A Portable Device for Simple Exosome Separation from Biological Samples
Abstract
:1. Introduction
2. Materials and Methods
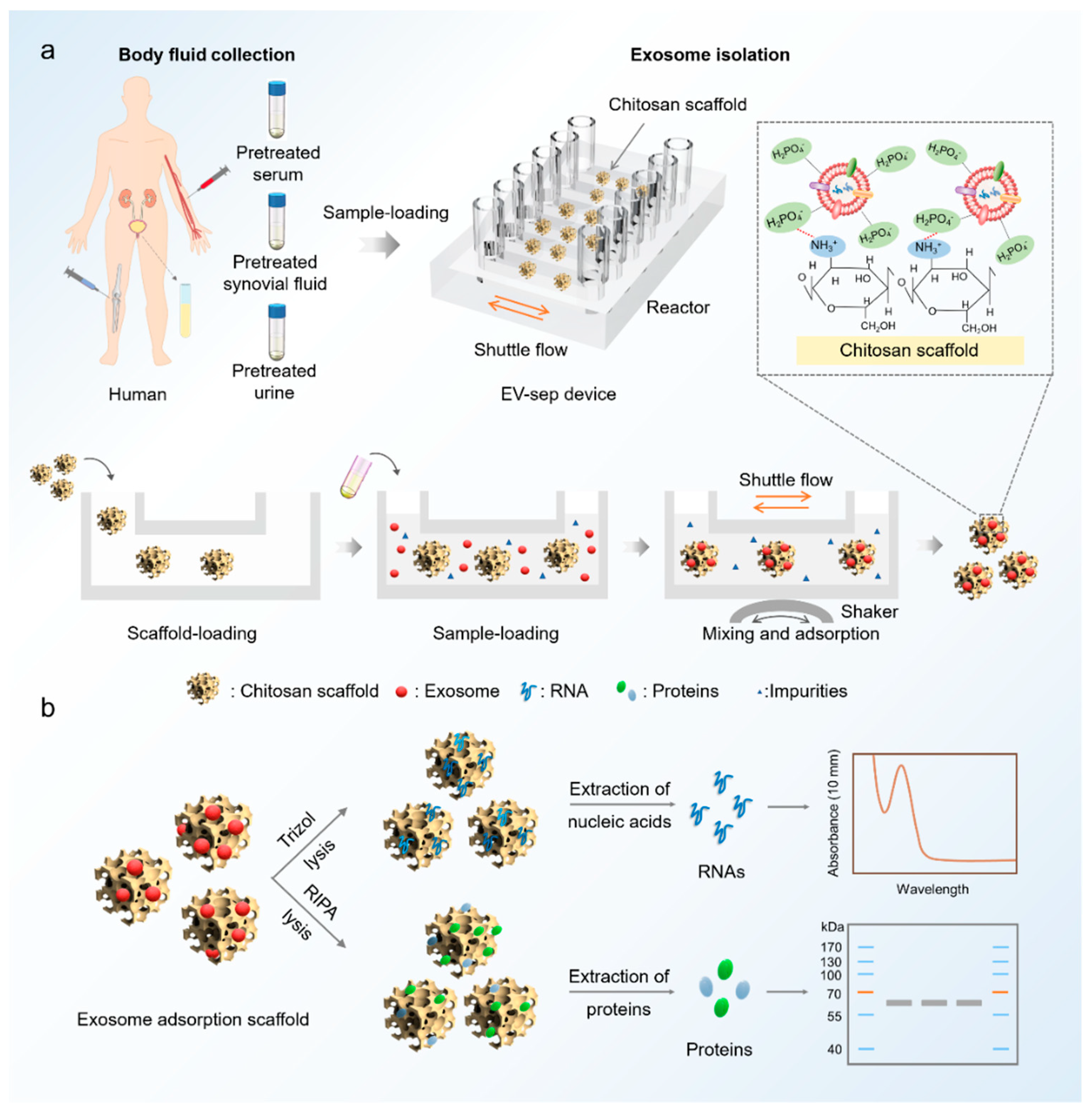
2.1. Design and Fabrication of the EV-Sep Device
2.2. Isolation of Exosomes on the EV-Sep Device
2.3. Exosome Preparation from Cell Culture Medium
2.4. Clinical Samples Collection and Pretreatment
2.5. Quantitative Analysis of Exosomes
2.6. Scanning Electron Microscope Imaging
2.7. Microscope Images and Photos
2.8. Aperture and Porosity Measurement
2.9. RNA Quantification
2.10. Western Blot
2.11. Particle Size Analysis
2.12. Statistical Analysis
3. Results and Discussion
3.1. Design and Operation of the EV-Sep Device
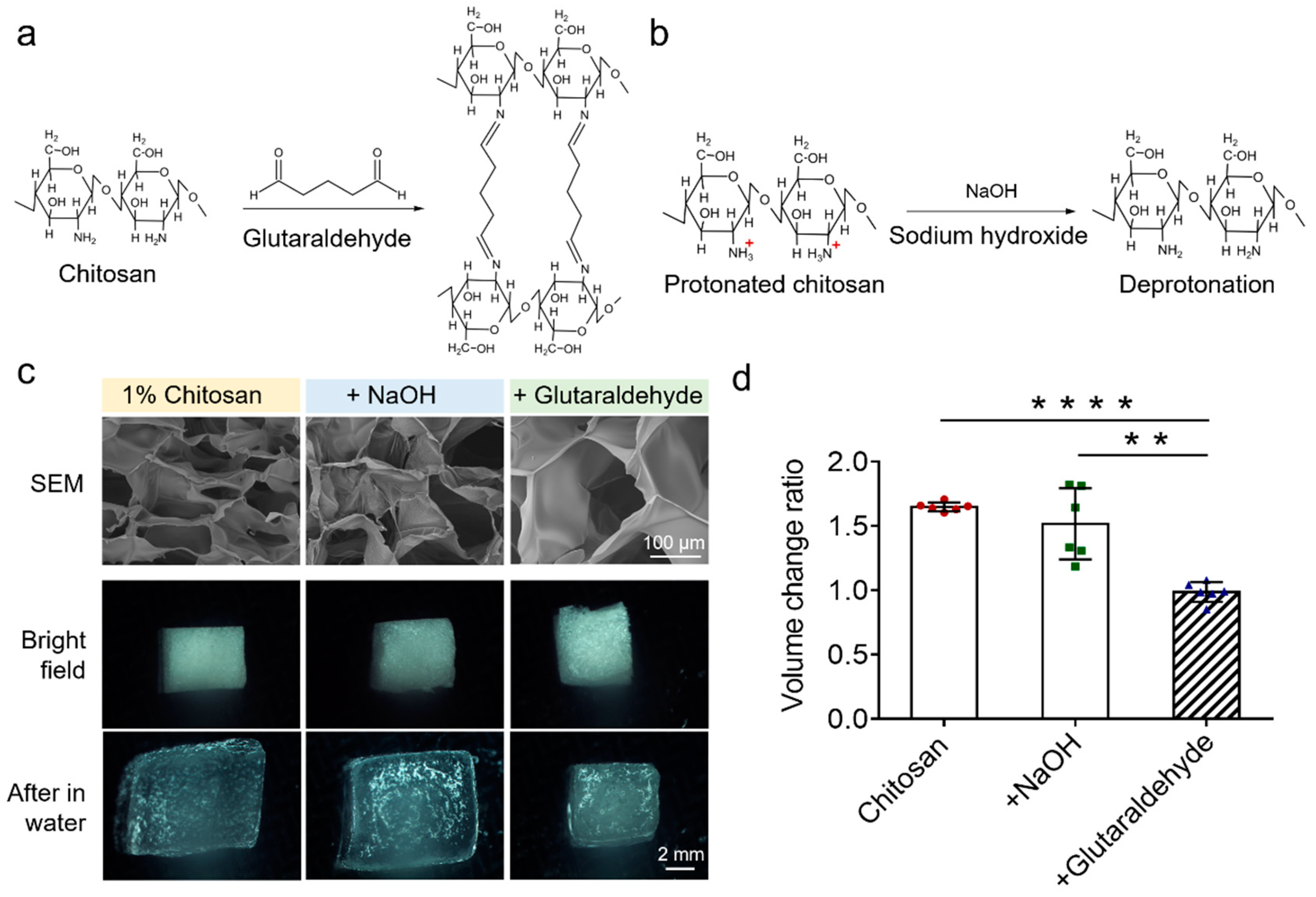
3.2. Optimizations of Chitosan Scaffolds of the EV-Sep Device
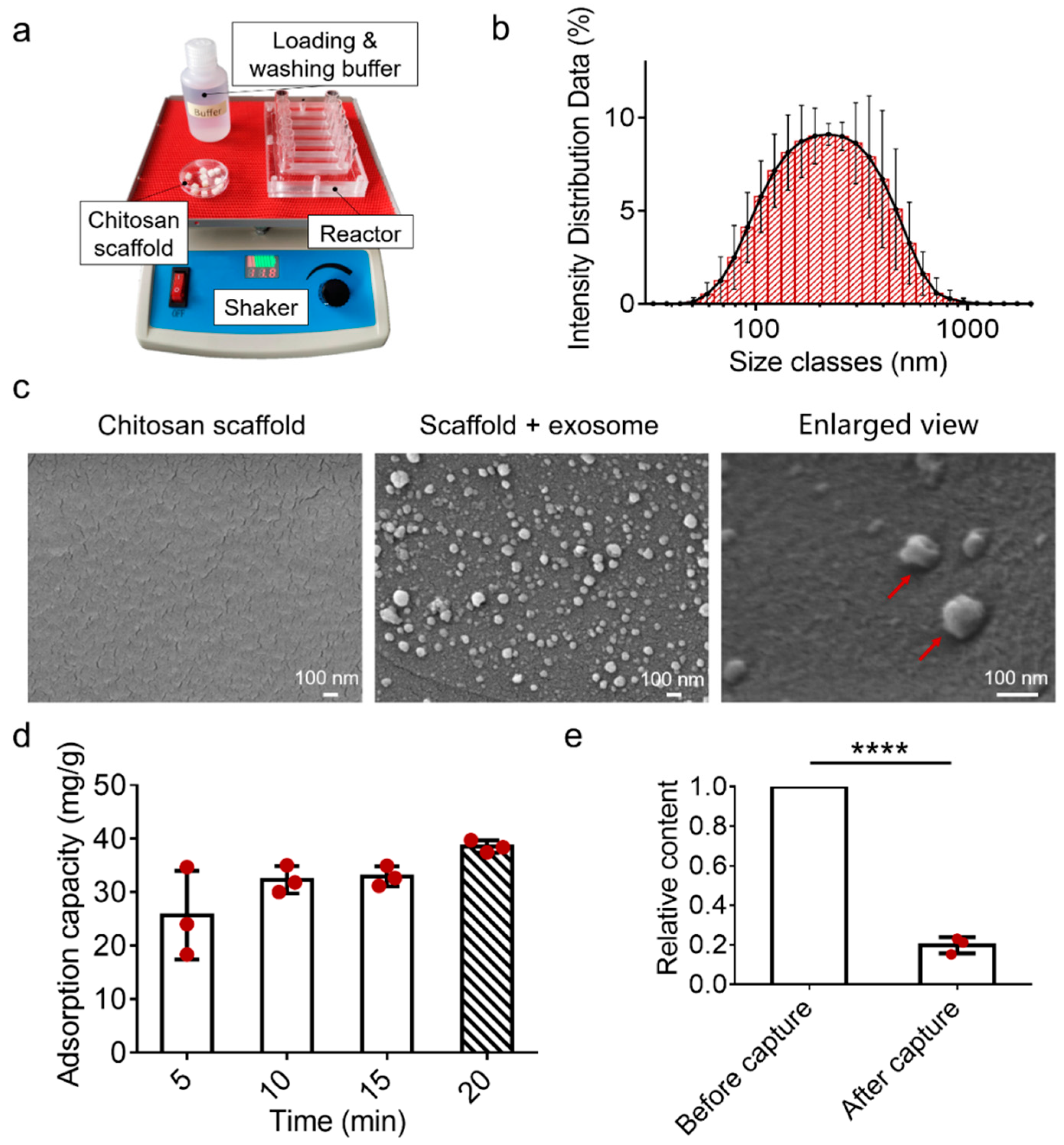
3.3. Characterizations of the EV-Sep Device for Exosome Capture
3.4. Performance of EV-Sep Device for Exosome Separation from Clinical Samples
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, 640. [Google Scholar] [CrossRef] [PubMed]
- Yanez-Mo, M.; Siljander, P.R.M.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tkach, M.; Thery, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexander, M.; Hu, R.; Runtsch, M.C.; Kagele, D.A.; Mosbruger, T.L.; Tolmachova, T.; Seabra, M.C.; Round, J.L.; Ward, D.M.; O’Connell, R.M. