Three-Dimensional Semantic Segmentation of Pituitary Adenomas Based on the Deep Learning Framework-nnU-Net: A Clinical Perspective
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Information
2.2. Magnetic Resonance Image Dataset
2.3. Tumor Segmentation
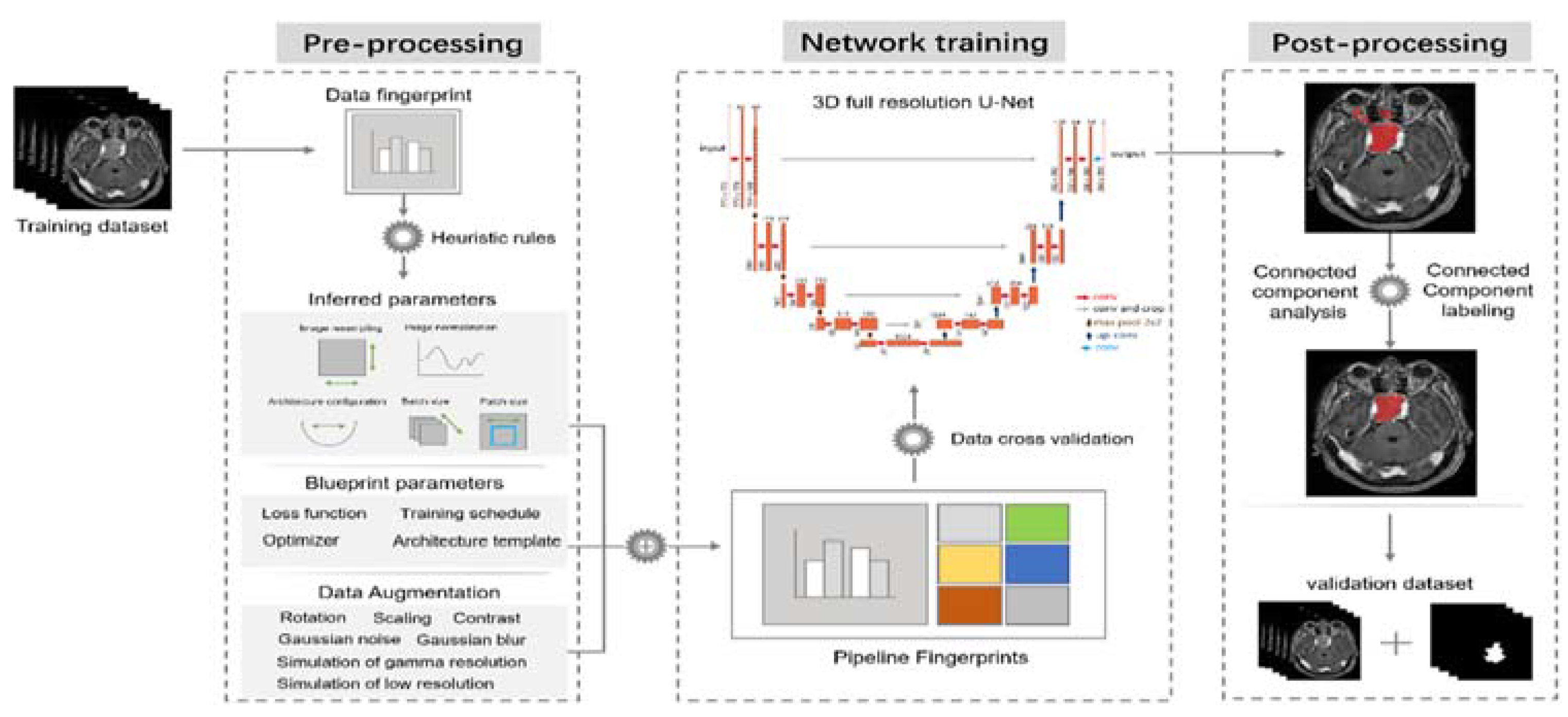
2.4. nnU-Net Framework
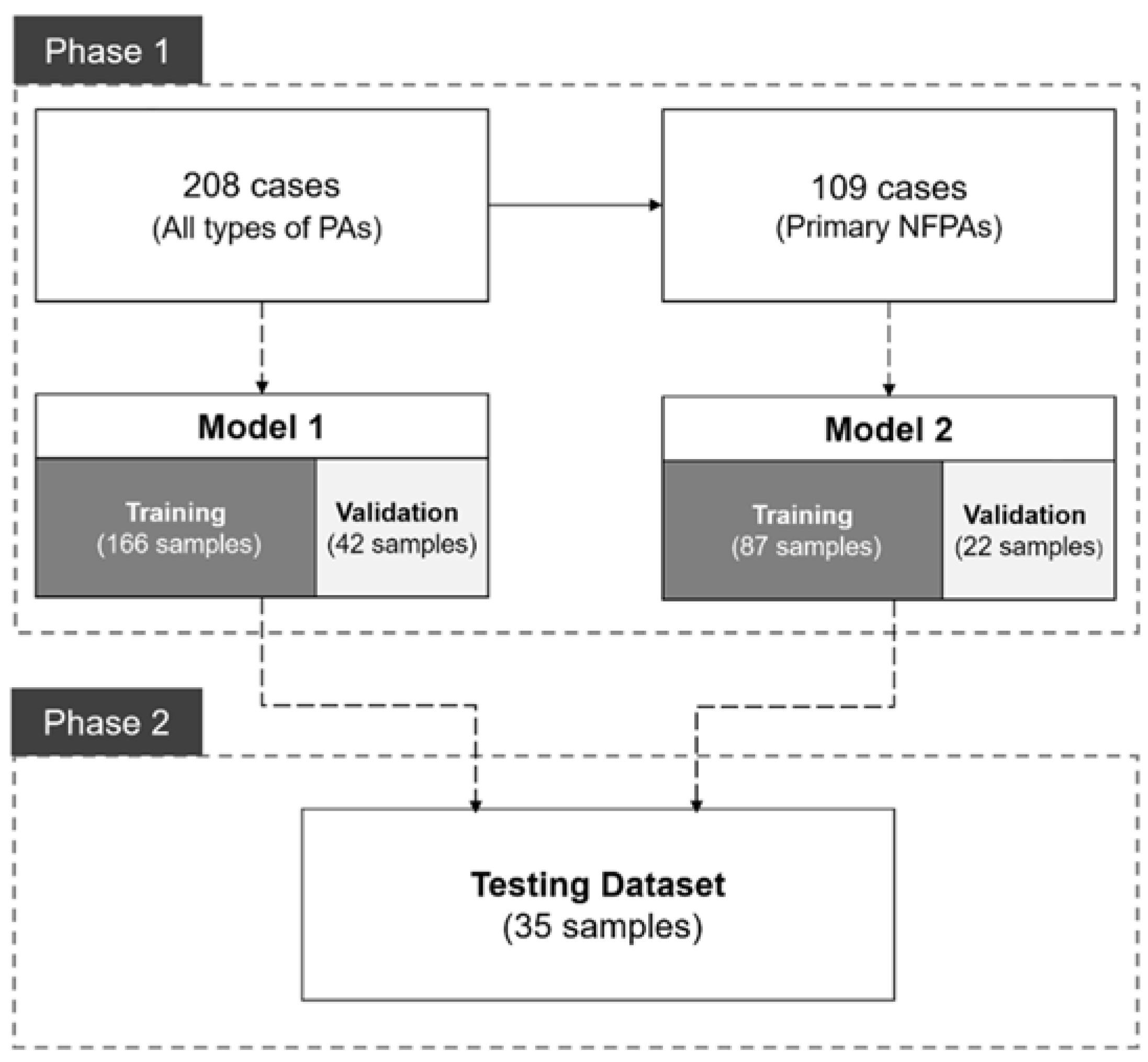
2.5. Study Design
3. Results
3.1. Patient Information and PA Characteristics
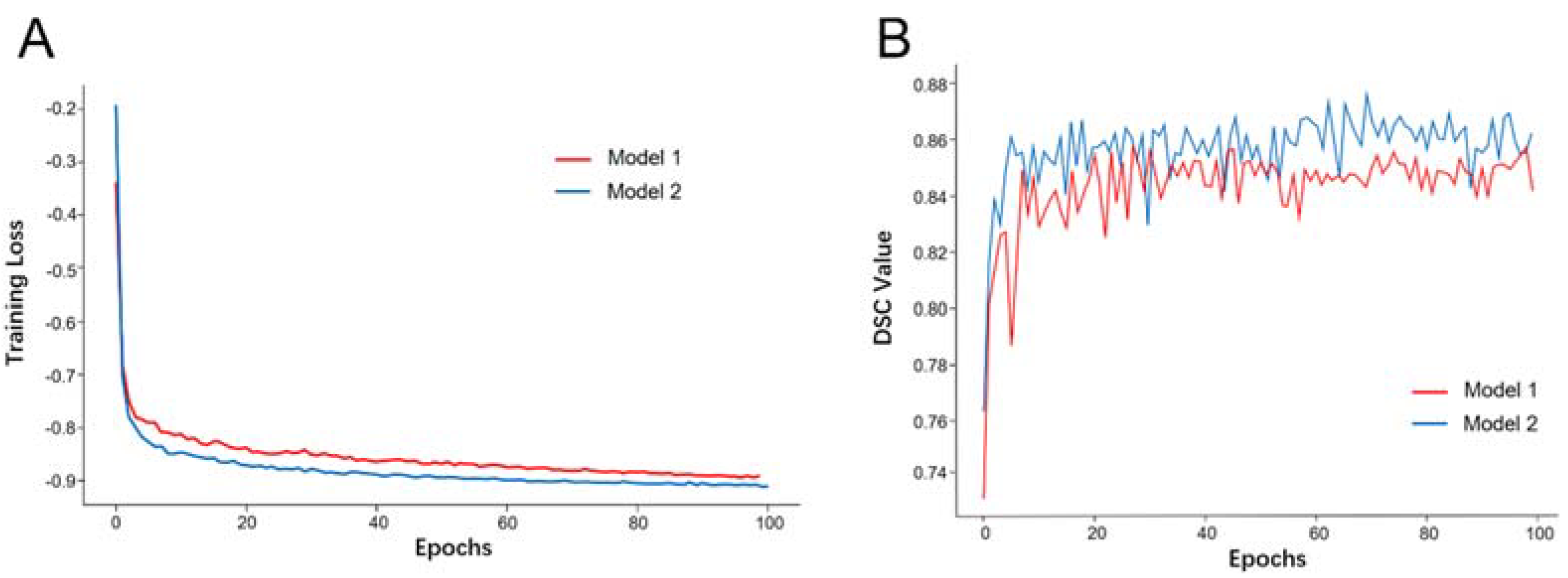
3.2. Model Training and Evaluation
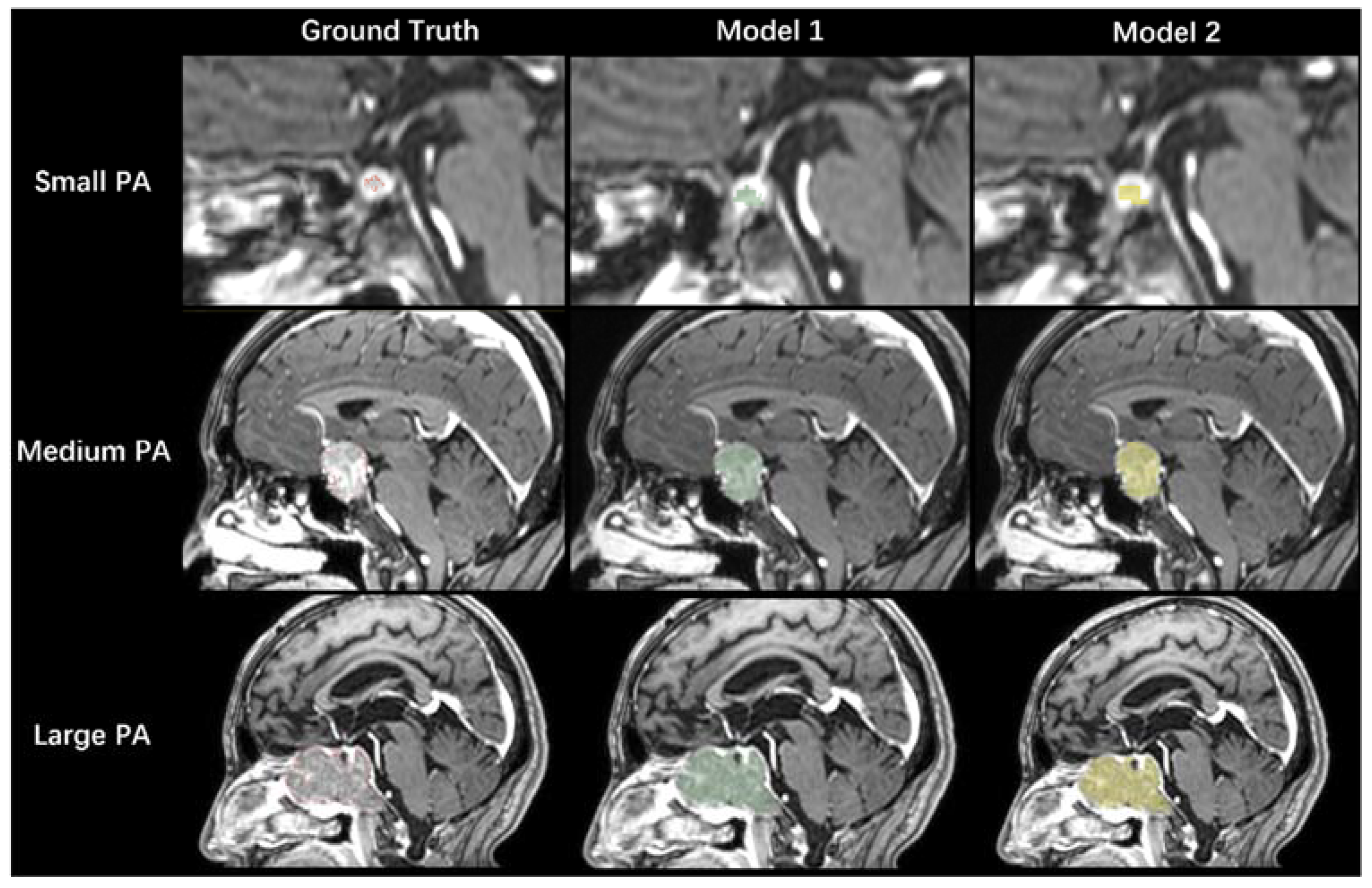
3.3. Model Performance in the Validation Dataset
3.4. Model Performance in the Testing Dataset
3.5. Performance Comparison between the Two Models
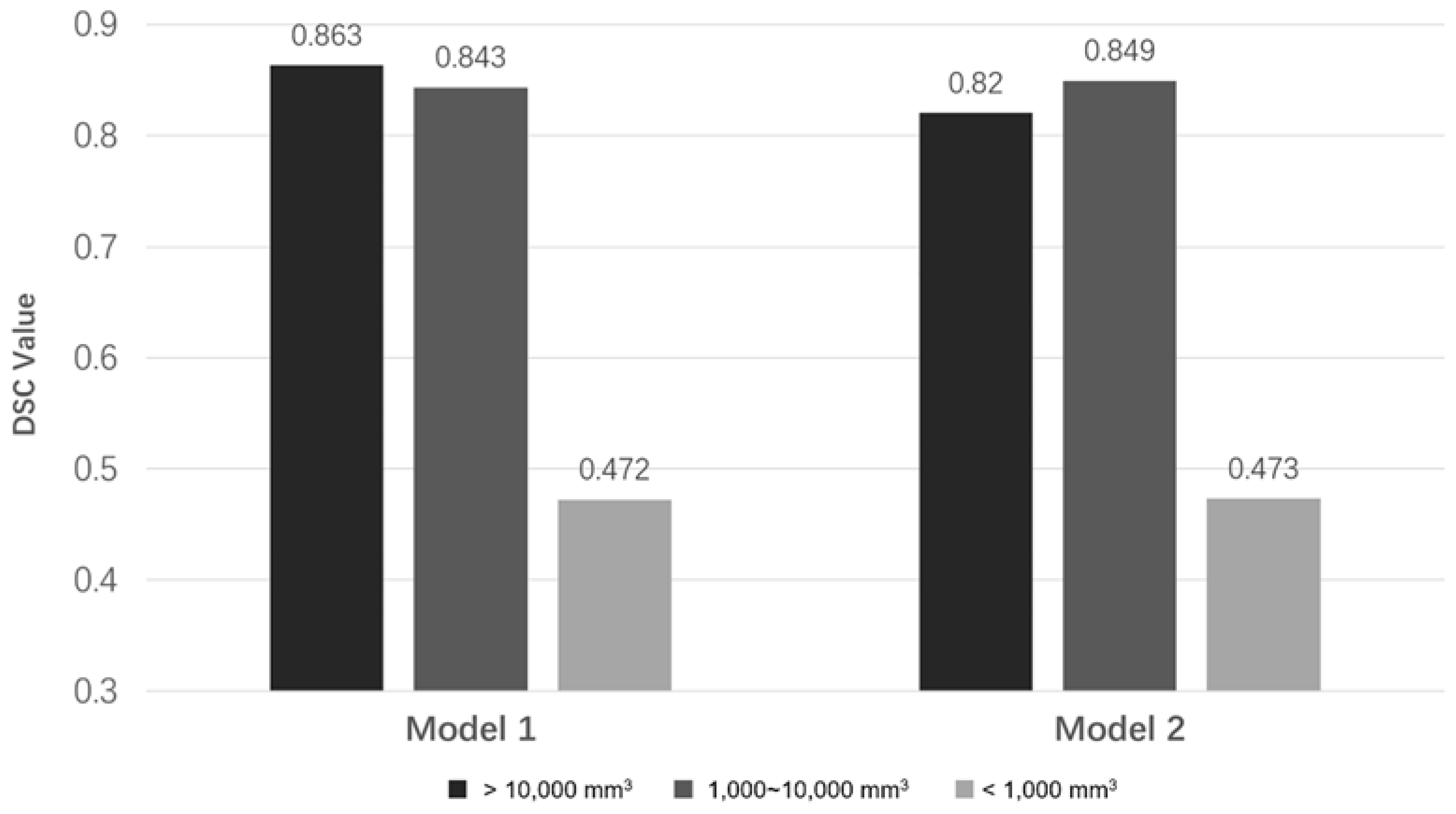
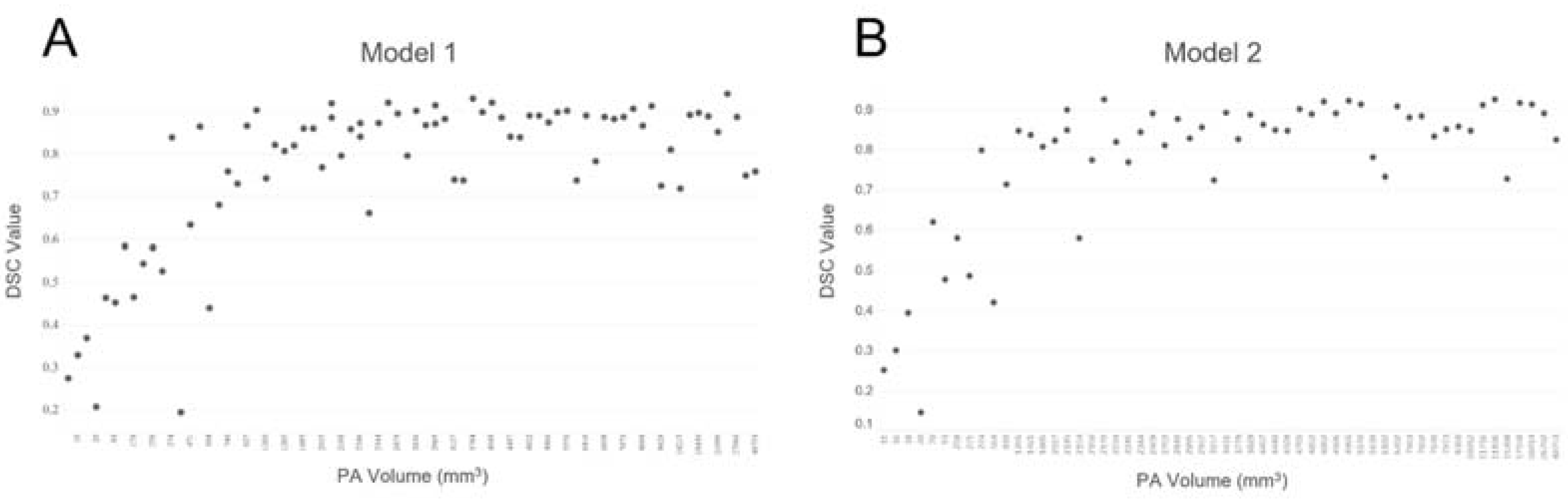
3.6. The Relationship between DSC Values and PA Volumes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jin, Z.; Wu, X.; Wang, Y. Clinical Study of Endoscopic Treatment of a Sellar Pituitary Adenomas with Sellar Diaphragm Defect. BMC Neurol. 