A Pillar-Free Diffusion Device for Studying Chemotaxis on Supported Lipid Bilayers
Abstract
1. Introduction
2. Materials and Methods
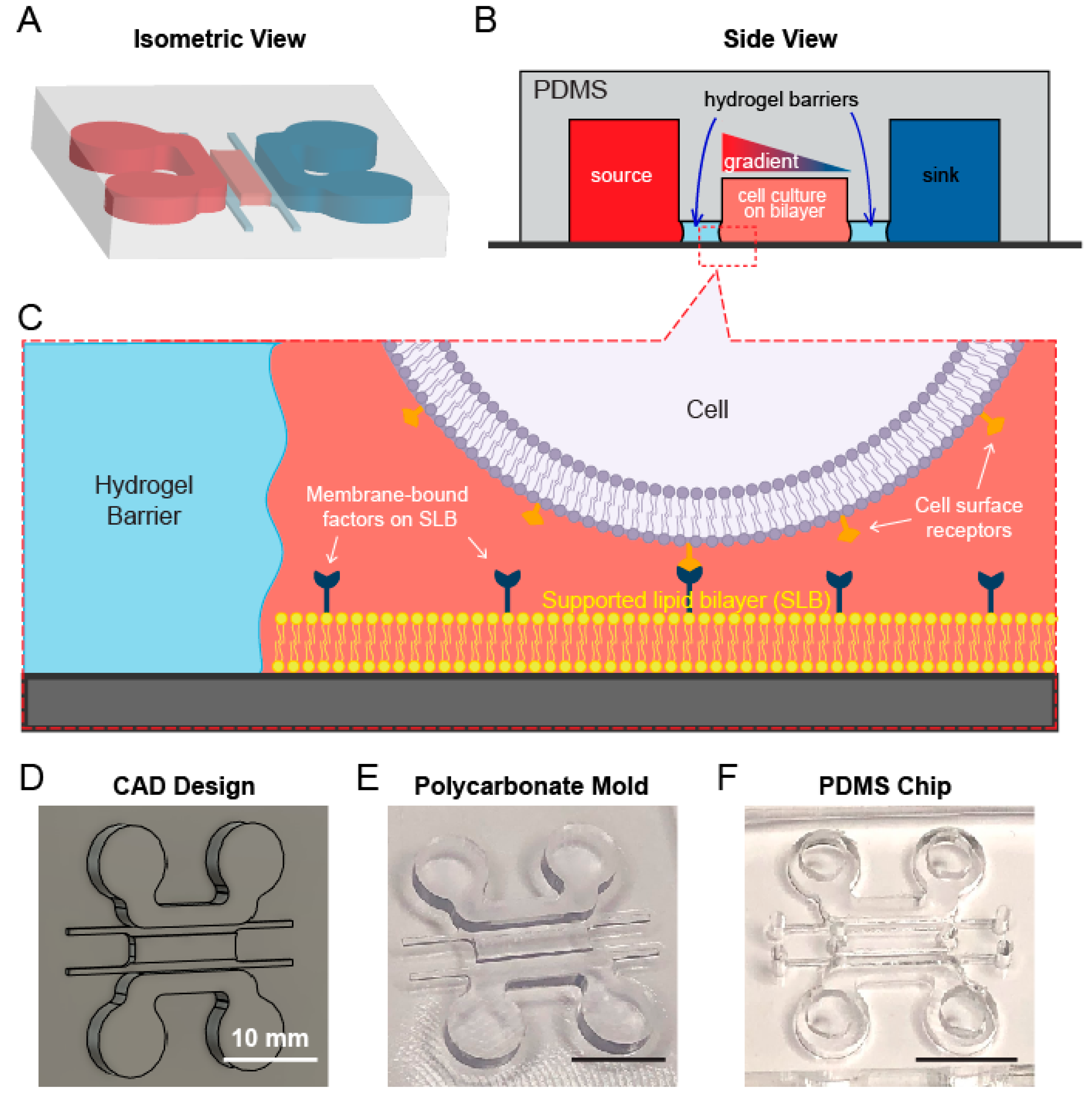
2.1. Design, Fabrication, and Assembly of the Microfluidic Device
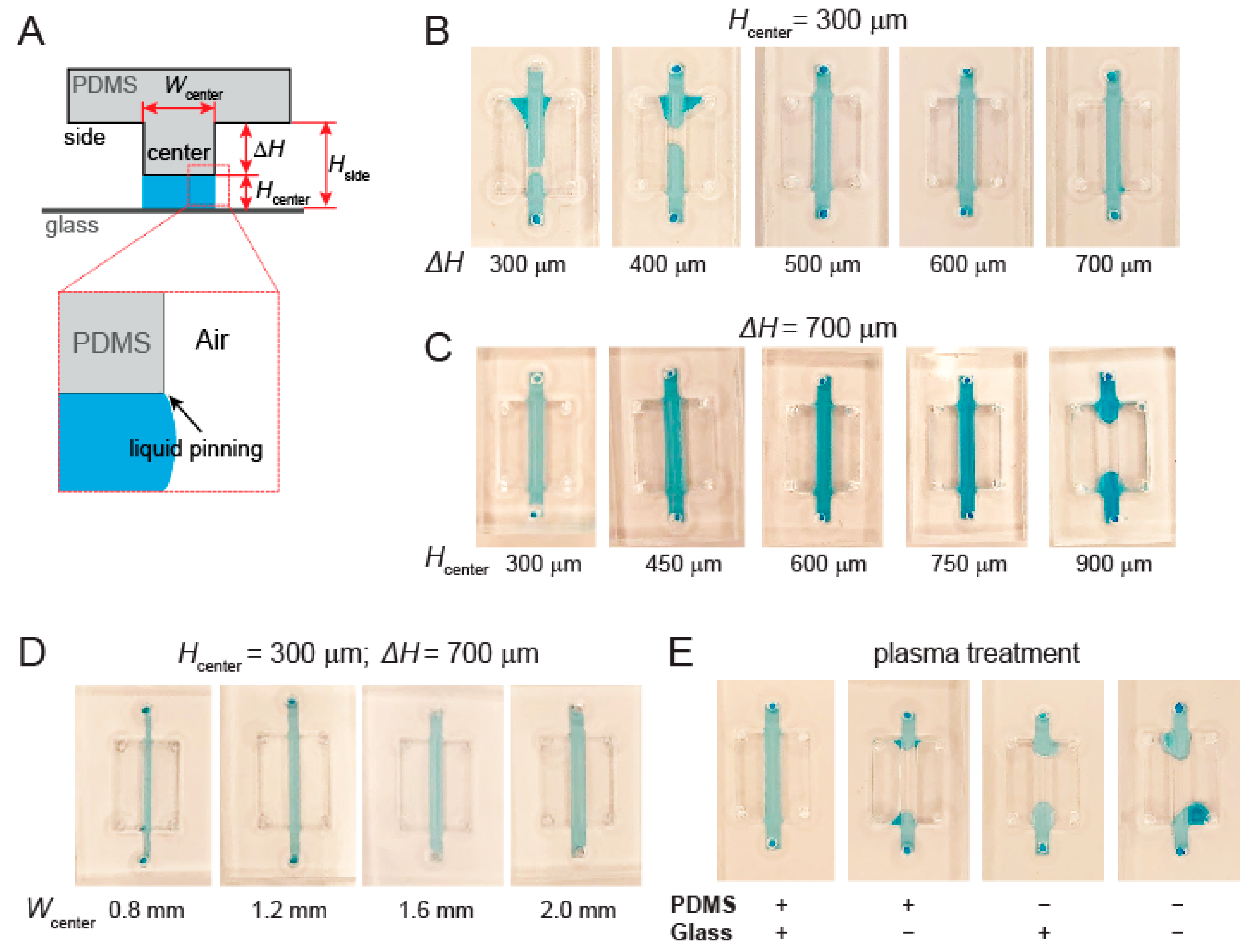
2.2. Formation of Gel Diffusion Barrier and Workflow of the Device
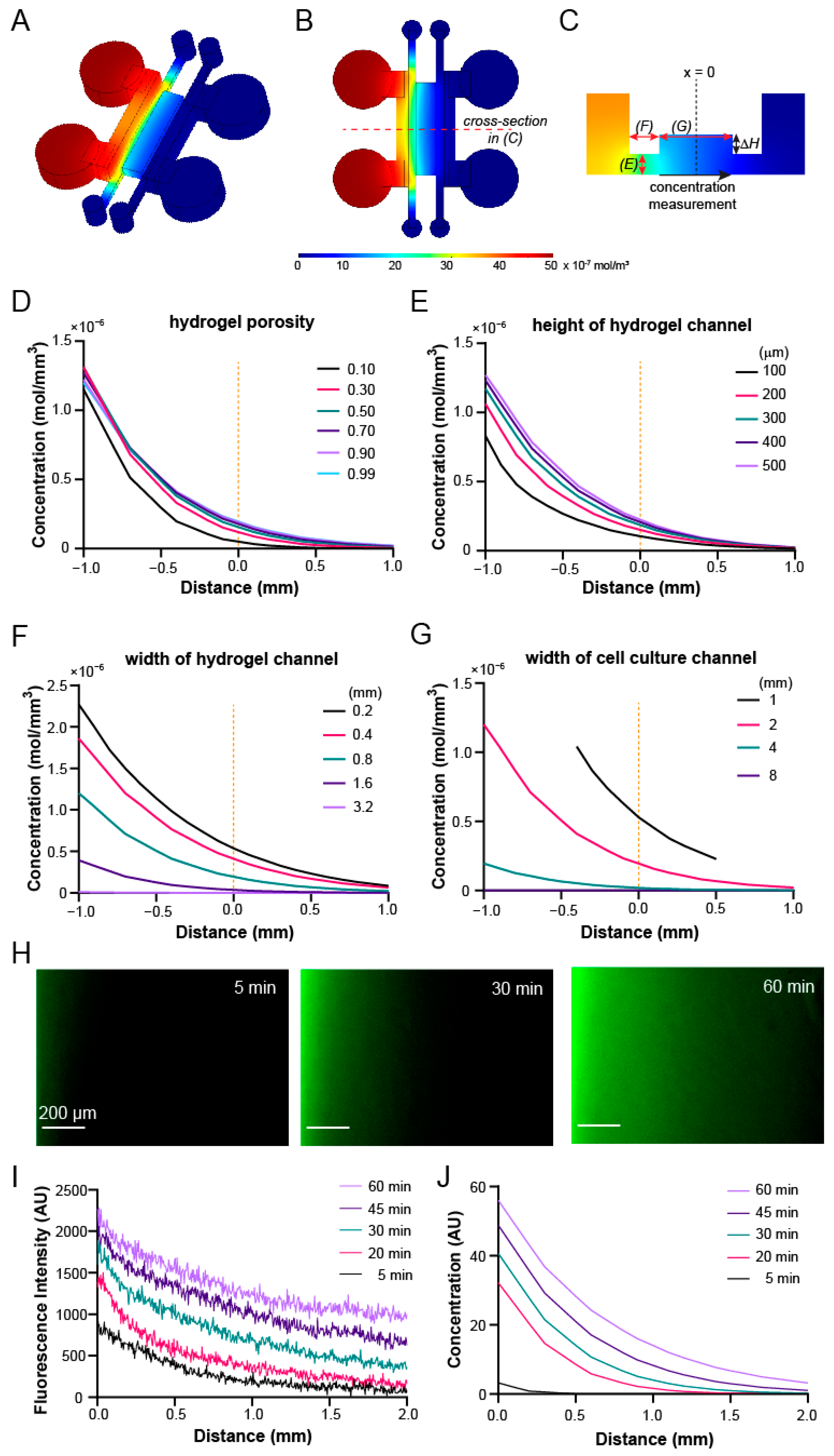
2.3. COMSOL Simulation and Characterization of Transport Phenomena with FITC-Dextran
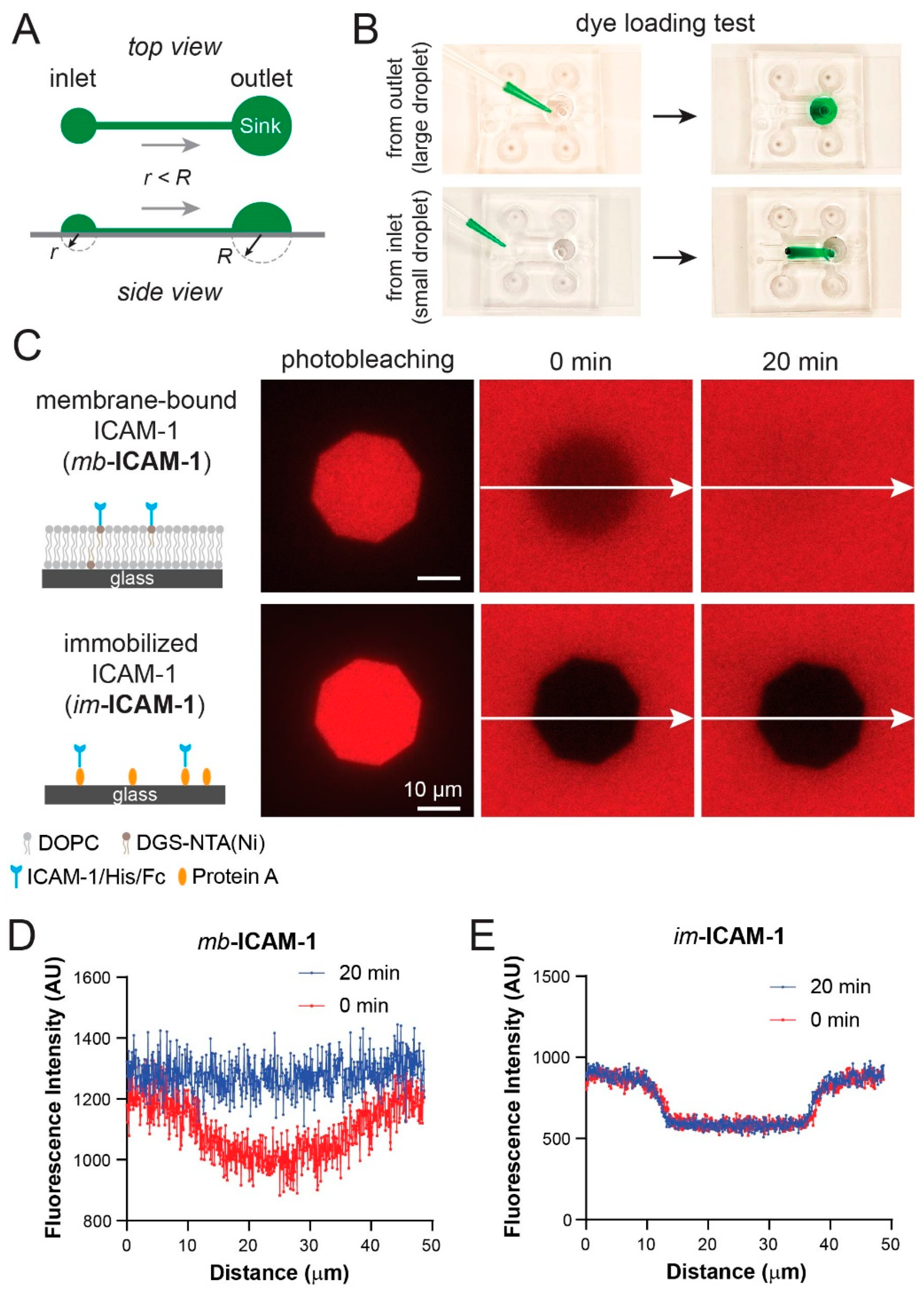
2.4. Preparation of Supported Lipid Bilayers and Protein Tethered Surfaces
2.5. ICAM-1 Capturing on Lipid Bilayer and Immobilization
2.6. Cell Seeding and Incubation
2.7. Imaging, Cell Tracking, and Data Analysis
2.8. Statistics
3. Results
3.1. A Multichannel Device Design Allows for Separate Lipid Bilayer and Chemoattractant Gradient Formation
3.2. Channel Height and Surface Treatment Are Key to Liquid Pinning-Based Hydrogel Barrier Formation
3.3. Coating Hydrogel Channel Is Necessary to Prevent Leakage of Soluble Factors
3.4. Gradient Profiles within the Device Can Be Optimized through COMSOL Simulation
3.5. Fluorescence Recovery after Photobleaching Confirms Lipid Bilayer Formation and Mobility in the Device
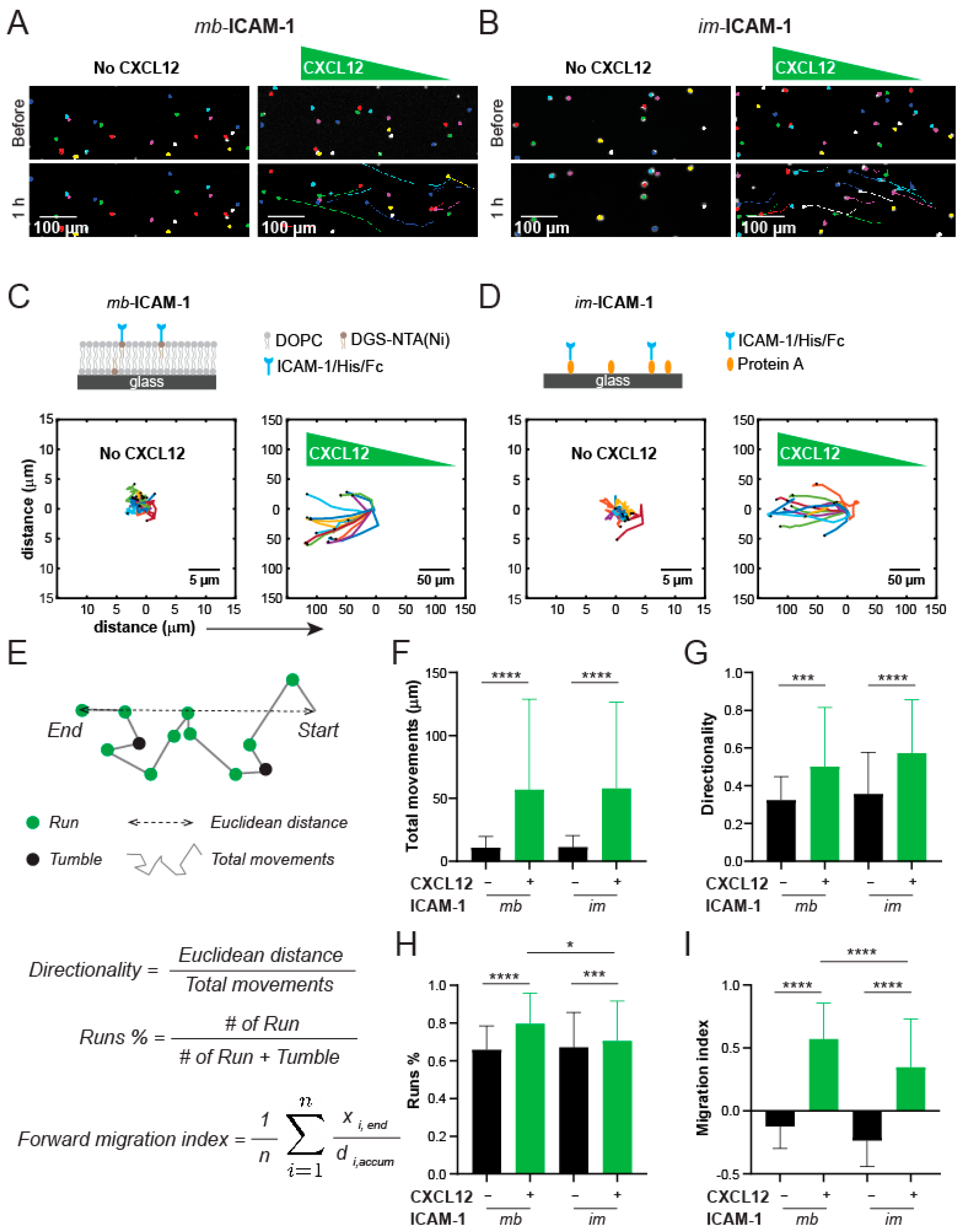
3.6. Jurkat Cells Have Different Chemotactic Profiles on Membrane-Bound vs. Immobilized ICAM-1
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fu, X.; Liu, G.; Halim, A.; Ju, Y.; Luo, Q.; Song, G. Mesenchymal Stem Cell Migration and Tissue Repair. Cells 2019, 8, 784. [Google Scholar] [CrossRef] [PubMed]
