Turntable Paper-Based Device to Detect Escherichia coli
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Suspensions Preparation
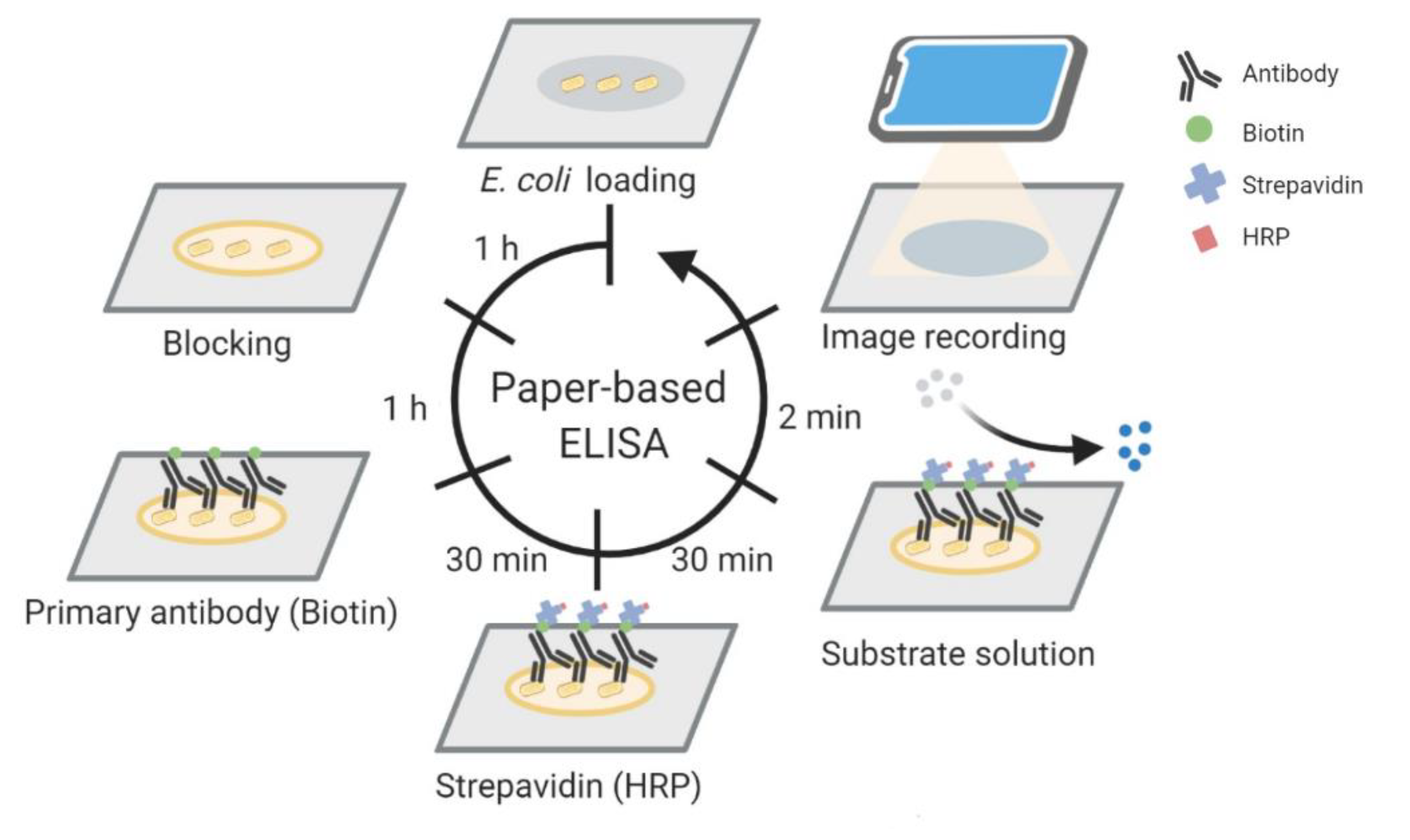
2.2. Paper-Based ELISA
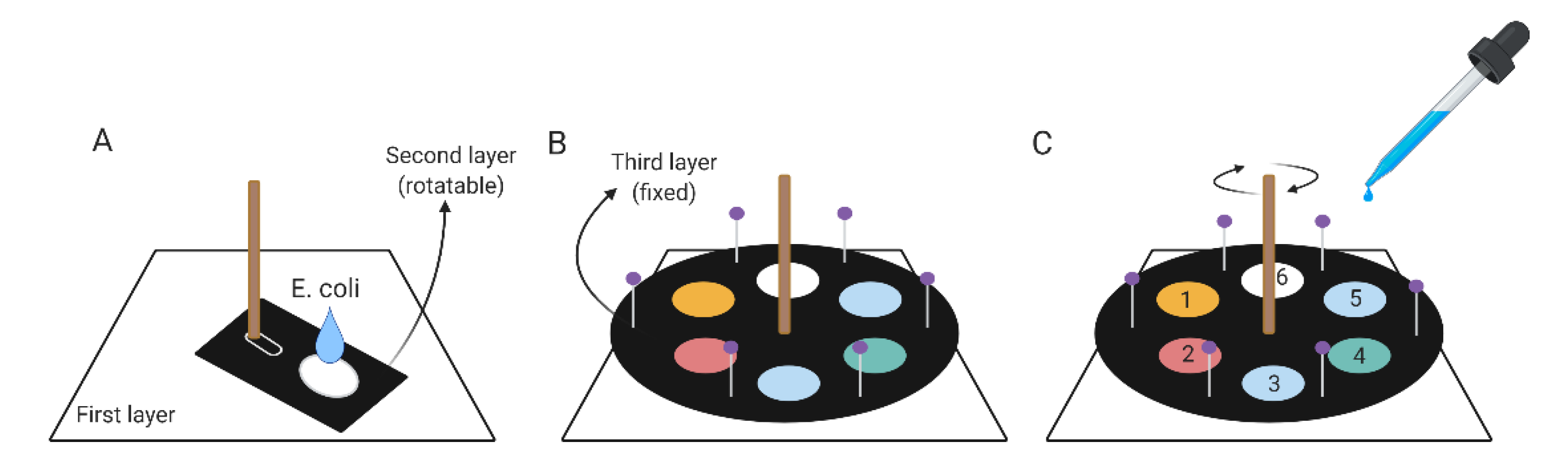
2.3. Components of the Turntable Paper-Based Device
2.4. Turntable Paper-Based Device Operating Protocol
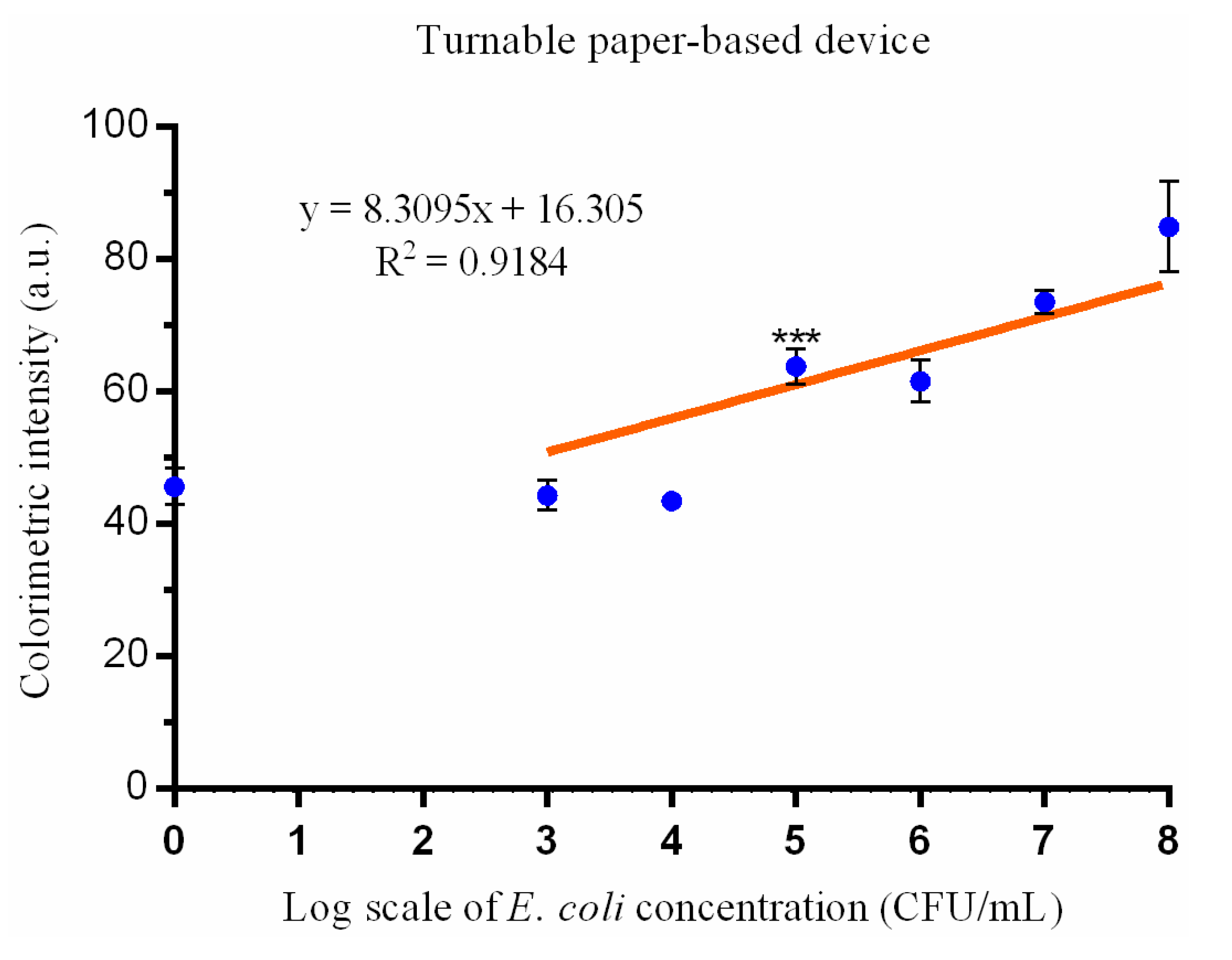
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lee, C.H.; Lee, Y.T.; Kung, C.H.; Ku, W.W.; Kuo, S.C.; Chen, T.L.; Fung, C.P. Risk factors of community-onset urinary tract infections caused by plasmid-mediated AmpC β-lactamase-producing Enterobacteriaceae. J. Microbiol. Immunol. Infect. 2015, 48, 269–275. [Google Scholar] [CrossRef]
- Olsvik, O.; Wasteson, Y.; Lund, A.; Hornes, E. Pathogenic Escherichia coli found in food. Int. J. Food Microbiol. 1991, 12, 103–113. [Google Scholar] [CrossRef]
- Lepelletier, D.; Caroff, N.; Reynaud, A.; Riehet, H. Escherichia coli: Epidemiology and analysis of risk factors for infections caused by resistant strains. Clin. Infect. Dis. 1999, 29, 548–552. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kuo, S.C.; Huang, W.C.; Wang, H.Y.; Shiau, Y.R.; Cheng, M.F.; Lauderdale, T.L. Colistin resistance gene mcr-1 in Escherichia coli isolates from humans and retail meats, Taiwan. J. Antimicrob. Chemother. 2016, 71, 2327–2329. [Google Scholar] [CrossRef]
- De Backer, D.; Dorman, T. Surviving Sepsis Guidelines: A continuous move toward better care of patients with sepsis. JAMA 2017, 317, 807–808. [Google Scholar] [CrossRef]
