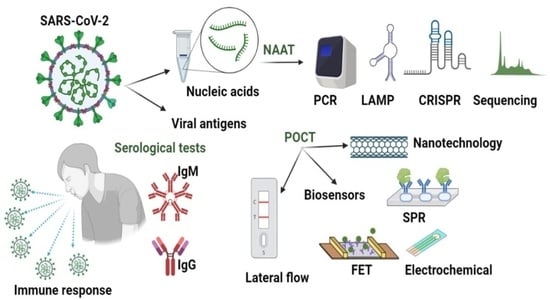
Point-of-Care Testing—The Key in the Battle against SARS-CoV-2 Pandemic
Abstract
:1. Introduction
2. Challenges in SARS-CoV-2 Detection
3. SARS-CoV-2—Structure and Characteristics
4. Specimen Collection and Sample Preparation
4.1. Nucleic Acids
4.2. Viral Antigens
5. Detection Methods
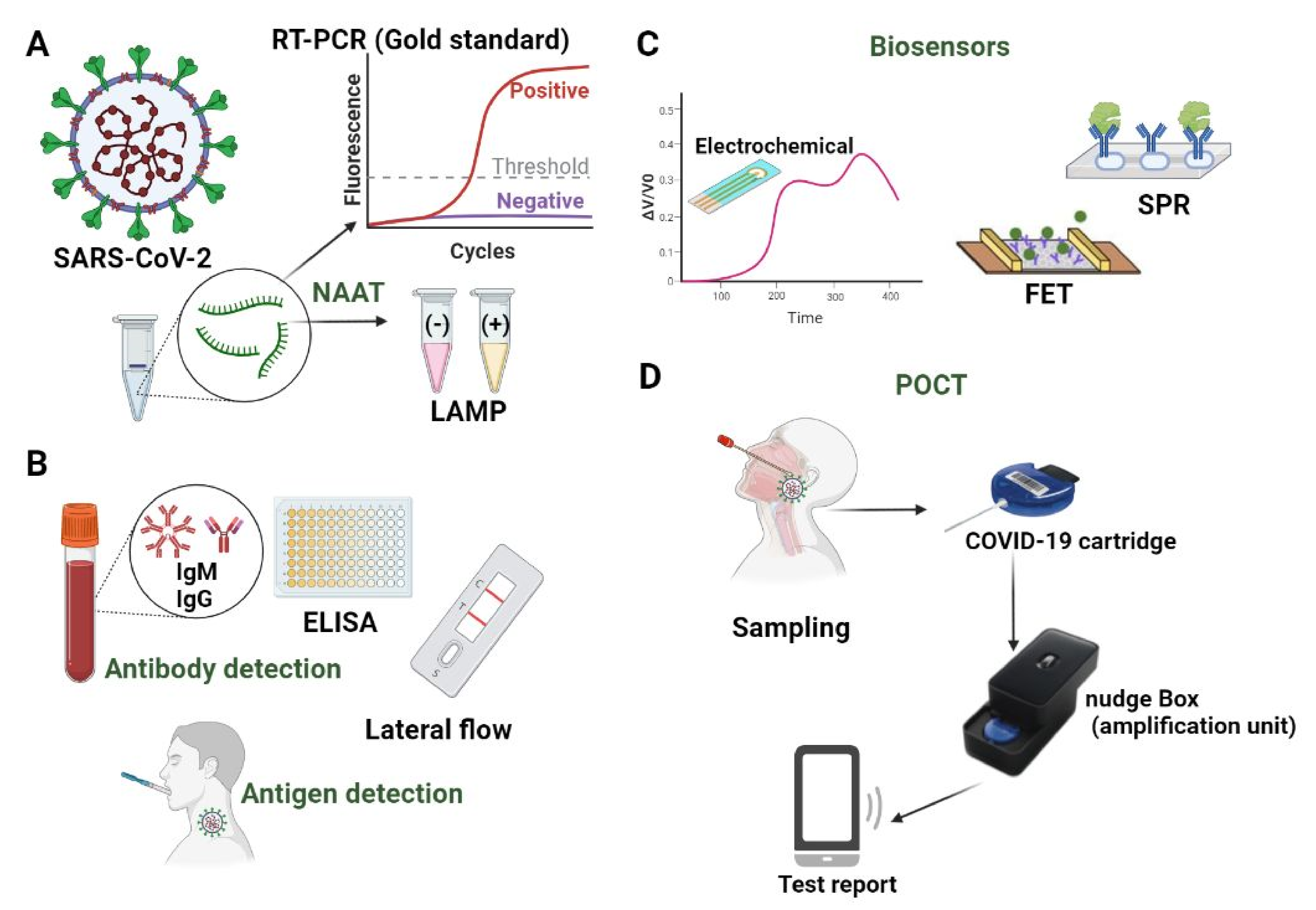
5.1. Nucleic Acids Amplification Testing (NAAT)
5.1.1. PCR Mediated Detection
5.1.2. Isothermal Amplification
5.1.3. Sequencing-Based Tests
5.1.4. CRISPR-Mediated Detection
5.1.5. Combined Methods
5.2. Serologic Tests
- Tests that detect a reaction and require trained personnel to interpret the results;
- Tests that detect only the presence of antibodies by a colourimetric change.
| Test Name | Method/Technology | Manufacturer | Ig | Time [min] | Sensitivity/Specificity [%] | Reference |
|---|---|---|---|---|---|---|
| SARS-CoV-2 IgG Assay | Chemiluminescent microparticle immunoassay | Abbott Lab. | IgG only against N protein | ~30 | 100/99.63 | [115] |
| COVID-19 IgG/IgM Rapid Test Cassette | Immunoassay colloidal gold | Acro Biotech | IgM, IgG | ~10 | IgG 100/98 IgM 85/96 | [116] |
| Anti-SARS-CoV-2 Rapid Test | Lateral flow immunoassay | Autobio Diagnostics | IgG and IgM only against S protein | ~15 | 99.0/99.04 | [117] |
| 2019-nCoV IgG/IgM detection kit (colloidal gold) | Solid-phase immuno-chromatographic | Biolodics | IgM and IgG | ~10 | 91.54/97.02 | [118] |
| Platelia SARS-CoV-2 Total Ab assay | Semiquantitative ELISA | Bio-Rad Lab | IgA, IgM, IgG against N protein | ~100 | 92.2/99.6 | [119] |
| COVISURE™ COVID-19 IgM/IgG Rapid Test | Lateral flow immunoassay | W.H.P.M., Inc. | IgM/IgG | ~15 | IgM 76.7/97.1 IgG 70/97.1 | [120] |
| qSARS-CoV-2 IgG/IgM Rapid Test | Lateral flow immunoassay | Cellex | IgG and IgM only against S and N proteins | 15–20 | 93.8/96 | [121] |
| Finecare TM 2019—nCoV Antobody Test | Lateral flow fluorescence immunoassay | Guanzhou Wondfo Biotech | IgM + IgG | ~15 | 86.43/99.57 | [122] |
| Clungene COVID-19 IgM/IgG rapid test cassette | Rapid immune antibody immunoassay test | Hangzhou Clongene Biotech | IgM, IgG | ~15 | 87.1/98.89 | [123] |
| LIAISON SARS-CoV-2 S1/S2 IgG | Chemiluminescent immunoassay | DiaSorin | IgG against S1/S2 protein | ~35 | 97.56/99.3 | [124] |
| Anti-SARS-CoV-2 ELISA IgG/IgA Anti-SARS-CoV-2 QuantiVac ELISA (IgG)/Anti-SARS-CoV-2 NCP ELISA IgG/IgM | ELISA for semi-quantitative and quantitative determination | Euroimmun (Perkin Elmer) | IgG, IgM, against S1 and nucleocapsid protein | 15–60 | 94.4/99.6 IgA 100/92.4 | [125] |
| VITROS Immunodiagnostic Products Anti-SARS-CoV-2 Total Reagent Pack | Chemiluminescent immunoassay | Ortho Clinical Diagnostics | Total antibody against S1 | ~50 | 100/100 | [126] |
| COVID-19 IgG/IgM rapid test device | Lateral flow | RayBiotech | 90.44/98.31 | [127] | ||
| Elecsys Anti-SARS-CoV-2 | Electrochemi-luminescence immunoassay | Roche | Total antibody against N protein | ~18 | 100/99.81 | [114] |
| Standard Q COVID-19 IgM/IgG Duo | Immunochromatography | SD Biosensor | IgM and IgG | ~10 | 90.6/96.1 | [128] |
| Atellica IM® SARS-CoV-2 Total (COV2T) | Chemiluminescent microparticle immunoassay | Siemens Healthcare | Total antibody against RBD of S1 protein | ~10 | 100/99.82 | [129] |
| MAGLUMI 2019-nCoV IgM/IgG (CLIA) | Immune-antibody assay quantitative | SNIBE Co. Ltd. | IgM, IgG | ~30 | IgM 78.7/97.5 IgG 91.2/96 | [130] |
| SGTi-flex COVID-19 IgM/IgG | Immunochromatography | Sugentech Inc. | IgM, IgG | ~10 | IgM 90.8/98.33 IgG 90.18/100 | [131] |
| SGT Anti-SARS-CoV-2 Total Ab ELISA | ELISA | IgM, IgA, IgG | ~150 | Higher than Rapid test |
5.3. Lateral Flow-Based Detection
5.4. Biosensors on Microfluidic Devices as POCT
5.4.1. Electrochemical Biosensors
5.4.2. Field-Effect Transistor (FET)
5.4.3. Plasmonic Biosensors
5.5. Nanotechnological Approaches
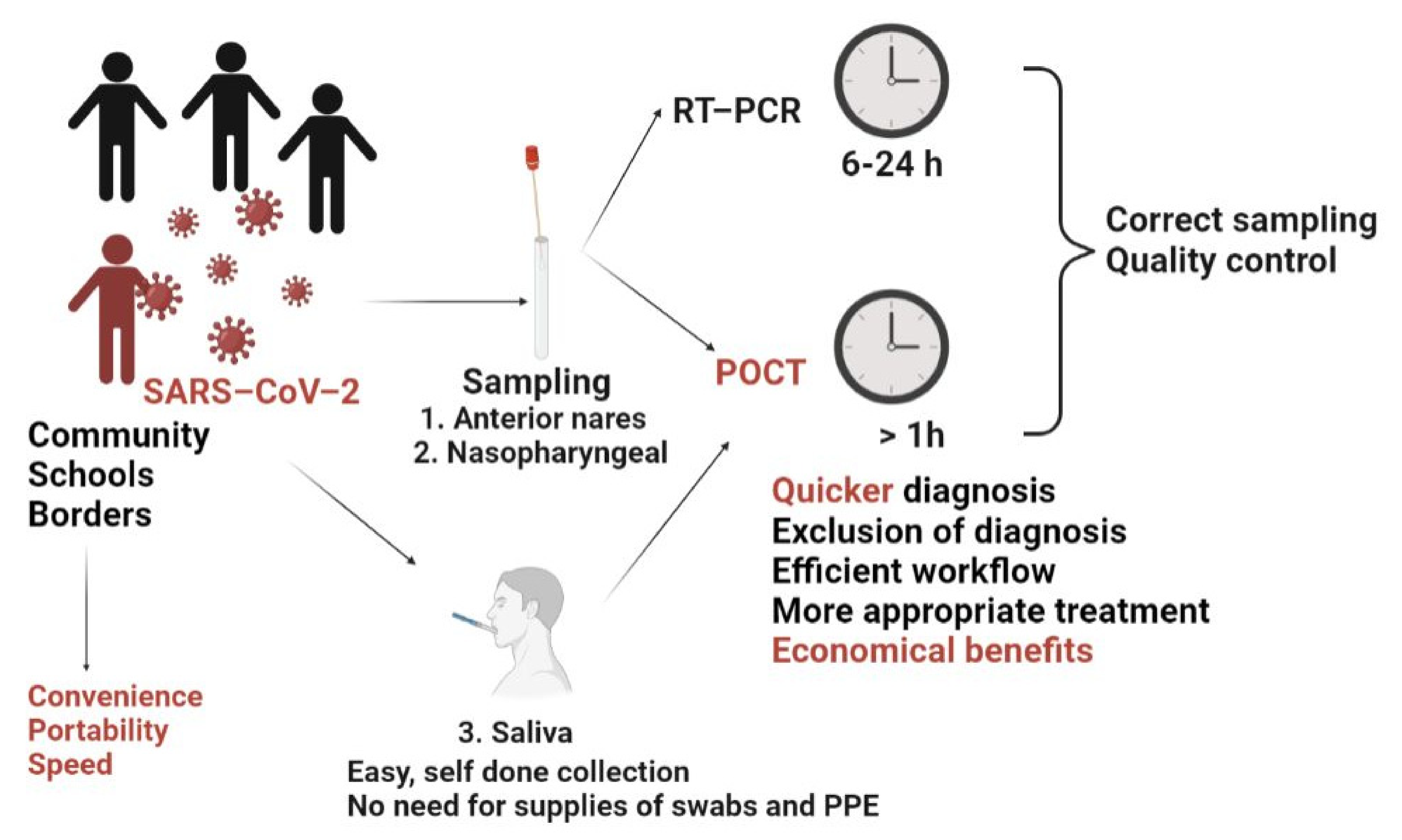
5.6. Point of Care Testing (POCT)
- -
- clinical benefits (i.e., quicker diagnostic, exclusion of diagnostic, more appropriate treatment and subsequent improved clinical outcome);
- -
- better access to testing in case of rural and remote sectors;
- -
- economic benefits—POCT enables the rapid identification and isolation of infected individuals, hence avoiding lockdown measures.
6. Interpretating the Tests’ Results for Clinical Applications
7. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Available online: https://covid19.who.int/ (accessed on 20 November 2021).
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://www.who.int/news-room/commentaries/detail/advice-on-the-use-of-point-of-care-immunodiagnostic-tests-for-covid-19 (accessed on 20 November 2021).
- Available online: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (accessed on 20 November 2021).
- Khachfe, H.H.; Chahrour, M.; Sammouri, J.; Salhab, H.; Makki, B.E.; Fares, M. An epidemiological study on COVID-19: A rapidly spreading disease. Cureus 2020, 12, e7313. [Google Scholar] [CrossRef] [Green Version]