- Cornwell, K.G.; Downing, B.R.; Pins, G.D. Characterizing Fibroblast Migration on Discrete Collagen Threads for Applications in Tissue Regeneration. J. Biomed. Mater. Res. A 2004, 71, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Simula, L.; Pacella, I.; Colamatteo, A.; Procaccini, C.; Cancila, V.; Bordi, M.; Tregnago, C.; Corrado, M.; Pigazzi, M.; Barnaba, V.; et al. Drp1 Controls Effective T Cell Immune-Surveillance by Regulating T Cell Migration, Proliferation, and Cmyc-Dependent Metabolic Reprogramming. Cell Rep. 2018, 25, 3059–3073 e10. [Google Scholar] [CrossRef] [PubMed]
- Rodero, M.P.; Poupel, L.; Loyher, P.L.; Hamon, P.; Licata, F.; Pessel, C.; Hume, D.V.; Combadière, C.; Boissonnas, A. Immune Surveillance of the Lung by Migrating Tissue Monocytes. Elife 2015, 4, e07847. [Google Scholar] [CrossRef]
- Jones, D.H.; Nakashima, T.; Sanchez, O.H.; Kozieradzki, I.; Komarova, S.V.; Sarosi, I.; Morony, S.; Rubin, E.; Sarao, R.; Hojilla, C.V.; et al. Regulation of Cancer Cell Migration and Bone Metastasis by Rankl. Nature 2006, 440, 692–696. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, J.; Yu, M.; Xie, Y.; Huang, Y.; Wolff, D.; Abel, W.; Tu, Y. Mitochondrial Dynamics Regulates Migration and Invasion of Breast Cancer Cells. Oncogene 2013, 32, 4814–4824. [Google Scholar] [CrossRef]
- Sribenja, S.; Wongkham, S.; Wongkham, C.; Yao, Q.; Chen, C. Roles and Mechanisms of Beta-Thymosins in Cell Migration and Cancer Metastasis: An Update. Cancer Invest. 2013, 31, 103–110. [Google Scholar] [CrossRef]