2020, 20, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berntsen, E.M.; Haukedal, M.D.; Håberg, A.K. Normative Data for Pituitary Size and Volume in the General Population between 50 and 66 Years. Pituitary 2021, 24, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Zukic, D.; Egger, J.; Bauer, M.H.A.; Kuhnt, D.; Carl, B.; Freisleben, B.; Kolb, A.; Nimsky, C. Preoperative Volume Determination for Pituitary Adenoma. In Proceedings of the SPIE Medical Imaging 2011, Lake Buena Vista, FL, USA, 12–17 February 2011; p. 79632T. [Google Scholar]
- Qian, Y.; Qiu, Y.; Li, C.-C.; Wang, Z.-Y.; Cao, B.-W.; Huang, H.-X.; Ni, Y.-H.; Chen, L.-L.; Sun, J.-Y. A Novel Diagnostic Method for Pituitary Adenoma Based on Magnetic Resonance Imaging Using a Convolutional Neural Network. Pituitary 2020, 23, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H.M.T.; Devadhas, D.; Heck, D.K.; Chacko, A.G.; Rebekah, G.; Oommen, R.; Samuel, E.J.J. Adaptive Threshold Segmentation of Pituitary Adenomas from FDG PET Images for Radiosurgery. J. Appl. Clin. Med. Phys. 2014, 15, 279–294. [Google Scholar] [CrossRef] [PubMed]
- Chuang, C.-C.; Lin, S.-Y.; Pai, P.-C.; Yan, J.-L.; Toh, C.-H.; Lee, S.-T.; Wei, K.-C.; Liu, Z.-H.; Chen, C.-M.; Wang, Y.-C.; et al. Different Volumetric Measurement Methods for Pituitary Adenomas and Their Crucial Clinical Significance. Sci. Rep. 2017, 7, 40792. [Google Scholar] [CrossRef] [PubMed]