- Rong, X.; Yang, L.; Chu, H.; Fan, M. Effect of delay in diagnosis on transmission of COVID-19. Math. Biosci. Eng. 2020, 17, 2725–2740. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Filipić, A.; Gutierrez-Aguirre, I.; Primc, G.; Mozetič, M.; Dobnik, D. Cold plasma, a new hope in the field of virus inactivation. Trends Biotechnol. 2020, 38, 1278–1291. [Google Scholar] [CrossRef]
- Zhu, H.; Podesva, P.; Liu, X.; Zhang, H.; Teply, T.; Xu, Y.; Chang, H.; Qian, A.; Lei, Y.; Li, Y. IoT PCR for pandemic disease detection and its spread monitoring. Sens. Actuators B Chem. 2020, 303, 127098. [Google Scholar] [CrossRef]
- Sun, Y.; Koh, V.; Marimuthu, K.; Ng, O.T.; Young, B.; Vasoo, S.; Chan, M.; Lee, V.J.; De, P.P.; Barkham, T. Epidemiological and clinical predictors of COVID-19. Clin. Infect. Dis. 2020, 71, 786–792. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.-L.; Tseng, W.-P.; Lin, C.-H.; Lee, T.-F.; Chung, M.-Y.; Huang, C.-H.; Chen, S.-Y.; Hsueh, P.-R.; Chen, S.-C. Four point-of-care lateral flow immunoassays for diagnosis of COVID-19 and for assessing dynamics of antibody responses to SARS-CoV-2. J. Infect. 2020, 81, 435–442. [Google Scholar] [CrossRef]
- Hopkins, C.; Smith, B. Widespread smell testing for COVID-19 has limited application. Lancet 2020, 396, 1630. [Google Scholar] [CrossRef]
- Menni, C.; Valdes, A.M.; Freidin, M.B.; Sudre, C.H.; Nguyen, L.H.; Drew, D.A.; Ganesh, S.; Varsavsky, T.; Cardoso, M.J.; Moustafa, J.S.E.-S. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat. Med. 2020, 26, 1037–1040. [Google Scholar] [CrossRef] [PubMed]
- Menni, C.; Sudre, C.H.; Steves, C.J.; Ourselin, S.; Spector, T.D. Quantifying additional COVID-19 symptoms will save lives. Lancet 2020, 395, e107–e108. [Google Scholar] [CrossRef]
- Joung, J.; Ladha, A.; Saito, M.; Segel, M.; Bruneau, R.; Huang, M.-l.W.; Kim, N.-G.; Yu, X.; Li, J.; Walker, B.D. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. MedRxiv 2020. [Google Scholar] [CrossRef]
- Tang, Y.-W.; Schmitz, J.E.; Persing, D.H.; Stratton, C.W. Laboratory diagnosis of COVID-19: Current issues and challenges. J. Clin. Microbiol. 2020, 58, e00512-20. [Google Scholar] [CrossRef] [Green Version]
- Deeks, J.J.; Raffle, A.E. Lateral flow tests cannot rule out SARS-CoV-2 infection. BMJ 2020, 371, m4787. [Google Scholar] [CrossRef]
- Lucia, C.; Federico, P.-B.; Alejandra, G.C. An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas12. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://www.fda.gov/media/141194/download (accessed on 20 November 2021).
- Cheung, K.S.; Hung, I.F.; Chan, P.P.; Lung, K.; Tso, E.; Liu, R.; Ng, Y.; Chu, M.Y.; Chung, T.W.; Tam, A.R. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: Systematic review and meta-analysis. Gastroenterology 2020, 159, 81–95. [Google Scholar] [CrossRef]
- Lin, L.; Jiang, X.; Zhang, Z.; Huang, S.; Zhang, Z.; Fang, Z.; Gu, Z.; Gao, L.; Shi, H.; Mai, L. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut 2020, 69, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitajima, M.; Ahmed, W.; Bibby, K.; Carducci, A.; Gerba, C.P.; Hamilton, K.A.; Haramoto, E.; Rose, J.B. SARS-CoV-2 in wastewater: State of the knowledge and research needs. Sci. Total. Environ. 2020, 739, 139076. [Google Scholar] [CrossRef] [PubMed]
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–516. [Google Scholar] [CrossRef]
- Heijnen, L.; Elsinga, G.; de Graaf, M.; Molenkamp, R.; Koopmans, M.P.; Medema, G. Droplet Digital RT-PCR to detect SARS-CoV-2 variants of concern in wastewater. medRxiv 2021. [Google Scholar] [CrossRef]
- Mihaescu, G.; Chifiriuc, M.C.; Iliescu, C.; Vrancianu, C.O.; Ditu, L.-M.; Marutescu, L.G.; Grigore, R.; Berteșteanu, Ș.; Constantin, M.; Gradisteanu Pircalabioru, G. SARS-CoV-2: From Structure to Pathology, Host Immune Response and Therapeutic Management. Microorganisms 2020, 8, 1468. [Google Scholar] [CrossRef]
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Neuman, B.W.; Adair, B.D.; Yoshioka, C.; Quispe, J.D.; Orca, G.; Kuhn, P.; Milligan, R.A.; Yeager, M.; Buchmeier, M.J. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J. Virol. 2006, 80, 7918. [Google Scholar] [CrossRef] [Green Version]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Pyrc, K.; Dijkman, R.; Deng, L.; Jebbink, M.F.; Ross, H.A.; Berkhout, B.; Van der Hoek, L. Mosaic structure of human coronavirus NL63, one thousand years of evolution. J. Mol. Biol. 2006, 364, 964–973. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.-W.; Kok, K.-H.; Zhu, Z.; Chu, H.; To, K.K.-W.; Yuan, S.; Yuen, K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef]
- Zhou, H.; Chen, X.; Hu, T.; Li, J.; Song, H.; Liu, Y.; Wang, P.; Liu, D.; Yang, J.; Holmes, E.C. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr. Biol. 2020, 30, 2196–2203.e3. [Google Scholar] [CrossRef] [PubMed]
- Loeffelholz, M.J.; Tang, Y.-W. Laboratory diagnosis of emerging human coronavirus infections—The state of the art. Emerg. Microbes Infect. 2020, 9, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html (accessed on 4 July 2021).