- van Buul, J.D.; Hordijk, P.L. Signaling in Leukocyte Transendothelial Migration. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 824–833. [Google Scholar] [CrossRef]
- Wright, D.E.; Bowman, E.P.; Wagers, A.J.; Butcher, E.C.; Weissman, I.L. Hematopoietic Stem Cells Are Uniquely Selective in Their Migratory Response to Chemokines. J. Exp. Med. 2002, 195, 1145–1154. [Google Scholar] [CrossRef]
- Hunter, M.C.; Teijeira, A.; Halin, C. T Cell Trafficking through Lymphatic Vessels. Front. Immunol. 2016, 7, 613. [Google Scholar] [CrossRef]
- Krummel, M.F.; Bartumeus, F.; Gérard, A. T cell migration, search strategies and mechanisms. Nat. Rev. Immunol. 2016, 16, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Mierke, C.T. Role of the Endothelium During Tumor Cell Metastasis: Is the Endothelium a Barrier or a Promoter for Cell Invasion and Metastasis? J. Biophys. 2008, 2008, 183516. [Google Scholar] [CrossRef] [PubMed]
- Orr, F.W.; Wang, H.H.; Lafrenie, R.; Scherbarth, S.; Nance, D.M. Interactions between cancer cells and the endothelium in metastasis. J. Pathol. 2000, 190, 310–329. [Google Scholar] [CrossRef]
- Kao, W.-T.; Lin, C.-Y.; Lee, L.-T.; Lee, P.-P.H.; Hung, C.-C.; Lin, Y.-S.; Chen, S.-H.; Ke, F.-C.; Hwang, J.-J.; Lee, M.-T. Investigation of MMP-2 and -9 in a highly invasive A431 tumor cell sub-line selected from a Boyden chamber assay. Anticancer. Res. 2008, 28, 2109–2120. [Google Scholar]
