Inertial Microfluidics-Based Separation of Microalgae Using a Contraction–Expansion Array Microchannel
Abstract
:1. Introduction
2. Materials and Methods
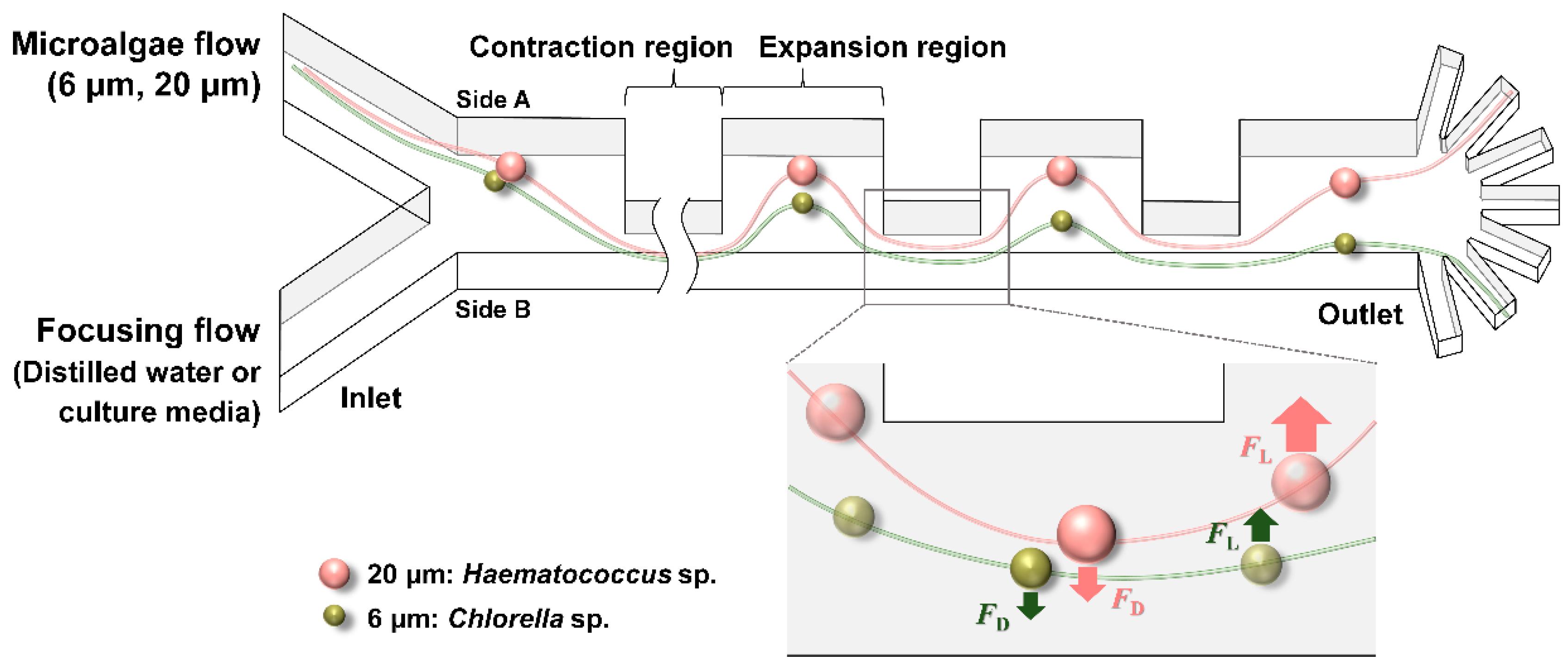
2.1. Design and Fabrication of Contraction–Expansion Array (CEA) Microchannel
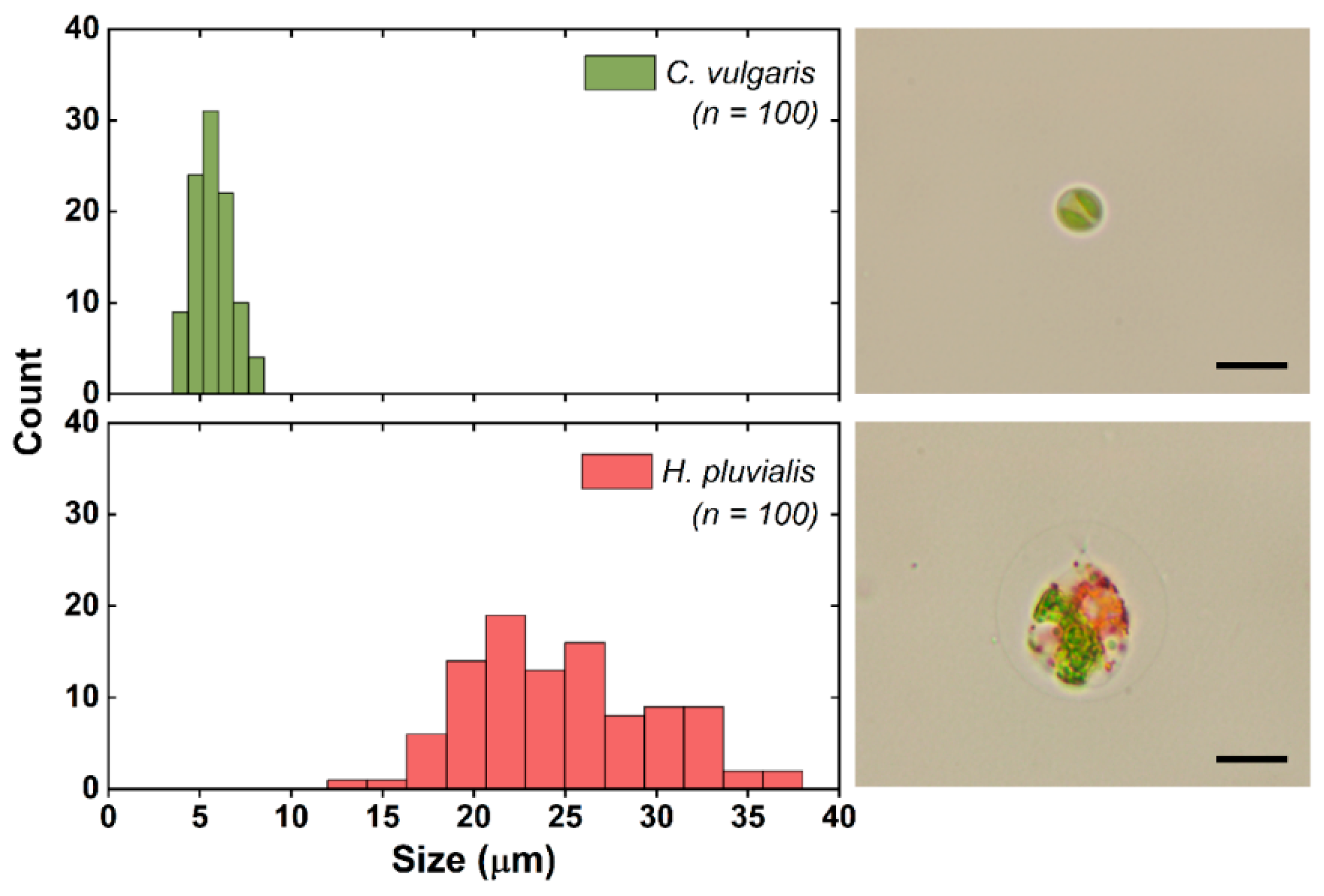
2.2. Fluorescent Microbeads and Microalgae Sample Preparation
2.3. Experimental Setup and Data Analysis
2.4. Purity and Reculturability of the Collected Algal Cells
3. Results and Discussion
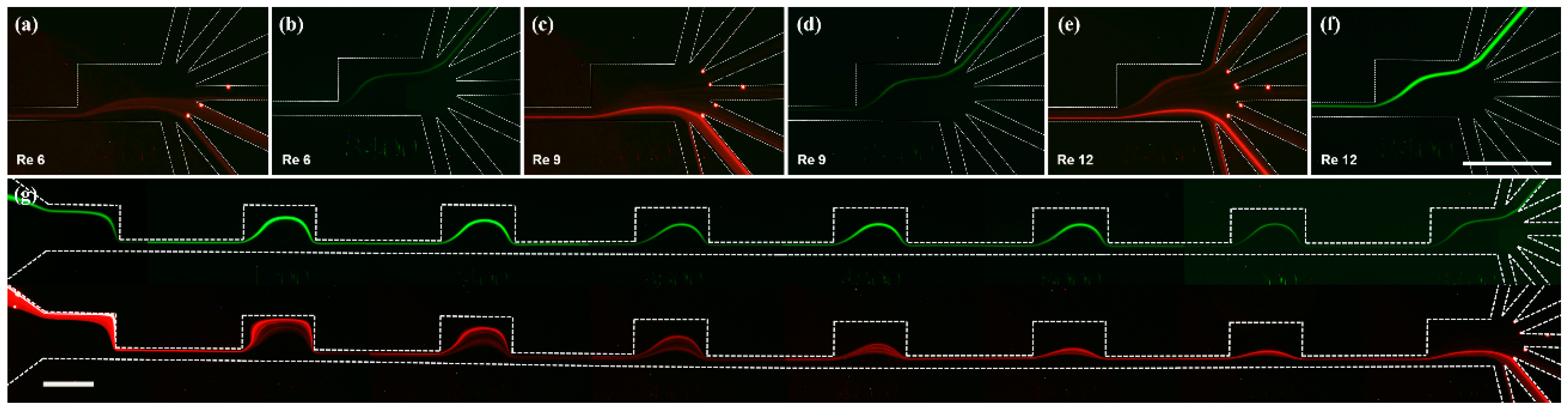
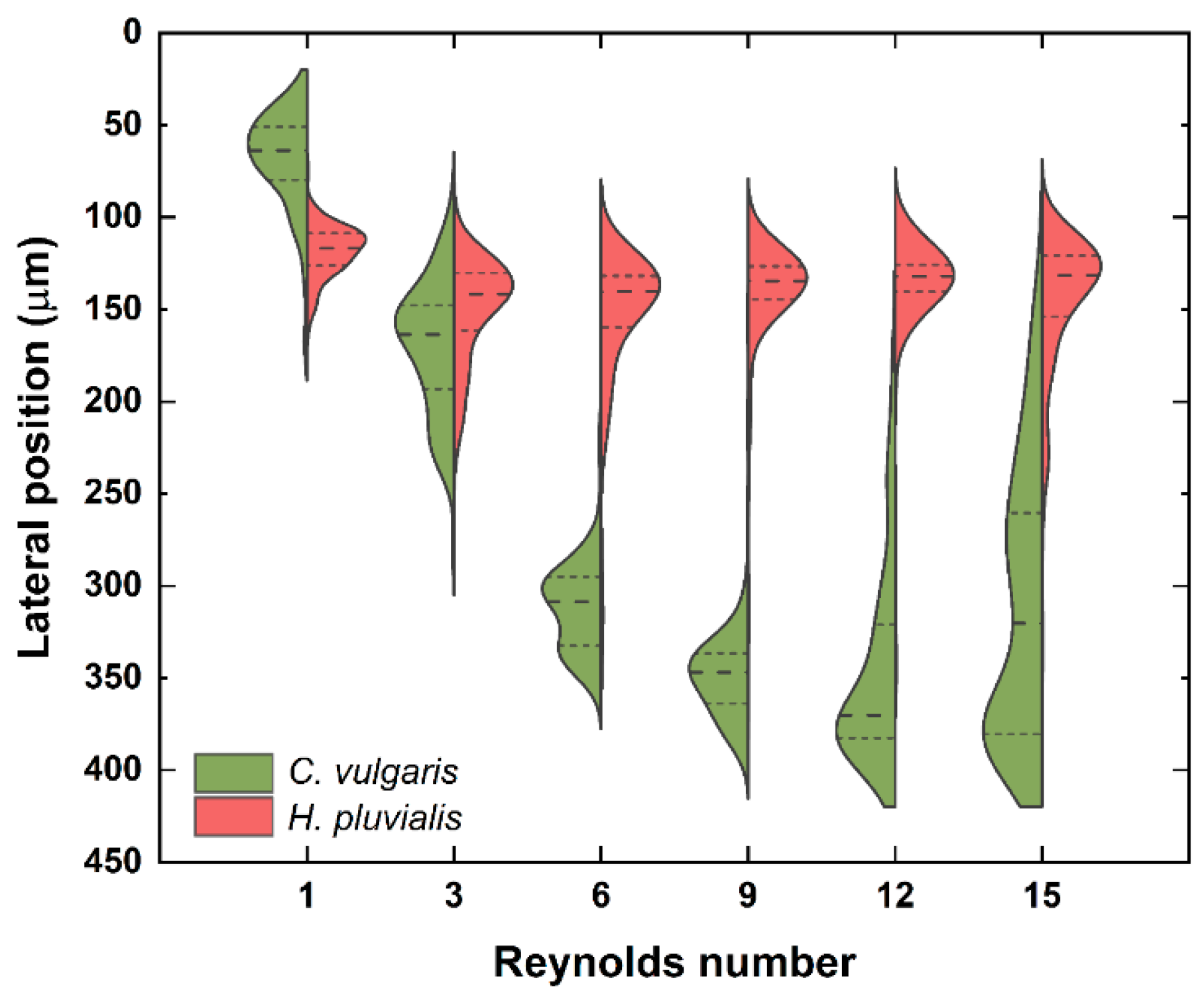
3.1. Separation of Fluorescent Microbeads
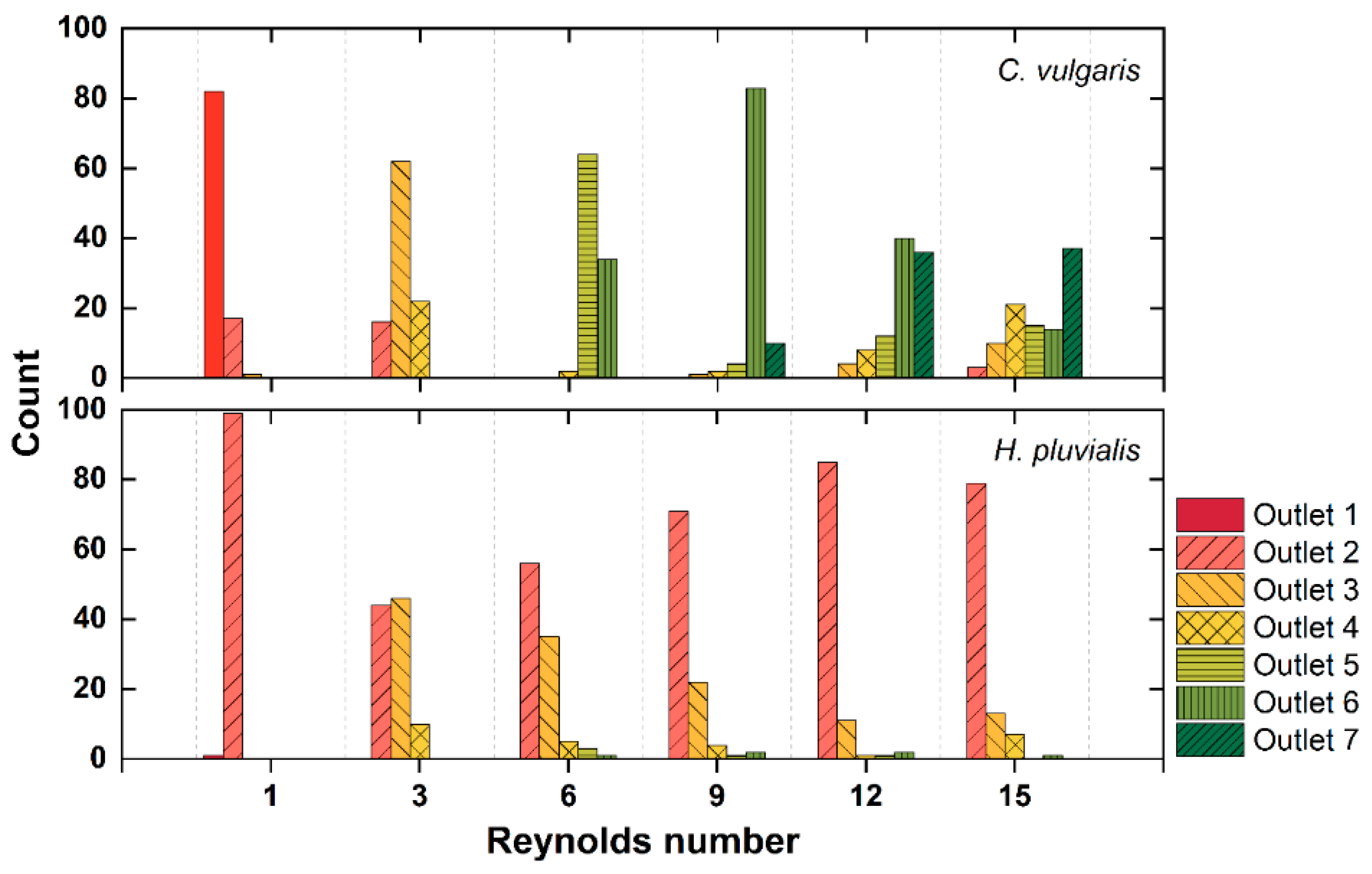
3.2. Separation of Microalgae
3.3. Purity and Reculturability of the Collected Algal Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Borowitzka, M.A. High-value products from microalgae—Their development and commercialisation. J. Appl. Phycol. 2013, 25, 743–756. [Google Scholar] [CrossRef]
