein determination using graphene oxide-aptamer modified gold nanoparticles in combination with Tween 80. Anal. Chim. Acta 2016, 941, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Tu, J.; Gan, Y.; Liang, T.; Hu, Q.; Wang, Q.; Ren, T.; Sun, Q.; Wan, H.; Wang, P. Graphene FET Array Biosensor Based on ssDNA Aptamer for Ultrasensitive Hg2+ Detection in Environmental Pollutants. Front. Chem. 2018, 6, 333. [Google Scholar] [CrossRef]
- Guo, H.; Li, J.; Li, Y.; Wu, D.; Ma, H.; Wei, Q.; Du, B. A turn-on fluorescent sensor for Hg2+ detection based on graphene oxide and DNA aptamers. New J. Chem. 2018, 42, 11147–11152. [Google Scholar] [CrossRef]
- Chang, D.; Li, L.; Shi, L.; Yang, Y. Hg2+ detection, pH sensing and cell imaging based on bright blue-fluorescent N-doped carbon dots. Analisys 2020, 145, 8030–8037. [Google Scholar] [CrossRef]
- Anichina, J.; Dobrusin, Z.; Bohme, D.K. Detection of T-T Mismatches Using Mass Spectrometry: Specific Interactions of Hg(II) with Oligonucleotides Rich in Thymine (T). J. Phys. Chem. B 2010, 114, 15106–15112. [Google Scholar] [CrossRef]
- Chen, J.; Liu, Y.; Ye, T.; Xiang, X.; Ji, X.; He, Z. A novel droplet dosing strategy-based versatile microscale biosensor for detection of DNA, protein and ion. Sensors Actuators B Chem. 2015, 215, 206–214. [Google Scholar] [CrossRef]
- Chen, K.; Lu, G.; Chang, J.; Mao, S.; Yu, K.; Cui, S.; Chen, J. Hg(II) Ion Detection Using Thermally Reduced Graphene Oxide Decorated with Functionalized Gold Nanoparticles. Anal. Chem. 2012, 84, 4057–4062. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, F.; Hormozi-Nezhad, M.R.; Mahmoudi, M. A new strategy to design colorful ratiometric probes and its application to fluorescent detection of Hg(II). Sens. Actuators B-Chem. 2018, B259, 894–899. [Google Scholar] [CrossRef]
- He, L.; Lu, Y.; Wang, F.; Gao, X.; Chen, Y.; Liu, Y. Bare eye detection of Hg(II) ions based on enzyme inhibition and using mercaptoethanol as a reagent to improve selectivity. Microchim. Acta 2018, 185, 174. [Google Scholar] [CrossRef] [PubMed]


| Sensing Method | Linear Range | Detection Limit (LOD) | References |
|---|---|---|---|
| Fluorescence | 0–1.0 μM | 0.05 μM | [1] |
| Fluorescence | 15–30 μM | 16 nM | [2] |
| Fluorescence | 0 Μm–400 μM | 0.124 μM | [38] |
| Colorimetric | 0–60 nM | 9.6 nM | [39] |
| 10–150 nM | 4.5 nM | ||
| 5–40 nM | 3.0 nM | ||
| Colorimetric | 25–750 nM | 50 nM | [27] |
| Surface plasmon resonance | - | 0.5 μM | [40] |
| Field effect transistor (FET) | - | 1 nM | [3] |
| Field effect transistor (FET) | 25 nM–14.2 μM | 25 nM | [20] |
| Fluorescence | 10 nM–1.4 μM | 4.6 nM | [41] |
| Colorimetric | 25–40 nM | 5 nM | [42] |
| Colorimetric | - | 0.316 μM | [43] |
| Fluorescence | 0.02–0.10 μM | 0.43 nM | This study |
| Samples | Added (nM) | Found (nM) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| 1 | 10 | 8.9 | 89 | 3.018 |
| 2 | 100 | 97.53 | 97 | 2.537 |
| 3 | 1000 | 1106.48 | 110 | 3.249 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Shi, H.; Li, R.; Liu, C.; Cheng, J.; Gao, L. Highly Sensitive Detection for Mercury Ions Using Graphene Oxide (GO) Sensors. Micromachines 2021, 12, 1070. https://doi.org/10.3390/mi12091070
Liu L, Shi H, Li R, Liu C, Cheng J, Gao L. Highly Sensitive Detection for Mercury Ions Using Graphene Oxide (GO) Sensors. Micromachines. 2021; 12(9):1070. https://doi.org/10.3390/mi12091070
Chicago/Turabian StyleLiu, Lei, Haixia Shi, Raoqi Li, Cheng Liu, Jia Cheng, and Li Gao. 2021. "Highly Sensitive Detection for Mercury Ions Using Graphene Oxide (GO) Sensors" Micromachines 12, no. 9: 1070. https://doi.org/10.3390/mi12091070
APA StyleLiu, L., Shi, H., Li, R., Liu, C., Cheng, J., & Gao, L. (2021). Highly Sensitive Detection for Mercury Ions Using Graphene Oxide (GO) Sensors. Micromachines, 12(9), 1070. https://doi.org/10.3390/mi12091070