- Bauer, K.A.; Perez, K.K.; Forrest, G.N.; Goff, D.A. Review of rapid diagnostic tests used by antimicrobial stewardship programs. Clin. Infect. Dis. 2014, 59, S134–S145. [Google Scholar] [CrossRef] [PubMed]
- Kozel, T.R.; Burnham-Marusich, A.R. Point-of-Care testing for infectious diseases: Past, present, and future. J. Clin. Microbiol. 2017, 55, 2313–2320. [Google Scholar] [CrossRef]
- Chen, H.; Liu, K.; Li, Z.; Wang, P. Point of care testing for infectious diseases. Clin. Chim. Acta 2019, 493, 138–147. [Google Scholar] [CrossRef]
- Pai, N.P.; Vadnais, C.; Denkinger, C.; Engel, N.; Pai, M. Point-of-care testing for infectious diseases: Diversity, complexity, and barriers in low- and middle-income countries. PLoS Med. 2012, 9, e1001306. [Google Scholar] [CrossRef] [PubMed]
- Payne, W.J.; Marshall, D.L.; Shockley, R.K.; Martin, W.J. Clinical laboratory applications of monoclonal antibodies. Clin. Microbiol. Rev. 1988, 1, 313–329. [Google Scholar] [CrossRef]
- Wright, P.F.; Nilsson, E.; Van Rooij, E.M.; Lelenta, M.; Jeggo, M.H. Standardisation and validation of enzyme-linked immunosorbent assay techniques for the detection of antibody in infectious disease diagnosis. Rev. Sci. Tech. 1993, 12, 435–450. [Google Scholar] [CrossRef]
- Sakamoto, S.; Putalun, W.; Vimolmangkang, S.; Phoolcharoen, W.; Shoyama, Y.; Tanaka, H.; Morimoto, S. Enzyme-linked immunosorbent assay for the quantitative/qualitative analysis of plant secondary metabolites. J. Nat. Med. 2018, 72, 32–42. [Google Scholar] [CrossRef]