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Thery, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Kanada, M.; Bachmann, M.H.; Contag, C.H. Signaling by Extracellular Vesicles Advances Cancer Hallmarks. Trends Cancer 2016, 2, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Jeong, K.; Yu, Y.J.; You, J.Y.; Rhee, W.J.; Kim, J.A. Exosome-mediated microRNA-497 delivery for anti-cancer therapy in a microfluidic 3D lung cancer model. Lab Chip 2020, 20, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Li, H.; Chen, W.; Qin, J. Microfluidic strategies for label-free exosomes isolation and analysis. Trac-Trends Anal. Chem. 2019, 118, 686–698. [Google Scholar] [CrossRef]
- Kirschner, M.B.; Edelman, J.J.B.; Kao, S.C.H.; Vallely, M.P.; van Zandwijk, N.; Reid, G. The Impact of Hemolysis on Cell-Free microRNA Biomarkers. Front. Genet. 2013, 4, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.K.; Pak, T.R. miRNA degradation in the mammalian brain. Am. J. Physiol.-Cell Physiol. 2020, 319, C624–C629. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Nalabotala, R.; Koo, K.M.; Bose, S.; Nayak, R.; Shiddiky, M.J.A. Separation of distinct exosome subpopulations: Isolation and characterization approaches and their associated challenges. Analyst 2021, 146, 3731–3749. [Google Scholar] [CrossRef] [PubMed]