- Brown, N.S.; Bicknell, R.; Brooks, S.A.; Schumacher, U. Cell Migration and the Boyden Chamber; Humana Press: Totowa, NJ, USA, 2001; pp. 47–54. [Google Scholar] [CrossRef]
- Lomakina, E.B.; Waugh, R.E. Micromechanical Tests of Adhesion Dynamics between Neutrophils and Immobilized ICAM-1. Biophys. J. 2004, 86, 1223–1233. [Google Scholar] [CrossRef]
- Feigelson, W.S.; Pasvolsky, R.; Cemerski, S.; Shulman, Z.; Grabovsky, V.; Ilani, T.; Sagiv, A.; Lemaitre, F.; Laudanna, C.; Shaw, A.S.; et al. Occupancy of Lymphocyte Lfa-1 by Surface-Immobilized Icam-1 Is Critical for Tcr- but Not for Chemokine-Triggered Lfa-1 Conversion to an Open Headpiece High-Affinity State. J. Immunol. 2010, 185, 7394–7404. [Google Scholar] [CrossRef]
- Jacobson, K.; Liu, P.; Lagerholm, C. The Lateral Organization and Mobility of Plasma Membrane Components. Cell 2019, 177, 806–819. [Google Scholar] [CrossRef]
- Ostrowski, M.A.; Huang, N.; Walker, T.W.; Verwijlen, T.; Poplawski, C.; Khoo, A.S.; Cooke, J.; Fuller, G.G.; Dunn, A.R. Microvascular Endothelial Cells Migrate Upstream and Align Against the Shear Stress Field Created by Impinging Flow. Biophys. J. 2014, 106, 366–374. [Google Scholar] [CrossRef]
- Bourget, J.M.; Kerouredan, O.; Medina, M.; Remy, M.; Thebaud, N.B.; Bareille, R.; Chassande, O.; Amedee, J.; Catros, S.; Devillard, R. Patterning of Endothelial Cells and Mesenchymal Stem Cells by Laser-Assisted Bioprinting to Study Cell Migration. Biomed. Res. Int. 2016, 2016, 3569843. [Google Scholar] [CrossRef]