- Taheri, S.; Ong, S.H.; Chong, V.F.H. Level-Set Segmentation of Brain Tumors Using a Threshold-Based Speed Function. Image Vis. Comput. 2010, 28, 26–37. [Google Scholar] [CrossRef]
- Egger, J.; Zukić, D.; Freisleben, B.; Kolb, A.; Nimsky, C. Segmentation of Pituitary Adenoma: A Graph-Based Method vs. a Balloon Inflation Method. Comput. Methods Programs Biomed. 2013, 110, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Meier, R.; Knecht, U.; Loosli, T.; Bauer, S.; Slotboom, J.; Wiest, R.; Reyes, M. Clinical Evaluation of a Fully-Automatic Segmentation Method for Longitudinal Brain Tumor Volumetry. Sci. Rep. 2016, 6, 23376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbosa, B.J.A.P.; Dimostheni, A.; Teixeira, M.J.; Tatagiba, M.; Lepski, G. Insular Gliomas and the Role of Intraoperative Assistive Technologies: Results from a Volumetry-Based Retrospective Cohort. Clin. Neurol. Neurosurg. 2016, 149, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Egger, J.; Kapur, T.; Nimsky, C.; Kikinis, R. Pituitary Adenoma Volumetry with 3D Slicer. PLoS ONE 2012, 7, e51788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, S.K.; Greenspan, H.; Davatzikos, C.; Duncan, J.S.; van Ginneken, B.; Madabhushi, A.; Prince, J.L.; Rueckert, D.; Summers, R.M. A Review of Deep Learning in Medical Imaging: Imaging Traits, Technology Trends, Case Studies with Progress Highlights, and Future Promises. Proc. IEEE 2021, 109, 820–838. [Google Scholar] [CrossRef]
- Akkus, Z.; Galimzianova, A.; Hoogi, A.; Rubin, D.L.; Erickson, B.J. Deep Learning for Brain MRI Segmentation: State of the Art and Future Directions. J. Digit. Imaging 2017, 30, 449–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zlochower, A.; Chow, D.S.; Chang, P.; Khatri, D.; Boockvar, J.A.; Filippi, C.G. Deep Learning AI Applications in the Imaging of Glioma. Top. Magn. Reson. Imaging 2020, 29, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Pernas, F.J.; Martínez-Zarzuela, M.; Antón-Rodríguez, M.; González-Ortega, D. A Deep Learning Approach for Brain Tumor Classification and Segmentation Using a Multiscale Convolutional Neural Network. Healthcare 2021, 9, 153. [Google Scholar] [CrossRef] [PubMed]