- Wang, C.-H.; Lien, K.-Y.; Wang, T.-Y.; Chen, T.-Y.; Lee, G.-B. An integrated microfluidic loop-mediated-isothermal-amplification system for rapid sample pre-treatment and detection of viruses. Biosens. Bioelectron. 2011, 26, 2045–2052. [Google Scholar] [CrossRef]
- Bokelmann, L.; Nickel, O.; Maricic, T.; Pääbo, S.; Meyer, M.; Borte, S.; Riesenberg, S. Point-of-care bulk testing for SARS-CoV-2 by combining hybridization capture with improved colorimetric LAMP. Nat. Commun. 2021, 12, 1467. [Google Scholar] [CrossRef]
- Rabe, B.A.; Cepko, C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc. Natl. Acad. Sci. USA 2020, 117, 24450–24458. [Google Scholar] [CrossRef]
- Available online: https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas-molecular-diagnostic-tests-sars-cov-2#individual-molecular (accessed on 20 November 2021).
- Alekseenko, A.; Barrett, D.; Pareja-Sanchez, Y.; Howard, R.J.; Strandback, E.; Ampah-Korsah, H.; Rovšnik, U.; Zuniga-Veliz, S.; Klenov, A.; Malloo, J. Direct detection of SARS-CoV-2 using non-commercial RT-LAMP reagents on heat-inactivated samples. Sci. Rep. 2021, 11, 1820. [Google Scholar] [CrossRef]
- Available online: https://www.fda.gov/media/144553/download (accessed on 8 November 2021).
- Available online: https://www.fda.gov/media/141948/download (accessed on 8 November 2021).
- Available online: https://www.fda.gov/media/139860/download (accessed on 8 November 2021).
- Available online: https://www.fda.gov/media/139293/download (accessed on 8 November 2021).
- Available online: https://www.fda.gov/media/138652/download (accessed on 8 November 2021).
- Dinnes, J.; Deeks, J.J.; Berhane, S.; Taylor, M.; Adriano, A.; Davenport, C.; Dittrich, S.; Emperador, D.; Takwoingi, Y.; Cunningham, J. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2021. [Google Scholar] [CrossRef]
- Deeks, J.J.; Dinnes, J.; Takwoingi, Y.; Davenport, C.; Spijker, R.; Taylor-Phillips, S.; Adriano, A.; Beese, S.; Dretzke, J.; di Ruffano, L.F. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst. Rev. 2020. [Google Scholar] [CrossRef]
- Corstjens, P.L.; Abrams, W.R.; Malamud, D. Saliva and viral infections. Periodontology 2000 2016, 70, 93–110. [Google Scholar] [CrossRef]
- Wyllie, A.L.; Fournier, J.; Casanovas-Massana, A.; Campbell, M.; Tokuyama, M.; Vijayakumar, P.; Warren, J.L.; Geng, B.; Muenker, M.C.; Moore, A.J. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 1283–1286. [Google Scholar] [CrossRef]
- Yu, C.Y.; Chan, K.G.; Yean, C.Y.; Ang, G.Y. Nucleic Acid-Based Diagnostic Tests for the Detection SARS-CoV-2: An Update. Diagnostics 2021, 11, 53. [Google Scholar] [CrossRef]
- Sheridan, C. Coronavirus and the race to distribute reliable diagnostics. Nat. Biotechnol. 2020, 38, 382. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [Green Version]
- Miller, S.; Chiu, C.; Rodino, K.G.; Miller, M.B. Point-Counterpoint: Should We Be Performing Metagenomic Next-Generation Sequencing for Infectious Disease Diagnosis in the Clinical Laboratory? J. Clin. Microbiol. 2020, 58, e01739-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, K.; Graziadio, S.; Turner, P.; Fanshawe, T.; Allen, J. Molecular and antibody point-of-care tests to support the screening, diagnosis and monitoring of COVID-19. Cent. Evid.-Based Med. 2020. Available online: https://www.cebm.net/wp-content/uploads/2020/04/POCT-Covid19.pdf (accessed on 9 November 2021).
- Available online: https://diagnostics.roche.com/us/en/landing-pages/roche-covid-19-updates.html (accessed on 3 June 2021).
- Dong, X.; Liu, L.; Tu, Y.; Zhang, J.; Miao, G.; Zhang, L.; Ge, S.; Xia, N.; Yu, D.; Qiu, X. Rapid PCR powered by microfluidics: A quick review under the background of COVID-19 pandemic. TrAC Trends Anal. Chem. 2021, 143, 116377. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhang, H.; Ni, S.; Korabečná, M.; Yobas, L.; Neuzil, P. The vision of point-of-care PCR tests for the COVID-19 pandemic and beyond. TrAC Trends Anal. Chem. 2020, 130, 115984. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Xu, Y.; Fohlerova, Z.; Chang, H.; Iliescu, C.; Neuzil, P. LAMP-on-a-chip: Revising microfluidic platforms for loop-mediated DNA amplification. TrAC Trends Anal. Chem. 2019, 113, 44–53. [Google Scholar] [CrossRef]
- Walker, G.T.; Fraiser, M.S.; Schram, J.L.; Little, M.C.; Nadeau, J.G.; Malinowski, D.P. Strand displacement amplification—An isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 1992, 20, 1691–1696. [Google Scholar] [CrossRef] [Green Version]
- Compton, J. Nucleic acid sequence-based amplification. Nature 1991, 350, 91–92. [Google Scholar] [CrossRef]
- Brentano, S.T.; Mcdonough, S.H. Isothermal amplification of RNA by transcription-mediated amplification (TMA). In Nonradioactive Analysis of Biomolecules; Springer: Berlin/Heidelberg, Germany, 2000; pp. 374–380. [Google Scholar]
- Yan, C.; Cui, J.; Huang, L.; Du, B.; Chen, L.; Xue, G.; Li, S.; Zhang, W.; Zhao, L.; Sun, Y. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020, 26, 773–779. [Google Scholar] [CrossRef]
- Zhang, Y.; Odiwuor, N.; Xiong, J.; Sun, L.; Nyaruaba, R.O.; Wei, H.; Tanner, N.A. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. MedRxiv 2020. [Google Scholar] [CrossRef]
- Chaibun, T.; Puenpa, J.; Ngamdee, T.; Boonapatcharoen, N.; Athamanolap, P.; O’Mullane, A.P.; Vongpunsawad, S.; Poovorawan, Y.; Lee, S.Y.; Lertanantawong, B. Rapid electrochemical detection of coronavirus SARS-CoV-2. Nat. Commun. 2021, 12, 802. [Google Scholar] [CrossRef] [PubMed]
- Behrmann, O.; Bachmann, I.; Spiegel, M.; Schramm, M.; Abd El Wahed, A.; Dobler, G.; Dame, G.; Hufert, F.T. Rapid detection of SARS-CoV-2 by low volume real-time single tube reverse transcription recombinase polymerase amplification using an exo probe with an internally linked quencher (exo-IQ). Clin. Chem. 2020, 66, 1047–1054. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Yaseen, A.B.; Kishi, J.Y.; Hong, F.; Saka, S.K.; Sheng, K.; Gopalkrishnan, N.; Schaus, T.E.; Yin, P. Single-strand RPA for rapid and sensitive detection of SARS-CoV-2 RNA. medRxiv 2020. [Google Scholar] [CrossRef]
- Jauset-Rubio, M.; Svobodová, M.; Mairal, T.; McNeil, C.; Keegan, N.; Saeed, A.; Abbas, M.N.; El-Shahawi, M.S.; Bashammakh, A.S.; Alyoubi, A.O. Ultrasensitive, rapid and inexpensive detection of DNA using paper based lateral flow assay. Sci. Rep. 2016, 6, 37732. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Suo, C.; Brown, T.; Wang, T.; Teichmann, S.A.; Bassett, A.R. INSIGHT: A scalable isothermal NASBA-based platform for COVID-19 diagnosis. bioRxiv 2020. [Google Scholar] [CrossRef]
- Schneider, M.; Iftner, T.; Ganzenmueller, T. Evaluation of the analytical performance and specificity of a SARS-CoV-2 transcription-mediated amplification assay. J. Virol. Methods 2021, 294, 114182. [Google Scholar] [CrossRef]
- Taki, K.; Yokota, I.; Fukumoto, T.; Iwasaki, S.; Fujisawa, S.; Takahashi, M.; Negishi, S.; Hayasaka, K.; Sato, K.; Oguri, S. SARS-CoV-2 detection by fluorescence loop-mediated isothermal amplification with and without RNA extraction. J. Infect. Chemother. 2021, 27, 410–412. [Google Scholar] [CrossRef]
- Lalli, M.A.; Langmade, J.S.; Chen, X.; Fronick, C.C.; Sawyer, C.S.; Burcea, L.C.; Wilkinson, M.N.; Fulton, R.S.; Heinz, M.; Buchser, W.J. Rapid and extraction-free detection of SARS-CoV-2 from saliva by colorimetric reverse-transcription loop-mediated isothermal amplification. Clin. Chem. 2021, 67, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Merindol, N.; Pépin, G.; Marchand, C.; Rheault, M.; Peterson, C.; Poirier, A.; Houle, C.; Germain, H.; Danylo, A. SARS-CoV-2 detection by direct rRT-PCR without RNA extraction. J. Clin. Virol. 2020, 128, 104423. [Google Scholar] [CrossRef]
- Thi, V.L.D.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.; Kirrmaier, D.; Freistaedter, A.; Papagiannidis, D.; Galmozzi, C.; Stanifer, M.L. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef]
- Sherrill-Mix, S.; Hwang, Y.; Roche, A.M.; Weiss, S.R.; Li, Y.; Graham-Wooten, J.; Taylor, L.J.; Collman, R.G.; Van Duyne, G.D.; Bushman, F.D. LAMP-BEAC: Detection of SARS-CoV-2 RNA Using RT-LAMP and Molecular Beacons. medRxiv 2020. [Google Scholar] [CrossRef]
- González-González, E.; Lara-Mayorga, I.M.; Rodríguez-Sánchez, I.P.; Zhang, Y.S.; Martínez-Chapa, S.O.; Trujillo-de Santiago, G.; Alvarez, M.M. Colorimetric loop-mediated isothermal amplification (LAMP) for cost-effective and quantitative detection of SARS-CoV-2: The change in color in LAMP-based assays quantitatively correlates with viral copy number. Anal. Methods 2021, 13, 169–178. [Google Scholar] [CrossRef]
- Thai, H.T.C.; Le, M.Q.; Vuong, C.D.; Parida, M.; Minekawa, H.; Notomi, T.; Hasebe, F.; Morita, K. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004, 42, 1956–1961. [Google Scholar] [CrossRef] [Green Version]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 2002, 16, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Francois, P.; Tangomo, M.; Hibbs, J.; Bonetti, E.-J.; Boehme, C.C.; Notomi, T.; Perkins, M.D.; Schrenzel, J. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol. Med. Microbiol. 2011, 62, 41–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellou, M.M.; Górska, A.; Mazzaferri, F.; Cremonini, E.; Gentilotti, E.; De Nardo, P.; Poran, I.; Leeflang, M.; Tacconelli, E.; Paul, M. Nucleic-acid-amplification tests from respiratory samples for the diagnosis of coronavirus infections: Systematic review and meta-analysis. Clin. Microbiol. Infect. 2020, 27, 341–351. [Google Scholar] [CrossRef]
- Available online: http://abbott.mediaroom.com.2020-03-27-Abbott-Launches-Molecular-Point-of-Care-Test-to-Detect-Novel-Coronavirus-in-as-Little-as-Five-Minutes (accessed on 20 November 2021).