- Lee, K.; Eisterhold, M.; Rindi, F.; Palanisami, S.; Nam, P. Isolation and screening of microalgae from natural habitats in the midwestern United States of America for biomass and biodiesel sources. J. Nat. Sci. Biol. Med. 2014, 5, 333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, P.; Gupta, S.K.; Guldhe, A.; Rawat, I.; Bux, F. Chapter 4. Microalgae isolation and basic culturing techniques. In Handbook of Marine Microalgae; Editor Kim, S.K., Ed.; Academic Press, Elsevier: Amsterdam, The Netherlands, 2015; pp. 43–54. [Google Scholar] [CrossRef]
- Spilling, K. Basic methods for isolating and culturing microalgae. In Biofuels from Algae. Methods in Molecular Biology; Spilling, K., Ed.; Humana Press: New York, NY, USA, 2017; Volume 1980, pp. 257–284. [Google Scholar] [CrossRef]
- Cellamare, M.; Rolland, A.; Jacquet, S. Flow cytometry sorting of freshwater phytoplankton. J. Appl. Phycol. 2010, 22, 87–100. [Google Scholar] [CrossRef]
- Graham, P.J.; Riordon, J.; Sinton, D. Microalgae on display: A microfluidic pixel-based irradiance assay for photosynthetic growth. Lab Chip 2015, 15, 3116–3124. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Ooms, M.D.; Sinton, D. Biomass-to-biocrude on a chip via hydrothermal liquefaction of algae. Lab Chip 2016, 16, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Wang, Y.; Wang, Z.; Zhong, W.; Wang, H.; Li, Y. An integrated microfluidic device in marine microalgae culture for toxicity screening application. Mar. Pollut. Bull. 2013, 72, 231–243. [Google Scholar] [CrossRef]
- Lee, H.; Choi, S. A micro-sized bio-solar cell for self-sustaining power generation. Lab Chip 2015, 15, 391–398. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Cheng, J.; Zhang, Y.; Ding, G.; Wang, X.; Chen, M.; Kang, Y.; Pan, X. Serial separation of microalgae in a microfluidic chip under inertial and dielectrophoretic forces. IEEE Sens. J. 2020, 20, 14607–14616. [Google Scholar] [CrossRef]