- Iuga, A.-I.; Carolus, H.; Höink, A.J.; Brosch, T.; Klinder, T.; Maintz, D.; Persigehl, T.; Baeßler, B.; Püsken, M. Automated Detection and Segmentation of Thoracic Lymph Nodes from CT Using 3D Foveal Fully Convolutional Neural Networks. BMC Med. Imaging 2021, 21, 69. [Google Scholar] [CrossRef] [PubMed]
- Shelhamer, E.; Long, J.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 640–651. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. NnU-Net: A Self-Configuring Method for Deep Learning-Based Biomedical Image Segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Taha, A.A.; Hanbury, A. Metrics for Evaluating 3D Medical Image Segmentation: Analysis, Selection, and Tool. BMC Med. Imaging 2015, 15, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ammu, R.; Sinha, N. Small Segment Emphasized Performance Evaluation Metric for Medical Images. In Proceedings of the 2020 International Conference on Signal Processing and Communications (SPCOM), Bangalore, India, 19–24 July 2020; pp. 1–5. [Google Scholar]
| Patient Information and PA Characteristics | No. of Cases (%) |
|---|---|
| Gender | |
| Male | 106 (51.0%) |
| Female | 102 (49.0%) |
| Primary/Recurrent PAs | |
| Primary | 168 (80.8%) |