- Available online: http://www.alere.com/en/home/product-details/id-now-covid-19.html (accessed on 20 November 2021).
- Zumla, A.; Al-Tawfiq, J.A.; Enne, V.I.; Kidd, M.; Drosten, C.; Breuer, J.; Muller, M.A.; Hui, D.; Maeurer, M.; Bates, M. Rapid point of care diagnostic tests for viral and bacterial respiratory tract infections—needs, advances, and future prospects. Lancet Infect. Dis. 2014, 14, 1123–1135. [Google Scholar] [CrossRef]
- Chandler-Brown, D.; Bueno, A.M.; Atay, O.; Tsao, D.S. A highly scalable and rapidly deployable RNA extraction-free COVID-19 assay by quantitative Sanger sequencing. Biorxiv 2020. [Google Scholar] [CrossRef]
- Bhoyar, R.C.; Jain, A.; Sehgal, P.; Divakar, M.K.; Sharma, D.; Imran, M.; Jolly, B.; Ranjan, G.; Rophina, M.; Sharma, S. High throughput detection and genetic epidemiology of SARS-CoV-2 using COVIDSeq next-generation sequencing. PLoS ONE 2021, 16, e0247115. [Google Scholar] [CrossRef]
- Available online: https://www.who.int/publications/i/item/10665-331501 (accessed on 2 June 2021).
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Singh, J.; Streithorst, J.; Granados, A.; Sotomayor-Gonzalez, A.; Zorn, K.; Gopez, A. Rapid detection of 2019 novel coronavirus SARS-CoV-2 using a CRISPR-based DETECTR lateral flow assay. MedRxiv 2020. [Google Scholar] [CrossRef]
- El-Tholoth, M.; Bau, H.H.; Song, J. A single and two-stage, closed-tube, molecular test for the 2019 novel coronavirus (COVID-19) at home, clinic, and points of entry. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Available online: https://www.fda.gov/media/137746/download (accessed on 15 June 2021).
- Ma, H.; Zeng, W.; He, H.; Zhao, D.; Jiang, D.; Zhou, P.; Cheng, L.; Li, Y.; Ma, X.; Jin, T. Serum IgA, IgM, and IgG responses in COVID-19. Cell. Mol. Immunol. 2020, 17, 773–775. [Google Scholar] [CrossRef] [PubMed]
- Long, Q.-X.; Liu, B.-Z.; Deng, H.-J.; Wu, G.-C.; Deng, K.; Chen, Y.-K.; Liao, P.; Qiu, J.-F.; Lin, Y.; Cai, X.-F. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020, 26, 845–848. [Google Scholar] [CrossRef]
- Amanat, F.; Stadlbauer, D.; Strohmeier, S.; Nguyen, T.H.; Chromikova, V.; McMahon, M.; Jiang, K.; Arunkumar, G.A.; Jurczyszak, D.; Polanco, J. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020, 26, 1033–1036. [Google Scholar] [CrossRef]
- Ni, L.; Ye, F.; Cheng, M.-L.; Feng, Y.; Deng, Y.-Q.; Zhao, H.; Wei, P.; Ge, J.; Gou, M.; Li, X. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity 2020, 52, 971–977.e3. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-point-care-antibody-test-covid-19 (accessed on 1 June 2021).
- Pan, Y.; Zhang, D.; Yang, P.; Poon, L.L.; Wang, Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020, 20, 411–412. [Google Scholar] [CrossRef]
- Okba, N.M.; Muller, M.A.; Li, W.; Wang, C.; GeurtsvanKessel, C.H.; Corman, V.M.; Lamers, M.M.; Sikkema, R.S.; de Bruin, E.; Chandler, F.D. SARS-CoV-2 specific antibody responses in COVID-19 patients. medRxiv 2020. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, M.; Yuan, J.; Wang, F.; Wang, Z.; Li, J.; Zhang, M.; Xing, L.; Wei, J.; Peng, L. Laboratory Diagnosis and Monitoring the Viral Shedding of SARS-CoV-2 Infection. Innovation 2020, 1, 100061. [Google Scholar] [CrossRef]
- Padoan, A.; Zuin, S.; Cosma, C.; Basso, D.; Plebani, M.; Bonfante, F. Clinical performances of an ELISA for SARS-CoV-2 antibody assay and correlation with neutralization activity. Clin. Chim. Acta Int. J. Clin. Chem. 2020, 510, 654. [Google Scholar] [CrossRef]
- Zhang, Z.L.; Hou, Y.L.; Li, D.T.; Li, F.Z. Diagnostic efficacy of anti-SARS-CoV-2 IgG/IgM test for COVID-19: A meta-analysis. J. Med Virol. 2021, 93, 366–374. [Google Scholar] [CrossRef]
- Chu, D.K.; Pan, Y.; Cheng, S.M.; Hui, K.P.; Krishnan, P.; Liu, Y.; Ng, D.Y.; Wan, C.K.; Yang, P.; Wang, Q. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020, 66, 549–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Yan, F. Patients with RT-PCR-confirmed COVID-19 and normal chest CT. Radiology 2020, 295, E3. [Google Scholar] [CrossRef] [PubMed]
- Whiting, P.; Singatullina, N.; Rosser, J. Computed tomography of the chest: I. Basic principles. BJA Educ. 2015, 15, 299–304. [Google Scholar] [CrossRef] [Green Version]
- Murphy, K.; Weaver, C. Janeway’s Immunobiology, 9th ed.; Garland Science, W.W. Norton & Company: New York, NY, USA, 2016. [Google Scholar]
- Dutta, N.K.; Mazumdar, K.; Gordy, J.T. The nucleocapsid protein of SARS–CoV-2: A target for vaccine development. J. Virol. 2020, 94, e00647-20. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.Y.; Ng, M.-Y.; Khong, P.-L. COVID-19 pneumonia: What has CT taught us? Lancet Infect. Dis. 2020, 20, 384–385. [Google Scholar] [CrossRef]
- Long, Q.-X.; Tang, X.-J.; Shi, Q.-L.; Li, Q.; Deng, H.-J.; Yuan, J.; Hu, J.-L.; Xu, W.; Zhang, Y.; Lv, F.-J. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020, 26, 1200–1204. [Google Scholar] [CrossRef]
- Sidiq, Z.; Hanif, M.; KumarDwivedi, K.; Chopra, K. Benefits and limitations of serological assays in COVID-19 infection. Indian J. Tuberc. 2020, 67, S163–S166. [Google Scholar] [CrossRef]
- Available online: https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/removal-lists-tests-should-no-longer-be-used-andor-distributed-covid-19-faqs-testing-sars-cov-2 (accessed on 8 November 2021).
- Available online: https://www.fda.gov/media/136625/download (accessed on 8 November 2021).
- Available online: https://www.fda.gov/media/137364/download (accessed on 8 November 2021).
- Available online: https://www.fda.gov/media/136963/download (accessed on 8 November 2021).
- Available online: https://diagnostics.roche.com/us/en/products/params/elecsys-anti-sars-cov-2.html (accessed on 7 June 2021).
- Available online: https://www.corelaboratory.abbott/us/en/offerings/segments/infectious-disease/sars-cov-2 (accessed on 5 June 2021).