- Wong, B.S.; Shah, S.R.; Yankaskas, C.L.; Bajpai, V.K.; Wu, P.H.; Chin, D.; Ifemembi, B.; ReFaey, K.; Schiapparelli, P.; Zheng, X.; et al. A Microfluidic Cell-Migration Assay for the Prediction of Progression-Free Survival and Recurrence Time of Patients with Glioblastoma. Nat. Biomed. Eng. 2021, 5, 26–40. [Google Scholar] [CrossRef]
- Pavesi, A.; Tan, A.T.; Koh, S.; Chia, A.; Colombo, M.; Antonecchia, E.; Miccolis, C.; Ceccarello, E.; Adriani, G.; Raimondi, M.T.; et al. A 3D microfluidic model for preclinical evaluation of TCR-engineered T cells against solid tumors. JCI Insight. 2017, 2, e89762. [Google Scholar] [CrossRef]
- Hao, J.; Zhou, H.; Nemes, K.; Yen, D.; Zhao, W.; Bramlett, C.; Wang, B.; Lu, R.; Shen, K. Membrane-bound SCF and VCAM-1 synergistically regulate the morphology of hematopoietic stem cells. J. Cell Biol. 2021, 220, e202010118. [Google Scholar] [CrossRef]
- Carrasco, Y.R.; Batista, F.D. B-cell activation by membrane-bound antigens is facilitated by the interaction of VLA-4 with VCAM-1. EMBO J. 2006, 25, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.J.; Contento, R.L.; Gordo, S.; Wucherpfennig, K.W.; Love, J.C. Functional single-cell analysis of T-cell activation by supported lipid bilayer-tethered ligands on arrays of nanowells. Lab Chip 2012, 13, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Yen, D.; Ando, Y.; Shen, K. A cost-effective micromilling platform for rapid prototyping of microdevices. Technology 2016, 4, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Truong, D.; Puleo, J.; Llave, A.; Mouneimne, G.; Kamm, R.D.; Nikkhah, M. Breast Cancer Cell Invasion into a Three Dimensional Tumor-Stroma Microenvironment. Sci. Rep. 2016, 6, 34094. [Google Scholar] [CrossRef]