| Recurrent | 40 (19.2%) |
| Nonfunctional PAs | 135 (64.9%) |
| Functional PAs | 73 (35.1%) |
| ACTH | 24 (11.5%) |
| GH | 28 (13.5%) |
| PRL | 16 (7.6%) |
| TSH | 5 (2.4%) |
| Size | |
| Giant-PAs (≥4 cm) | 21 (10.1%) |
| Macro-PAs (1 cm~4 cm); | 164 (78.8%) |
| Micro-PAs (≤1 cm) | 23 (11.1%) |
| Volume | |
| Large (≥10,000 mm3) | 23 (11.1%) |
| Medium (10,000~1000 mm3) | 132 (63.5%) |
| Small (≤1000 mm3) | 53 (25.5%) |
| Total | 208 |
| Model 1 | Model 2 | |||||
|---|---|---|---|---|---|---|
| Train | Validation | Mean DSC | Train | Validation | Mean DSC | |
| Gender | ||||||
| Male | 82 | 24 | 0.838 | 58 | 14 | 0.864 |
| Female | 84 | 18 | 0.756 | 29 | 8 | 0.833 |
| Primary/Recurrent PAs | ||||||
| Primary | 136 | 32 | 0.808 | 87 | 22 | 0.853 |
| Recurrent | 30 | 10 | 0.787 | - | - | - |
| Nonfunctional PAs | 107 | 28 | 0.828 | 87 | 22 | 0.853 |
| Functional PAs | 59 | 14 | 0.753 | - | - | - |
| ACTH | 20 | 4 | 0.709 | - | - | - |
| GH | 22 | 6 | 0.729 | - | - | - |
| PRL | 14 | 2 | 0.768 | - | - | - |
| TSH | 3 | 2 | 0.896 | - | - | - |
| Size | ||||||
| Giant PAs | 13 | 8 | 0.832 | 6 | 2 | 0.820 |
| Macroadenomas | 132 | 32 | 0.811 | 78 | 20 | 0.856 |