- Available online: https://www.acrobiotech.com/covid-19-tests (accessed on 1 July 2021).
- Available online: https://www.cardinalhealth.com/en/cmp/ext/med/med-lab/hardy-diagnostics-autobio-anti-sars-cov-2-rapid-test.html (accessed on 3 June 2021).
- Available online: https://www.biolidics.com/2019-ncov-igg-igm-antibody-detection-kit (accessed on 28 June 2021).
- Available online: https://www.bio-rad.com/en-ro/sku/72710-platelia-sars-cov-2-total-ab-assay?ID=72710 (accessed on 29 June 2021).
- Available online: https://www.accessdata.fda.gov/cdrh_docs/presentations/maf/maf3267-a001.pdf (accessed on 28 June 2021).
- Available online: https://cellexcovid.com/ (accessed on 3 June 2021).
- Available online: https://en.wondfo.com.cn/pt/41index.html (accessed on 3 June 2021).
- Available online: https://covid-19-diagnostics.jrc.ec.europa.eu/devices/detail/675 (accessed on 28 June 2021).
- Available online: https://www.diasorin.com/en/node/11756/ (accessed on 1 July 2021).
- Available online: https://www.coronavirus-diagnostics.com/antibody-detection-tests-for-covid-19.html (accessed on 1 July 2021).
- Available online: https://www.orthoclinicaldiagnostics.com/global/covid19/antibody-test (accessed on 1 July 2021).
- Available online: https://doc.raybiotech.com/pdf/Manual/CG-CoV-IgM-IgG-RUO-FP.pdf (accessed on 1 July 2021).
- Available online: http://sdbiosensor.com/xe/product/7662# (accessed on 1 July 2021).
- Available online: https://www.siemens-healthineers.com/en-us/laboratory-diagnostics/assays-by-diseases-conditions/infectious-disease-assays/cov2t-assay (accessed on 1 July 2021).
- Available online: https://covid-19-diagnostics.jrc.ec.europa.eu/literature/detail/640 (accessed on 28 June 2021).
- Available online: http://sugentech.com/products/products-view.php?ct=7&target=32 (accessed on 29 June 2021).
- Available online: https://covid-19-diagnostics.jrc.ec.europa.eu/literature/detail/1023 (accessed on 28 June 2021).
- Guo, L.; Ren, L.; Yang, S.; Xiao, M.; Chang, D.; Yang, F.; Dela Cruz, C.S.; Wang, Y.; Wu, C.; Xiao, Y. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin. Infect. Dis. 2020, 71, 778–785. [Google Scholar] [CrossRef] [Green Version]
- Wang, P. Combination of serological total antibody and RT-PCR test for detection of SARS-CoV-2 infections. J. Virol. Methods 2020, 283, 113919. [Google Scholar] [CrossRef]
- Available online: https://www.coronavirus-diagnostics.com (accessed on 2 July 2021).
- Available online: https://www.acrobiotech.com/covid-19-test (accessed on 8 November 2021).
- Available online: https://www.snibe.com/zh_en/en_newsView.aspx?id=746 (accessed on 23 June 2021).
- Available online: https://www.biocompare.com/Life-Science-News/562900-SARS-CoV-2-COVID-19-Research-News-Latest-Updates (accessed on 6 November 2021).
- Bortz, R.H., III; Florez, C.; Laudermilch, E.; Wirchnianski, A.S.; Lasso, G.; Malonis, R.J.; Georgiev, G.I.; Vergnolle, O.; Herrera, N.G.; Morano, N.C. Single-Dilution COVID-19 Antibody Test with Qualitative and Quantitative Readouts. Msphere 2021, 6, e00224-21. [Google Scholar] [CrossRef]
- Pohanka, M. Point-of-Care Diagnoses and Assays Based on Lateral Flow Test. Int. J. Anal. Chem. 2021, 2021, 6685619. [Google Scholar] [CrossRef]
- Yadav, S.; Sadique, M.A.; Ranjan, P.; Kumar, N.; Singhal, A.; Srivastava, A.K.; Khan, R. SERS based lateral flow immunoassay for point-of-care detection of SARS-CoV-2 in clinical samples. ACS Appl. Bio Mater. 2021, 4, 2974–2995. [Google Scholar] [CrossRef]
- Kudr, J.; Michalek, P.; Ilieva, L.; Adam, V.; Zitka, O. COVID-19: A challenge for electrochemical biosensors. TrAC Trends Anal. Chem. 2021, 136, 116192. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Huang, C.; Huang, Z.; Lin, F.; He, Q.; Tao, D.; Jaffrezic-Renault, N.; Guo, Z. Advancements in electrochemical biosensing for respiratory virus detection: A review. TrAC Trends Anal. Chem. 2021, 139, 116253. [Google Scholar] [CrossRef]
- Ma, K.-S.; Zhou, H.; Zoval, J.; Madou, M. DNA hybridization detection by label free versus impedance amplifying label with impedance spectroscopy. Sens. Actuators B Chem. 2006, 114, 58–64. [Google Scholar] [CrossRef]
- Zhu, Z.; Shi, L.; Feng, H.; Zhou, H.S. Single domain antibody coated gold nanoparticles as enhancer for Clostridium difficile toxin detection by electrochemical impedance immunosensors. Bioelectrochemistry 2015, 101, 153–158. [Google Scholar] [CrossRef] [Green Version]
- Cui, F.; Zhou, H.S. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens. Bioelectron. 2020, 165, 112349. [Google Scholar] [CrossRef] [PubMed]
- Rashed, M.Z.; Kopechek, J.A.; Priddy, M.C.; Hamorsky, K.T.; Palmer, K.E.; Mittal, N.; Valdez, J.; Flynn, J.; Williams, S.J. Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. Biosens. Bioelectron. 2021, 171, 112709. [Google Scholar] [CrossRef]
- Chandra, P. Miniaturized label-free smartphone assisted electrochemical sensing approach for personalized COVID-19 diagnosis. Sens. Int. 2020, 1, 100019. [Google Scholar] [CrossRef]
- Imran, S.; Ahmadi, S.; Kerman, K. Electrochemical Biosensors for the Detection of SARS-CoV-2 and Other Viruses. Micromachines 2021, 12, 174. [Google Scholar] [CrossRef]
- Raziq, A.; Kidakova, A.; Boroznjak, R.; Reut, J.; Öpik, A.; Syritski, V. Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen. Biosens. Bioelectron. 2021, 178, 113029. [Google Scholar] [CrossRef]
- Singhal, A.; Parihar, A.; Kumar, N.; Khan, R. High throughput molecularly imprinted polymers based electrochemical nanosensors for point-of-care diagnostics of COVID-19. Mater. Lett. 2021, 306, 130898. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, P.; Singhal, A.; Yadav, S.; Kumar, N.; Murali, S.; Sanghi, S.K.; Khan, R. Rapid diagnosis of SARS-CoV-2 using potential point-of-care electrochemical immunosensor: Toward the future prospects. Int. Rev. Immunol. 2021, 40, 126–142. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Lu, G.; Yu, K.; Bo, Z.; Chen, J. Specific protein detection using thermally reduced graphene oxide sheet decorated with gold nanoparticle-antibody conjugates. Adv. Mater. 2010, 22, 3521–3526. [Google Scholar] [CrossRef] [PubMed]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef] [Green Version]
- Sengupta, J.; Hussain, C.M. Graphene-based field-effect transistor biosensors for the rapid detection and analysis of viruses: A perspective in view of COVID-19. Carbon Trends 2020, 2, 100011. [Google Scholar] [CrossRef]
- Mejía-Salazar, J.; Oliveira, O.N., Jr. Plasmonic biosensing: Focus review. Chem. Rev. 2018, 118, 10617–10625. [Google Scholar] [CrossRef]
- Liu, J.; Jalali, M.; Mahshid, S.; Wachsmann-Hogiu, S. Are plasmonic optical biosensors ready for use in point-of-need applications? Analyst 2020, 145, 364–384. [Google Scholar] [CrossRef] [Green Version]
- Iravani, S. Nano-and biosensors for the detection of SARS-CoV-2: Challenges and opportunities. Mater. Adv. 2020, 1, 3092–3103. [Google Scholar] [CrossRef]
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano 2020, 14, 5268–5277. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.R. Development of point-of-care biosensors for COVID-19. Front. Chem. 2020, 8, 517. [Google Scholar] [CrossRef] [PubMed]
- Berkenbrock, J.A.; Grecco-Machado, R.; Achenbach, S. Microfluidic devices for the detection of viruses: Aspects of emergency fabrication during the COVID-19 pandemic and other outbreaks. Proc. R. Soc. A 2020, 476, 20200398. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gong, Z.; Tang, W.; Row, K.H.; Qiu, H. Carbon dots in sample preparation and chromatographic separation: Recent advances and future prospects. TrAC Trends Anal. Chem. 2020, 134, 116135. [Google Scholar] [CrossRef]
- Mokhtarzadeh, A.; Eivazzadeh-Keihan, R.; Pashazadeh, P.; Hejazi, M.; Gharaatifar, N.; Hasanzadeh, M.; Baradaran, B.; de la Guardia, M. Nanomaterial-based biosensors for detection of pathogenic virus. TrAC Trends Anal. Chem. 2017, 97, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Nikaeen, G.; Abbaszadeh, S.; Yousefinejad, S. Application of nanomaterials in treatment, anti-infection and detection of coronaviruses. Nanomedicine 2020, 15, 1501–1512. [Google Scholar] [CrossRef]