- Shih, C.M.; Chang, C.L.; Hsu, M.Y.; Lin, J.Y.; Kuan, C.M.; Wang, H.K.; Huang, C.T.; Chung, M.C.; Huang, K.C.; Hsu, C.E.; et al. Paper-based ELISA to rapidly detect Escherichia coli. Talanta 2015, 145, 2–5. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, A.; Evans, L.E.; Alhazzani, W.; Levy, M.M.; Antonelli, M.; Ferrer, R.; Kumar, A.; Sevransky, J.E.; Sprung, C.L.; Nunnally, M.E.; et al. Surviving Sepsis Campaign: International guidelines for management of sepsis and septic shock: 2016. Intensive Care Med. 2017, 43, 304–377. [Google Scholar] [CrossRef]
- Wang, P.; Kricka, L.J. Current and emerging trends in point-of-care technology and strategies for clinical validation and implementation. Clin. Chem. 2018, 64, 1439–1452. [Google Scholar] [CrossRef] [PubMed]
- Aydin, S. A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides 2015, 72, 4–15. [Google Scholar] [CrossRef]
- Adel Ahmed, H.; Azzazy, H.M. Power-free chip enzyme immunoassay for detection of prostate specific antigen (PSA) in serum. Biosens. Bioelectron. 2013, 49, 478–484. [Google Scholar] [CrossRef]
- Costantini, F.; Sberna, C.; Petrucci, G.; Manetti, C.; de Cesare, G.; Nascetti, A.; Caputo, D. Lab-on-chip system combining a microfluidic-ELISA with an array of amorphous silicon photosensors for the detection of celiac disease epitopes. Sens. Bio-Sens. Res. 2015, 6, 51–58. [Google Scholar] [CrossRef]
- Thiha, A.; Ibrahim, F. A colorimetric enzyme-linked immunosorbent assay (ELISA) detection platform for a point-of-care dengue detection system on a lab-on-compact-disc. Sensors 2015, 15, 11431–11441. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, Y.; Bernard, P.E.; Reuben, J.M.; Ueno, N.T.; Arlinghaus, R.B.; Zu, Y.; Qin, L. Multiplexed volumetric bar-chart chip for point-of-care diagnostics. Nat. Commun. 2012, 3, 1283. [Google Scholar] [CrossRef] [PubMed]