- Haessler, U.; Teo, J.C.M.; Foretay, D.; Renaud, P.; Swartz, M.A. Migration dynamics of breast cancer cells in a tunable 3D interstitial flow chamber. Integr. Biol. 2011, 4, 401–409. [Google Scholar] [CrossRef]
- Polacheck, W.J.; German, A.E.; Mammoto, A.; Ingber, D.E.; Kamm, R.D. Mechanotransduction of Fluid Stresses Governs 3d Cell Migration. Proc. Natl. Acad. Sci. USA 2014, 111, 2447–2452. [Google Scholar] [CrossRef]
- Vickerman, V.; Kamm, R.D. Mechanism of a flow-gated angiogenesis switch: Early signaling events at cell–matrix and cell–cell junctions. Integr. Biol. 2012, 4, 863–874. [Google Scholar] [CrossRef]
- Tung, C.K.; Krupa, O.; Apaydin, E.; Liou, J.J.; Diaz-Santana, A.; Kim, B.J.; Wu, M. A Contact Line Pinning Based Microfluidic Platform for Modelling Physiological Flows. Lab Chip 2013, 13, 3876–3885. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Datta, A.; Berg, J.M.; Gangopadhyay, S. Studies on Surface Wettability of Poly(Dimethyl) Siloxane (Pdms) and Glass under Oxygen-Plasma Treatment and Correlation with Bond Strength. J. Microelectromech. Syst. 2005, 14, 590–597. [Google Scholar] [CrossRef]
- Mazia, D.; Schatten, G.; Sale, W. Adhesion of Cells to Surfaces Coated with Polylysine. Applications to Electron Microscopy. J. Cell Biol. 1975, 66, 198–200. [Google Scholar] [CrossRef]