| Microadenomas | 21 | 2 | 0.563 | 3 | 0 | - |
| Volume | ||||||
| Large (≥10,000 mm3) | 16 | 7 | 0.847 | 10 | 5 | 0.873 |
| Medium (1000~10,000 mm3) | 107 | 25 | 0.852 | 62 | 17 | 0.847 |
| Small (≤1000 mm3) | 43 | 10 | 0.649 | 15 | 0 | - |
| Total | 166 | 42 | 0.803 | 87 | 22 | 0.853 |
| Testing Dataset | Model 1 | Model 2 | |
|---|---|---|---|
| Mean DSC | Mean DSC | ||
| Gender | |||
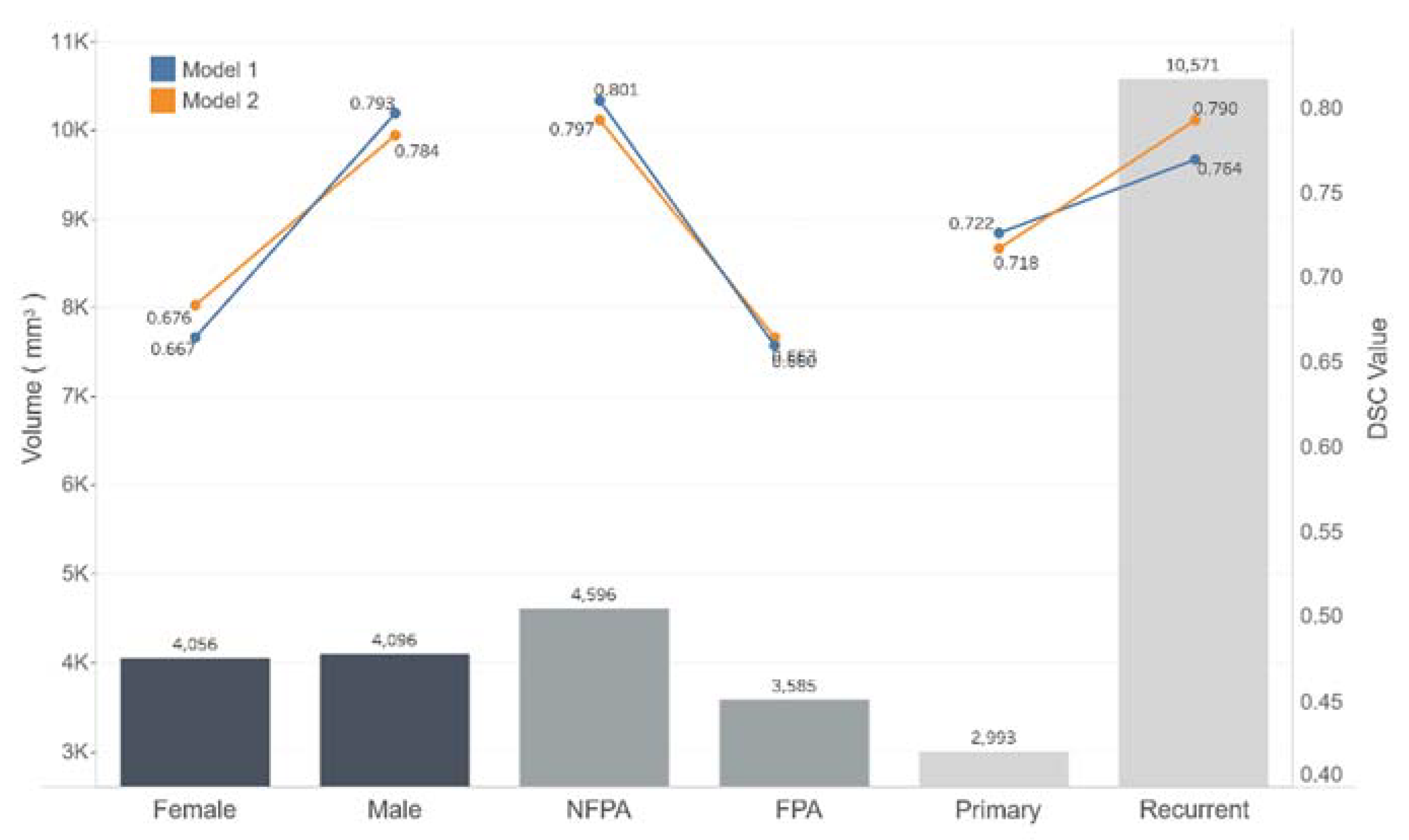
| Male | 17 | 0.793 | 0.784 |
| Female | 18 | 0.667 | 0.676 |
| Primary/Recurrent PAs | |||
| Primary | 30 | 0.722 | 0.718 |
| Recurrent | 5 | 0.764 | 0.790 |
| Nonfunctional PAs | 17 | 0.801 | 0.797 |
| Functional PAs | 18 | 0.660 | 0.663 |
| ACTH | 10 | 0.679 | 0.672 |
| GH | 4 | 0.659 | 0.688 |
| PRL | 4 | 0.614 | 0.615 |
| TSH | - | - | - |
| Size | |||
| Giant PAs | 4 | 0.843 | 0.799 |
| Macroadenomas | 22 | 0.821 | 0.830 |
| Microadenomas | 9 | 0.451 | 0.448 |
| Volume | |||
| Large (≥10,000 mm3) | 3 | 0.863 | 0.820 |
| Medium (1000~10,000 mm3) | 21 | 0.843 | 0.849 |
| Small (≤1000 mm3) | 11 | 0.472 | 0.473 |