- Sadique, M.A.; Yadav, S.; Ranjan, P.; Verma, S.; Salammal, S.T.; Khan, M.A.; Kaushik, A.; Khan, R. High-performance antiviral nano-systems as a shield to inhibit viral infections: SARS-CoV-2 as a model case study. J. Mater. Chem. B 2021. [Google Scholar] [CrossRef]
- Draz, M.S.; Shafiee, H. Applications of gold nanoparticles in virus detection. Theranostics 2018, 8, 1985. [Google Scholar] [CrossRef] [PubMed]
- Layqah, L.A.; Eissa, S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim. Acta 2019, 186, 224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Jeerapan, I.; Imani, S.; Cho, T.N.; Bandodkar, A.; Cinti, S.; Mercier, P.P.; Wang, J. Noninvasive alcohol monitoring using a wearable tattoo-based iontophoretic-biosensing system. ACS Sens. 2016, 1, 1011–1019. [Google Scholar] [CrossRef]
- Talebian, S.; Wallace, G.G.; Schroeder, A.; Stellacci, F.; Conde, J. Nanotechnology-based disinfectants and sensors for SARS-CoV-2. Nat. Nanotechnol. 2020, 15, 618–621. [Google Scholar] [CrossRef]
- Moitra, P.; Alafeef, M.; Dighe, K.; Frieman, M.B.; Pan, D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano 2020, 14, 7617–7627. [Google Scholar] [CrossRef]
- Zhong, J.; Rosch, E.L.; Viereck, T.; Schilling, M.; Ludwig, F. Toward rapid and sensitive detection of SARS-CoV-2 with functionalized magnetic nanoparticles. ACS Sens. 2021, 6, 976–984. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wen, T.; Shi, F.-J.; Zeng, X.-Y.; Jiao, Y.-J. Rapid detection of IgM antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticle-based lateral-flow assay. ACS Omega 2020, 5, 12550–12556. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Z.; Zhai, X.; Li, Y.; Lin, L.; Zhao, H.; Bian, L.; Li, P.; Yu, L.; Wu, Y. Rapid and sensitive detection of anti-SARS-CoV-2 IgG, using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal. Chem. 2020, 92, 7226–7231. [Google Scholar] [CrossRef]
- Chen, R.; Ren, C.; Liu, M.; Ge, X.; Qu, M.; Zhou, X.; Liang, M.; Liu, Y.; Li, F. Early Detection of SARS-CoV-2 Seroconversion in Humans with Aggregation-Induced Near-Infrared Emission Nanoparticle-Labeled Lateral Flow Immunoassay. ACS Nano 2021, 15, 8996–9004. [Google Scholar] [CrossRef] [PubMed]
- Gibani, M.M.; Toumazou, C.; Sohbati, M.; Sahoo, R.; Karvela, M.; Hon, T.-K.; De Mateo, S.; Burdett, A.; Leung, K.F.; Barnett, J. Assessing a novel, lab-free, point-of-care test for SARS-CoV-2 (CovidNudge): A diagnostic accuracy study. Lancet Microbe 2020, 1, e300–e307. [Google Scholar] [CrossRef]
- Yin, J.; Zou, Z.; Hu, Z.; Zhang, S.; Zhang, F.; Wang, B.; Lv, S.; Mu, Y. A “sample-in-multiplex-digital-answer-out” chip for fast detection of pathogens. Lab Chip 2020, 20, 979–986. [Google Scholar] [CrossRef]
- Ma, Y.-D.; Li, K.-H.; Chen, Y.-H.; Lee, Y.-M.; Chou, S.-T.; Lai, Y.-Y.; Huang, P.-C.; Ma, H.-P.; Lee, G.-B. A sample-to-answer, portable platform for rapid detection of pathogens with a smartphone interface. Lab Chip 2019, 19, 3804–3814. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Wu, X.; Wan, Z.; Li, Y.; Zuo, L.; Qin, J.; Jin, X.; Zhang, C. Development of a novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Virol. Sin. 2020, 35, 344–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, T.; Duong Bang, D.; Wolff, A. 2019 novel coronavirus disease (COVID-19): Paving the road for rapid detection and point-of-care diagnostics. Micromachines 2020, 11, 306. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.R.; Nilghaz, A.; Chen, L.; Chou, K.C.; Lu, X. Modification of thread-based microfluidic device with polysiloxanes for the development of a sensitive and selective immunoassay. Sens. Actuators B Chem. 2018, 260, 1043–1051. [Google Scholar] [CrossRef]
- Böhm, A.; Trosien, S.; Avrutina, O.; Kolmar, H.; Biesalski, M. Covalent attachment of enzymes to paper fibers for paper-based analytical devices. Front. Chem. 2018, 6, 214. [Google Scholar] [CrossRef]
- Hu, J.; Choi, J.R.; Wang, S.; Gong, Y.; Feng, S.; Pingguan-Murphy, B.; Lu, T.J.; Xu, F. Multiple test zones for improved detection performance in lateral flow assays. Sens. Actuators B Chem. 2017, 243, 484–488. [Google Scholar] [CrossRef]
- Rodriguez, N.M.; Wong, W.S.; Liu, L.; Dewar, R.; Klapperich, C.M. A fully integrated paperfluidic molecular diagnostic chip for the extraction, amplification, and detection of nucleic acids from clinical samples. Lab Chip 2016, 16, 753–763. [Google Scholar] [CrossRef]
- Choi, J.R.; Hu, J.; Tang, R.; Gong, Y.; Feng, S.; Ren, H.; Wen, T.; Li, X.; Abas, W.A.B.W.; Pingguan-Murphy, B. An integrated paper-based sample-to-answer biosensor for nucleic acid testing at the point of care. Lab Chip 2016, 16, 611–621. [Google Scholar] [CrossRef]
- Tang, R.; Yang, H.; Choi, J.R.; Gong, Y.; Hu, J.; Wen, T.; Li, X.; Xu, B.; Mei, Q.; Xu, F. based device with on-chip reagent storage for rapid extraction of DNA from biological samples. Microchim. Acta 2017, 184, 2141–2150. [Google Scholar] [CrossRef]
- Reboud, J.; Xu, G.; Garrett, A.; Adriko, M.; Yang, Z.; Tukahebwa, E.M.; Rowell, C.; Cooper, J.M. based microfluidics for DNA diagnostics of malaria in low resource underserved rural communities. Proc. Natl. Acad. Sci. USA 2019, 116, 4834–4842. [Google Scholar] [CrossRef] [Green Version]
- Trinh, T.N.D.; La, H.C.; Lee, N.Y. Fully integrated and foldable microdevice encapsulated with agarose for long-term storage potential for point-of-care testing of multiplex foodborne pathogens. ACS Sens. 2019, 4, 2754–2762. [Google Scholar] [CrossRef]
- Kukhtin, A.C.; Sebastian, T.; Golova, J.; Perov, A.; Knickerbocker, C.; Linger, Y.; Bueno, A.; Qu, P.; Villanueva, M.; Holmberg, R.C. Lab-on-a-Film disposable for genotyping multidrug-resistant Mycobacterium tuberculosis from sputum extracts. Lab Chip 2019, 19, 1217–1225. [Google Scholar] [CrossRef]
- Aalberts, M.; van Dissel-Emiliani, F.M.F.; van Adrichem, N.P.H.; van Wijnen, M.; Wauben, M.H.; Stout, T.A.; Stoorvogel, W. Identification of distinct populations of prostasomes that differentially express prostate stem cell antigen, annexin A1, and GLIPR2 in humans. Biol. Reprod. 2012, 86. [Google Scholar] [CrossRef]
- Available online: http://www.cepheid.com/coronavirus (accessed on 3 June 2021).
- Available online: https://www.fda.gov/news-events/press-announcements/covid-19-update-fda-authorizes-first-diagnostic-test-where-results-can-be-read-directly-testing-card (accessed on 3 June 2021).
- Pollock, N.R.; Jacobs, J.R.; Tran, K.; Cranston, A.; Smith, S.; O’Kane, C.; Roady, T.; Moran, A.; Scarry, A.; Carroll, M. Performance and Implementation Evaluation of the Abbott BinaxNOW Rapid Antigen Test in a High-throughput Drive-through Community Testing Site in Massachusetts. J. Clin. Microbiol. 2021, 59, e00083-21. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.fda.gov/media/136525/download (accessed on 20 June 2021).
- Available online: https://www.cuehealth.com/news-listing/2020/3/26/cuehealthbardaCOVID-19 (accessed on 20 June 2021).
- Available online: https://www.fda.gov/news-events/press-announcements/fda-permits-marketing-first-sars-cov-2-diagnostic-test-using-traditional-premarket-review-process (accessed on 20 June 2021).
- Available online: https://www.fda.gov/media/144656/download (accessed on 20 June 2021).
- Available online: https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas (accessed on 20 June 2021).
- Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-continues-advance-over-counter-and-other-screening-test-developmen (accessed on 25 June 2021).
- Available online: https://bdveritor.bd.com/en-us/bd-veritor-plus-system (accessed on 25 June 2021).
- Available online: https://www.fda.gov/media/147265/download (accessed on 25 June 2021).
- Available online: https://www.fda.gov/media/147254/download (accessed on 26 June 2021).
- Available online: https://www.fda.gov/media/147259/download (accessed on 25 June 2021).
- Available online: https://www.fda.gov/media/147264/download (accessed on 26 June 2021).
- Available online: https://www.fda.gov/media/144659/download (accessed on 27 June 2021).
- Available online: https://www.fda.gov/media/141951/download (accessed on 25 June 2021).
- Available online: https://www.fda.gov/media/146499/download (accessed on 25 June 2021).
- Available online: https://www.fda.gov/media/138654/download (accessed on 28 June 2021).