- Lequin, R.M. Enzyme Immunoassay (EIA)/Enzyme-Linked Immunosorbent Assay (ELISA). Clin Chem. 2005, 51, 2415–2418. [Google Scholar] [CrossRef]
- Vashist, S.K.; van Oordt, T.; Schneider, E.M.; Zengerle, R.; von Stetten, F.; Luong, J.H. A smartphone-based colorimetric reader for bioanalytical applications using the screen-based bottom illumination provided by gadgets. Biosens. Bioelectron. 2015, 67, 248–255. [Google Scholar] [CrossRef]
- McGeough, C.M.; O’Driscoll, S. Camera phone-based quantitative analysis of c-reactive protein ELISA. IEEE Trans. Biomed. Circuits Syst. 2013, 7, 655–659. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.K.; Huang, H.Y.; Chen, W.R.; Nishie, W.; Ujiie, H.; Natsuga, K.; Fan, S.T.; Wang, H.K.; Lee, J.Y.; Tsai, W.L.; et al. Paper-based ELISA for the detection of autoimmune antibodies in body fluid-the case of bullous pemphigoid. Anal. Chem. 2014, 86, 4605–4610. [Google Scholar] [CrossRef] [PubMed]
- Hsu, M.Y.; Hung, Y.C.; Hwang, D.K.; Lin, S.C.; Lin, K.H.; Wang, C.Y.; Choi, H.Y.; Wang, Y.P.; Cheng, C.M. Detection of aqueous VEGF concentrations before and after intravitreal injection of anti-VEGF antibody using low-volume sampling paper-based ELISA. Sci. Rep. 2016, 6, 34631. [Google Scholar] [CrossRef]
- Ranney, G.B.; Thigpen, C.C. The sample coefficient of determination in simple linear regression. Am. Stat. 1981, 35, 152–153. [Google Scholar]