- Renn, D.W. Agar and Agarose: Indispensable Partners in Biotechnology. Ind. Eng. Chem. Prod. Res. Dev. 1984, 23, 17–21. [Google Scholar] [CrossRef]
- Davidson, S.M.; Selvaraj, P.; He, D.; Boi-Doku, C.; Yellon, R.L.; Vicencio, J.M.; Yellon, D.M. Remote Ischaemic Preconditioning Involves Signalling through the Sdf-1alpha/Cxcr4 Signalling Axis. Basic Res. Cardiol. 2013, 108, 377. [Google Scholar] [CrossRef] [PubMed]
- Ju, J.; Park, J.Y.; Kim, K.C.; Kim, H.; Berthier, E.; Beebe, D.J.; Lee, S.H. Backward Flow in a Surface Tension Driven Micropump. J. Micromech. Microeng. 2008, 18, 087002. [Google Scholar] [CrossRef]
- Amadi, O.C.; Steinhauser, M.L.; Nishi, Y.; Chung, S.; Kamm, R.D.; McMahon, A.P.; Lee, R.T. A low resistance microfluidic system for the creation of stable concentration gradients in a defined 3D microenvironment. Biomed. Microdevices 2010, 12, 1027–1041. [Google Scholar] [CrossRef][Green Version]
- Coluccio, M.L.; D’Attimo, M.A.; Cristiani, C.M.; Candeloro, P.; Parrotta, E.; Dattola, E.; Guzzi, F.; Cuda, G.; Lamanna, E.; Carbone, E.; et al. A Passive Microfluidic Device for Chemotaxis Studies. Micromachines 2019, 10, 551. [Google Scholar] [CrossRef]
- Pietrosimone, K.M.; Yin, X.; Knecht, D.A.; Lynes, M.A. Measurement of cellular chemotaxis with ECIS/Taxis. J. Vis. Exp. 2012, 62, e3840. [Google Scholar] [CrossRef] [PubMed]
- Sonmez, U.M.; Wood, A.; Justus, K.; Jiang, W.; Syed-Picard, F.; LeDuc, P.R.; Kalinski, P.; Davidson, L.A. Chemotactic Responses of Jurkat Cells in Microfluidic Flow-Free Gradient Chambers. Micromachines 2020, 11, 384. [Google Scholar] [CrossRef]