| Total | 35 | 0.7279 | 0.7284 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shu, X.; Zhou, Y.; Li, F.; Zhou, T.; Meng, X.; Wang, F.; Zhang, Z.; Pu, J.; Xu, B. Three-Dimensional Semantic Segmentation of Pituitary Adenomas Based on the Deep Learning Framework-nnU-Net: A Clinical Perspective. Micromachines 2021, 12, 1473. https://doi.org/10.3390/mi12121473
Shu X, Zhou Y, Li F, Zhou T, Meng X, Wang F, Zhang Z, Pu J, Xu B. Three-Dimensional Semantic Segmentation of Pituitary Adenomas Based on the Deep Learning Framework-nnU-Net: A Clinical Perspective. Micromachines. 2021; 12(12):1473. https://doi.org/10.3390/mi12121473
Chicago/Turabian StyleShu, Xujun, Yijie Zhou, Fangye Li, Tao Zhou, Xianghui Meng, Fuyu Wang, Zhizhong Zhang, Jian Pu, and Bainan Xu. 2021. "Three-Dimensional Semantic Segmentation of Pituitary Adenomas Based on the Deep Learning Framework-nnU-Net: A Clinical Perspective" Micromachines 12, no. 12: 1473. https://doi.org/10.3390/mi12121473
APA StyleShu, X., Zhou, Y., Li, F., Zhou, T., Meng, X., Wang, F., Zhang, Z., Pu, J., & Xu, B. (2021). Three-Dimensional Semantic Segmentation of Pituitary Adenomas Based on the Deep Learning Framework-nnU-Net: A Clinical Perspective. Micromachines, 12(12), 1473. https://doi.org/10.3390/mi12121473