- Momen-Heravi, F.; Balaj, L.; Alian, S.; Mantel, P.-Y.; Halleck, A.E.; Trachtenberg, A.J.; Soria, C.E.; Oquin, S.; Bonebreak, C.M.; Saracoglu, E.; et al. Current methods for the isolation of extracellular vesicles. Biol. Chem. 2013, 394, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Lamparski, H.G.; Metha-Damani, A.; Yao, J.Y.; Patel, S.; Hsu, D.H.; Ruegg, C.; Le Pecq, J.B. Production and characterization of clinical grade exosomes derived from dendritic cells. J. Immunol. Methods 2002, 270, 211–226. [Google Scholar] [CrossRef]
- Chen, W.W.; Cao, R.K.; Su, W.T.; Zhang, X.; Xu, Y.H.; Wang, P.; Gan, Z.Q.; Xie, Y.Y.; Li, H.J.; Qin, J.H. Simple and fast isolation of circulating exosomes with a chitosan modified shuttle flow microchip for breast cancer diagnosis. Lab Chip 2021, 21, 1759–1770. [Google Scholar] [CrossRef] [PubMed]
- Pivarciova, L.; Rosskopfova, O.G.; Galambos, M.; Rajec, P. Sorption of technetium on glutaraldehyde crosslinked chitosan. J. Radioanal. Nucl. Chem. 2016, 309, 1251–1256. [Google Scholar] [CrossRef]
- Frick, J.M.; Ambrosi, A.; Pollo, L.D.; Tessaro, I.C. Influence of Glutaraldehyde Crosslinking and Alkaline Post-treatment on the Properties of Chitosan-Based Films. J. Polym. Environ. 2018, 26, 2748–2757. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Y.Y.; Cai, J.; Ren, L.; Wang, L.; Wang, Y.J. Preparation and characterization of bottle-brush polymer via host-guest self-assembly between beta-cyclodextrin and adamantane. Polym. Int. 2014, 63, 1930–1935. [Google Scholar] [CrossRef]