- Giaever, I.; Keese, C.R. Monitoring fibroblast behavior in tissue culture with an applied electric field. Proc. Natl. Acad. Sci. USA 1984, 81, 3761–3764. [Google Scholar] [CrossRef] [PubMed]
- Bangasser, B.L.; Shamsan, G.A.; Chan, C.E.; Opoku, K.N.; Tüzel, E.; Schlichtmann, B.W.; Kasim, J.A.; Fuller, B.J.; McCullough, B.R.; Rosenfeld, S.S.; et al. Shifting the optimal stiffness for cell migration. Nat. Commun. 2017, 8, 15313. [Google Scholar] [CrossRef] [PubMed]
- Shoaib, T.; Nalam, P.C.; He, Y.; Chen, Y.; Espinosa-Marzal, R.M. Assembly, Morphology, Diffusivity, and Indentation of Hydrogel-Supported Lipid Bilayers. Langmuir 2017, 33, 7105–7117. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, J.; Zhao, W.; Oh, J.M.; Shen, K. A Pillar-Free Diffusion Device for Studying Chemotaxis on Supported Lipid Bilayers. Micromachines 2021, 12, 1254. https://doi.org/10.3390/mi12101254
Hao J, Zhao W, Oh JM, Shen K. A Pillar-Free Diffusion Device for Studying Chemotaxis on Supported Lipid Bilayers. Micromachines. 2021; 12(10):1254. https://doi.org/10.3390/mi12101254
Chicago/Turabian StyleHao, Jia, Winfield Zhao, Jeong Min Oh, and Keyue Shen. 2021. "A Pillar-Free Diffusion Device for Studying Chemotaxis on Supported Lipid Bilayers" Micromachines 12, no. 10: 1254. https://doi.org/10.3390/mi12101254
APA StyleHao, J., Zhao, W., Oh, J. M., & Shen, K. (2021). A Pillar-Free Diffusion Device for Studying Chemotaxis on Supported Lipid Bilayers. Micromachines, 12(10), 1254. https://doi.org/10.3390/mi12101254