- Kim, H.E.; Schuck, A.; Lee, S.H.; Lee, Y.; Kang, M.; Kim, Y.-S. Sensitive electrochemical biosensor combined with isothermal amplification for point-of-care COVID-19 tests. Biosens. Bioelectron. 2021, 182, 113168. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Haage, V.C.; Bleicker, T.; Schmidt, M.L.; Mühlemann, B.; Zuchowski, M.; Jo, W.K.; Tscheak, P.; Möncke-Buchner, E.; Müller, M.A. Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests: A single-centre laboratory evaluation study. Lancet Microbe 2021, 2, e311–e319. [Google Scholar] [CrossRef]
- Moeller, M.E.; Fock, J.; Pah, P.; Veras, A.D.L.C.; Bade, M.; Donolato, M.; Israelsen, S.B.; Eugen-Olsen, J.; Benfield, T.; Engsig, F.N. Evaluation of commercially available immuno-magnetic agglutination in comparison to enzyme-linked immunosorbent assays for rapid point-of-care diagnostics of COVID-19. J. Med Virol. 2021, 93, 3084–3091. [Google Scholar] [CrossRef] [PubMed]
- Whitman, J.D.; Hiatt, J.; Mowery, C.T.; Shy, B.R.; Yu, R.; Yamamoto, T.N.; Rathore, U.; Goldgof, G.M.; Whitty, C.; Woo, J.M. Test performance evaluation of SARS-CoV-2 serological assays. Nat. Biotechnol. 2020, 38, 1174. [Google Scholar] [CrossRef]
- Kost, G.J. Designing and Interpreting Coronavirus Disease 2019 (COVID-19) Diagnostics: Mathematics, Visual Logistics, and Low Prevalence. Arch. Pathol. Lab. Med. 2021, 145, 291–307. [Google Scholar] [CrossRef]
- Kost, G.J. Diagnostic strategies for endemic coronavirus disease 2019 (COVID-19): Rapid antigen tests, repeat testing, and prevalence boundaries. Arch. Pathol. Lab. Med. 2021. [Google Scholar] [CrossRef]
- Kost, G.J. The Impact of Increasing Disease Prevalence, False Omissions, and Diagnostic Uncertainty on Coronavirus Disease 2019 (COVID-19) Test Performance. Arch. Pathol. Lab. Med. 2021, 145, 797–813. [Google Scholar] [CrossRef] [PubMed]
- McGuinness, B.; Troncone, M.; James, L.P.; Bisch, S.P.; Iyer, V. Reassessing the operative threshold for abdominal aortic aneurysm repair in the context of COVID-19. J. Vasc. Surg. 2021, 73, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Kalish, H.; Klumpp-Thomas, C.; Hunsberger, S.; Baus, H.A.; Fay, M.P.; Siripong, N.; Wang, J.; Hicks, J.; Mehalko, J.; Travers, J. Undiagnosed SARS-CoV-2 Seropositivity During the First Six Months of the COVID-19 Pandemic in the United States. Sci. Transl. Med. 2021, 13. [Google Scholar] [CrossRef]
- Available online: https://www.idsociety.org/practice-guideline/covid-19-guideline-diagnostics (accessed on 18 June 2021).
- Datta, M.; Singh, D.D.; Naqvi, A.R. Molecular Diagnostic Tools for the Detection of SARS-CoV-2. Int. Rev. Immunol. 2021, 40, 143–156. [Google Scholar] [CrossRef]
- Jalandra, R.; Yadav, A.K.; Verma, D.; Dalal, N.; Sharma, M.; Singh, R.; Kumar, A.; Solanki, P.R. Strategies and perspectives to develop SARS-CoV-2 detection methods and diagnostics. Biomed. Pharmacother. 2020, 129, 110446. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waggoner, J.J.; Stittleburg, V.; Pond, R.; Saklawi, Y.; Sahoo, M.K.; Babiker, A.; Hussaini, L.; Kraft, C.S.; Pinsky, B.A.; Anderson, E.J. Triplex real-time RT-PCR for severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020, 26, 1633. [Google Scholar] [CrossRef] [PubMed]
- Khalilov, R.; Hosainzadegan, M.; Eftekhari, A. Necessity of different countries to deal with similar phenomena of COVID-19 coronavirus. Adv. Biol. Earth Sci 2020, 5, 5–6. [Google Scholar]
- Döhla, M.; Boesecke, C.; Schulte, B.; Diegmann, C.; Sib, E.; Richter, E.; Eschbach-Bludau, M.; Aldabbagh, S.; Marx, B.; Eis-Hübinger, A.-M. Rapid point-of-care testing for SARS-CoV-2 in a community screening setting shows low sensitivity. Public Health 2020, 182, 170–172. [Google Scholar] [CrossRef]
- Arevalo-Rodriguez, I.; Buitrago-Garcia, D.; Simancas-Racines, D.; Zambrano-Achig, P.; Del Campo, R.; Ciapponi, A.; Sued, O.; Martinez-Garcia, L.; Rutjes, A.W.; Low, N. False-negative results of initial RT-PCR assays for COVID-19: A systematic review. PLoS ONE 2020, 15, e0242958. [Google Scholar] [CrossRef]
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.-Y.; Chen, L.; Wang, M. Presumed asymptomatic carrier transmission of COVID-19. JAMA 2020, 323, 1406–1407. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.; Liu, T.; Huang, L.; Liu, H.; Lei, M.; Xu, W.; Hu, X.; Chen, J.; Liu, B. Use of chest CT in combination with negative RT-PCR assay for the 2019 novel coronavirus but high clinical suspicion. Radiology 2020, 295, 22–23. [Google Scholar] [CrossRef] [PubMed]
- Veyrenche, N.; Bolloré, K.; Pisoni, A.; Bedin, A.S.; Mondain, A.M.; Ducos, J.; Segondy, M.; Montes, B.; Pastor, P.; Morquin, D. Diagnosis value of SARS-CoV-2 antigen/antibody combined testing using rapid diagnostic tests at hospital admission. J. Med Virol. 2021, 93, 3069–3076. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- To, K.K.-W.; Tsang, O.T.-Y.; Yip, C.C.-Y.; Chan, K.-H.; Wu, T.-C.; Chan, J.M.-C.; Leung, W.-S.; Chik, T.S.-H.; Choi, C.Y.-C.; Kandamby, D.H. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020, 71, 841–843. [Google Scholar] [CrossRef] [Green Version]
- Ismail, S.A.; Huntley, C.; Post, N.; Rigby, S.; Shrotri, M.; Williams, S.V.; Peacock, S.J. Horses for courses? Assessing the potential value of a surrogate, point-of-care test for SARS-CoV-2 epidemic control. Influenza Other Respir. Viruses 2021, 15, 3–6. [Google Scholar] [CrossRef]
- Dhawan, A.P.; Heetderks, W.J.; Pavel, M.; Acharya, S.; Akay, M.; Mairal, A.; Wheeler, B.; Dacso, C.C.; Sunder, T.; Lovell, N. Current and future challenges in point-of-care technologies: A paradigm-shift in affordable global healthcare with personalized and preventive medicine. IEEE J. Transl. Eng. Health Med. 2015, 3, 1–10. [Google Scholar] [CrossRef]
- Magro, L.; Escadafal, C.; Garneret, P.; Jacquelin, B.; Kwasiborski, A.; Manuguerra, J.-C.; Monti, F.; Sakuntabhai, A.; Vanhomwegen, J.; Lafaye, P. Paper microfluidics for nucleic acid amplification testing (NAAT) of infectious diseases. Lab Chip 2017, 17, 2347–2371. [Google Scholar] [CrossRef] [Green Version]
- Phan, Q.A.; Truong, L.B.; Medina-Cruz, D.; Dincer, C.; Mostafavi, E. CRISPR/Cas-powered nanobiosensors for diagnostics. Biosens. Bioelectron. 2021, 197, 113732. [Google Scholar] [CrossRef]
- Antiochia, R. Nanobiosensors as new diagnostic tools for SARS, MERS and COVID-19: From past to perspectives. Microchim. Acta 2020, 187, 639. [Google Scholar] [CrossRef]
- Ishikawa, F.N.; Chang, H.-K.; Curreli, M.; Liao, H.-I.; Olson, C.A.; Chen, P.-C.; Zhang, R.; Roberts, R.W.; Sun, R.; Cote, R.J. Label-free, electrical detection of the SARS virus N-protein with nanowire biosensors utilizing antibody mimics as capture probes. ACS Nano 2009, 3, 1219–1224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunea, A.-C.; Dediu, V.; Laszlo, E.A.; Pistriţu, F.; Carp, M.; Iliescu, F.S.; Ionescu, O.N.; Iliescu, C. E-Skin: The Dawn of a New Era of On-Body Monitoring Systems. Micromachines 2021, 12, 1091. [Google Scholar] [CrossRef] [PubMed]
- Sadique, M.A.; Yadav, S.; Ranjan, P.; Khan, M.A.; Kumar, A.; Khan, R. Rapid detection of SARS-CoV-2 using graphene-based IoT integrated advanced electrochemical biosensor. Mater. Lett. 2021, 305, 130824. [Google Scholar] [CrossRef]
| Assay & Company. | RNA Extraction | Isothermal Reaction | Type of Detection (LoD) | Required Platform |
|---|---|---|---|---|
| Color SARS-CoV-2 RT-LAMP Diagnostic Assay (Color Health, Inc.) | Require RNA extraction | RT-LAMP | Colourimetric (0.75 copies/μL) | Microplates read by spectrophotometry |
| Lucira CHECK-IT COVID-19 Test Kit (Lucira Health, Inc.) | Includes RNA extraction | RT-LAMP | Colourimetric (2700 copies/swab) | Microfluidic, over-the-counter device |
| Aptima SARS-CoV-2 assay (Hologic, Inc.) | Includes RNA extraction | TMA multiplex | Chemiluminiscence (0.1 TCID50/mL) | Automate system Aptima Panther (Hologic Inc.) |
| Cue COVID-19 Test for Home and Over The Counter (OTC) Use (Cue Health Inc.) | Does not require RNA extraction | unspecified | Electrochemical (20 copies/swab or 1.3 copies/μL) | Portable device Cue Health Inc. |
| Solana SARS-CoV-2 Assay (Quidel Corporation) | sample prep. (heat treatment) | RT-HDA | Fluorescence (1.16 × 104 copies/mL) | Solana Instrument (Quidel Corporation) |