- Jiang, S.; Lyu, C.; Zhao, P.; Li, W.; Kong, W.; Huang, C.; Genin, G.M.; Du, Y. Cryoprotectant enables structural control of porous scaffolds for exploration of cellular mechano-responsiveness in 3D. Nat. Commun. 2019, 10, 3491. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.; Xie, Y.; Chang, Y.; Xu, Y.; Zhao, M.; Deng, P.; Qin, J.; Li, H. A Portable Device for Simple Exosome Separation from Biological Samples. Micromachines 2021, 12, 1182. https://doi.org/10.3390/mi12101182
Chen W, Xie Y, Chang Y, Xu Y, Zhao M, Deng P, Qin J, Li H. A Portable Device for Simple Exosome Separation from Biological Samples. Micromachines. 2021; 12(10):1182. https://doi.org/10.3390/mi12101182
Chicago/Turabian StyleChen, Wenwen, Yingying Xie, Yuang Chang, Yuhai Xu, Mengqian Zhao, Pengwei Deng, Jianhua Qin, and Hongjing Li. 2021. "A Portable Device for Simple Exosome Separation from Biological Samples" Micromachines 12, no. 10: 1182. https://doi.org/10.3390/mi12101182
APA StyleChen, W., Xie, Y., Chang, Y., Xu, Y., Zhao, M., Deng, P., Qin, J., & Li, H. (2021). A Portable Device for Simple Exosome Separation from Biological Samples. Micromachines, 12(10), 1182. https://doi.org/10.3390/mi12101182