- Wagner, M.; Adamczak, R.; Porollo, A.; Meller, J. Linear regression models for solvent accessibility prediction in proteins. J. Comput. Biol. 2005, 12, 355–369. [Google Scholar] [CrossRef]
- Zor, T.; Selinger, Z. Linearization of the Bradford protein assay increases its sensitivity: Theoretical and experimental studies. Anal. Biochem. 1996, 236, 302–308. [Google Scholar] [CrossRef]

| Characteristics | Paper-Based ELISA Turntable Paper-Based Device | |
|---|---|---|
| Filter paper | Whatman Fusion 5 | Whatman Grade 3 |
| Time | About 4 h | About 4 h |
| Reagents volume | 200 μL | 300 μL |
| Limit of detection | 103 CFU/mL | 105 CFU/mL |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.-C.; Tsai, Y.-H.; Shen, C.-F.; He, M.-Y.; Fu, Y.-C.; Sang, C.-Y.; Lee, Y.-T.; Cheng, C.-M. Turntable Paper-Based Device to Detect Escherichia coli. Micromachines 2021, 12, 194. https://doi.org/10.3390/mi12020194
Wang Y-C, Tsai Y-H, Shen C-F, He M-Y, Fu Y-C, Sang C-Y, Lee Y-T, Cheng C-M. Turntable Paper-Based Device to Detect Escherichia coli. Micromachines. 2021; 12(2):194. https://doi.org/10.3390/mi12020194
Chicago/Turabian StyleWang, Yung-Chih, Yao-Hung Tsai, Ching-Fen Shen, Ming-Yao He, Yi-Chen Fu, Chen-Yu Sang, Yi-Tzu Lee, and Chao-Min Cheng. 2021. "Turntable Paper-Based Device to Detect Escherichia coli" Micromachines 12, no. 2: 194. https://doi.org/10.3390/mi12020194
APA StyleWang, Y.-C., Tsai, Y.-H., Shen, C.-F., He, M.-Y., Fu, Y.-C., Sang, C.-Y., Lee, Y.-T., & Cheng, C.-M. (2021). Turntable Paper-Based Device to Detect Escherichia coli. Micromachines, 12(2), 194. https://doi.org/10.3390/mi12020194