| Sherlock CRISPR SARS-CoV-2 Kit (Sherlock BioSciences, Inc.) | Require RNA extraction | RT-LAMP | Fluorescence aided by the enzymatic system CRISP/Cas (LoD = 6.75 copies/μL VTM) | BioTek NEO2 microplate reader |
| iAMP COVID-19 Detection Kit (Atila BioSystems, In.) | Does not require RNA extraction | OEMGA | Fluorescence (2000 copies/swab) | Real-time PCR instrument |
| MobileDetect Bio BCC19 (MD-Bio BCC19) Test Kit (Detectachem Inc.) | Does not require RNA extraction | RT-LAMP | Colourimetric (75 copies/μL) | In tube reaction. |
| ID NOW COVID-19 (Abbott Diagnostics Inc.) | sample prep. (heat treatment) | unspecified | Fluorescence (125 copies/mL) | ID NOW instrument |
| SARS-CoV-2 DETECTR Reagent Kit (Mammoth Biosciences, Inc.) | Require RNA extraction | RT-LAMP | Fluorescence aided by the enzymatic system CRISP/Cas (20 copies/μL VTM) | rtPCR instrument |
| ProAmpRT SARS-CoV-2 TEST (Pro-Lab Diagnostics) | Require RNA extraction | unspecified | Fluorescence (LoD = 125 copies/swab) | Genie HT Instrument (OptiGene) |
| SARS-CoV-2 RNA DETECTR Assay (USCF Health Clinical Laboratories) | Require RNA extraction | RT-LAMP | Fluorescence aided by the enzymatic system CRISP/Cas CRISPR/Cas (20 copies/μL) | Real-time PCR instrument |
| Name/Manufacturer | Type of Test | Detection/Turnaround Time | Sample | Intended Use | Performance | Sensitivity LoD | Reference |
|---|---|---|---|---|---|---|---|
| BD VeritorTM/Becton, Dickinson | Chromatographic digital immunoassay | Rapid test Qualitative det. Of viral nucleocapsid antigens 15 min | direct anterior nasal swabs from symptomatic, asymptomatic individuals for processing within 60 min | For prescription use as POCT/ screening with a prescription under CLIA with the BD Veritor™ Plus Analyzer | PPA: 84% (95% CI: 67%–93%) NPA: 100% (95% CI: 98%-100%) | 1.4 × 102 TCID50/mL | [189,199] |
| QuickVue At-Home OTC COVID-19 Test and Kit/Quidel Corp. | LFIA | Rapid test, Qualitative det. of viral nucleocapsid antigens 10 min | direct anterior nasal swabs | OTC at-home serial screening for symptomatic, asymptomatic individuals | PPA: 83% (95% CI: 74.9–89.6) NPA: 99.2% (95% CI: 97.2–99.8) does not differentiate between SARS-CoV and SARS-CoV-2 | 1.91 × 104 TCID50/mL | [200] |
| BinaxNOW COVID-19 Antigen Self Test/Abbott Diagnostics | LFIC membrane assay | Qualitative detection of nucleocapsid protein antigen w/o viral transport media 15 min | direct anterior nasal swabs | OTC at-home serial screening for symptomatic, asymptomatic individuals | PPA: 77.2% (95% CI: 70.1–83.4) NPA: 98% (95% CI: 96.6–99.5) Sensitivity of the assay decreases over time. The presence of mupirocin may interfere with the BinaxNOW COVID-19 Antigen Self Test and may cause false negative results | 140.6 TCID50/mL | [201] |
| BinaxNOW COVID-19 Ag Card 2 Home Test/Abbott Diagnostics | LFIC membrane assay | Qualitative detection of nucleocapsid protein antigen w/o viral transport media 15 min | direct anterior nasal swabs from symptomatic, asymptomatic individuals | OTC at-home serial screening only with the supervision of a telehealth proctor. The results are reported to the user and to the relevant public health authorities | PPA: 78.3% (95% CI: 71.1–84.4) NPA: 98% (95% CI: 96.6–99.5) Sensitivity of the assay decreases over time does not differentiate between SARS-CoV and SARSCoV-2. | 140.6 TCID50/mL | [202] |
| BinaxNOW COVID-19 Ag 2 Card kit/Abbott Diagnostics | LFIC membrane assay | Qualitative detection of nucleocapsid protein antigen w/o viral transport media 15 min | direct anterior nasal swabs | POCT screening in patient care settings operating under a CLIA Certificate for symptomatic, asymptomatic individuals | PPA: 77.2% (95% CI: 70.1–84.4%) NPA: 98% (95% CI: 96.6–99.5%) sensitivity of the assay decreases over time does not differentiate between SARS-CoV and SARSCoV-2 | 140.6 TCID50/mL | [203] |
| MatMaCorp COVID-19 2SF Test/ DBA MatmaCorp, | RT-PCR + isothermal NAA | Qualitative detection of nucleic acid from SARS-CoV-2 | nasopharyngeal, mid-turbinate, anterior nasal swabs | POCT screening in patient care settings. for symptomatic, asymptomatic individuals; it comprises sample preparation and amplification and detection. | PPA: 88.5% (95% CI: 79.5–93.8%) NPA: 100% (95% CI: 88.7–100%). | 2000 genome equivalents per ml (100 genome equivalents per 50 µL reaction) | [204] |
| GENETWORx COVID-19 Nasal Swab Test and Kit/RCA Lab. Services LLC | rt RT-PCR (Home Collection) | Qualitative detection of nucleic acid from SARS-CoV-2 | nasal swab | unsupervised at home self-collected samples, by qualified laboratory personnel | PPA: 99.6% NPA as expected with the correct collected samples | 1.8 × 104 NDU/mL ** | [43] |
| Verily COVID-19 RT-PCR Test and Kit/Verily Life Sciences | rt RT-PCR | qualitative detection of nucleic acid from SARS-CoV-2 | anterior nasal, mid-turbinate, nasopharyngeal, and oropharyngeal swab | Pooling, Home Collection, unsupervised at home self-collected | PPA: 100% (95% CI: 89.9–100%) NPA: 100% (95% CI: 88.7–100%) low viral loads may not be detected in sample pools due to the decreased sensitivity of pooled testing | 60 GCE/mL | [205] |
| CRSP SARS-CoV-2 RRT-PCR Diagnostic Assay/CRSP, LLC at MIT & Harvard | Real-time RT-PCR | qualitative detection of nucleic acid from SARS-CoV-2 | unsupervised at home self-collected nasal swab | individuals suspected of COVID-19 by their healthcare provider | PPA: 98.3% (95% CI: 91–99.7%) NPA: 100.0% (95% CI: 96.6–100) Improper collection, transport, or storage of specimens may lower the efficiency | 1600 copies/mL | [206] |
| Phosphorus COVID-19 RT-qPCR/Phosphorus Diagnostics | Real-time RT-PCR | the qualitative detection of nucleic acid from SARS-CoV-2 in | Upper respiratory tract swabs, washes/ aspirates, bronchoalveolar lavage (BAL) specimens from (2) saliva specimens | For individuals suspected of COVID-19 by their healthcare provider; | PPA: 95.0% (95% CI: 76.4–99.1) NPA: 99.2% (95% CI: 95.7–99.8) | 5 copies/ µL in NP swab; 1.0 copy/µL in saliva | [207] |
| Test Type | Recommending for | Comments |
|---|---|---|
| Serological | the evaluation of patients with a high clinical suspicion for COVID-19 and with negative molecular testing at at least two weeks since symptom onset | The certainty of available evidence supporting the use of serology for either diagnosis or epidemiology was, however, graded as very low to moderate. [211] |
| he assessment of multisystem inflammatory syndrome in children | ||
| the serosurveillance studies | ||
| Molecular | all symptomatic individuals suspected of having COVID-19 | prioritization of testing will depend on institutional-specific resources and the needs of different patient populations. [211] |
| asymptomatic individuals with known or suspected contact with a COVID-19 case | ||
| asymptomatic individuals without known exposure is suggested when the results will impact isolation/quarantine/personal protective equipment (PPE) usage decisions, dictate eligibility for surgery, or inform solid organ or hematopoietic stem cell transplantation timing |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iliescu, F.S.; Ionescu, A.M.; Gogianu, L.; Simion, M.; Dediu, V.; Chifiriuc, M.C.; Pircalabioru, G.G.; Iliescu, C. Point-of-Care Testing—The Key in the Battle against SARS-CoV-2 Pandemic. Micromachines 2021, 12, 1464. https://doi.org/10.3390/mi12121464
Iliescu FS, Ionescu AM, Gogianu L, Simion M, Dediu V, Chifiriuc MC, Pircalabioru GG, Iliescu C. Point-of-Care Testing—The Key in the Battle against SARS-CoV-2 Pandemic. Micromachines. 2021; 12(12):1464. https://doi.org/10.3390/mi12121464
Chicago/Turabian StyleIliescu, Florina Silvia, Ana Maria Ionescu, Larisa Gogianu, Monica Simion, Violeta Dediu, Mariana Carmen Chifiriuc, Gratiela Gradisteanu Pircalabioru, and Ciprian Iliescu. 2021. "Point-of-Care Testing—The Key in the Battle against SARS-CoV-2 Pandemic" Micromachines 12, no. 12: 1464. https://doi.org/10.3390/mi12121464
APA StyleIliescu, F. S., Ionescu, A. M., Gogianu, L., Simion, M., Dediu, V., Chifiriuc, M. C., Pircalabioru, G. G., & Iliescu, C. (2021). Point-of-Care Testing—The Key in the Battle against SARS-CoV-2 Pandemic. Micromachines, 12(12), 1464. https://doi.org/10.3390/mi12121464