- Park, J.-W.; Kim, S.H.; Ito, T.; Fujii, T.; Kim, S.Y.; Laurell, T.; Lee, S.W.; Goda, K. Acoustofluidic harvesting of microalgae on a single chip. Biomicrofluidics 2016, 10, 034119. [Google Scholar] [CrossRef] [Green Version]
- Di Carlo, D. Inertial microfluidics. Lab Chip 2009, 9, 3038–3046. [Google Scholar] [CrossRef]
- Amini, H.; Lee, W.; Di Carlo, D. Inertial microfluidic physics. Lab Chip 2014, 14, 2739–2761. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yan, S.; Yuan, D.; Alici, G.; Nguyen, N.-T.; Ebrahimi Warkiani, M.; Li, W. Fundamentals and applications of inertial microfluidics: A review. Lab Chip 2016, 16, 10–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, D.; Zhao, Q.; Yan, S.; Tang, S.-Y.; Zhang, Y.; Yun, G.; Nguyen, N.-T.; Zhang, J.; Li, M.; Li, W. Sheathless separation of microalgae from bacteria using a simple straight channel based on viscoelastic microfluidics. Lab Chip 2019, 19, 2811–2821. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Lee, M.G.; Choi, S.; Park, J.-K. Inertia-activated cell sorting of immune-specifically labeled cells in a microfluidic device. RSC Adv. 2014, 4, 39140–39144. [Google Scholar] [CrossRef]
- Lee, M.G.; Shin, J.H.; Choi, S.; Park, J.-K. Enhanced blood plasma separation by modulation of inertial lift force. Sens. Actuators B Chem. 2014, 190, 311–317. [Google Scholar] [CrossRef]
- Lee, M.G.; Choi, S.; Park, J.-K. Inertial separation in a contraction-expansion array microchannel. J. Chromatogr. A 2011, 1218, 4138–4143. [Google Scholar] [CrossRef]
- Kim, G.-Y.; Han, J.-I.; Park, J.-K. Inertial microfluidics-based cell sorting. BioChip J. 2018, 12, 257–267. [Google Scholar] [CrossRef]
- Kim, G.-Y.; Yun, Y.-M.; Shin, H.-S.; Kim, H.-S.; Han, J.-I. Scenedesmus-based treatment of nitrogen and phosphorus from effluent of anaerobic digester and bio-oil production. Bioresour. Technol. 2015, 196, 235–240. [Google Scholar] [CrossRef]
- Schaap, A.; Dumon, J.; Toonder, J. Den Sorting algal cells by morphology in spiral microchannels using inertial microfluidics. Microfluid. Nanofluid. 2016, 20, 125. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.G.; Shin, J.H.; Bae, C.Y.; Choi, S.; Park, J.-K. Label-free cancer cell separation from human whole blood using inertial microfluidics at low shear stress. Anal. Chem. 2013, 85, 6213–6218. [Google Scholar] [CrossRef]
- Syed, M.S.; Rafeie, M.; Vandamme, D.; Asadnia, M.; Henderson, R.; Taylor, R.A.; Warkiani, M.E. Selective separation of microalgae cells using inertial microfluidics. Bioresour. Technol. 2018, 252, 91–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, G.-Y.; Son, J.; Han, J.-I.; Park, J.-K. Inertial Microfluidics-Based Separation of Microalgae Using a Contraction–Expansion Array Microchannel. Micromachines 2021, 12, 97. https://doi.org/10.3390/mi12010097
Kim G-Y, Son J, Han J-I, Park J-K. Inertial Microfluidics-Based Separation of Microalgae Using a Contraction–Expansion Array Microchannel. Micromachines. 2021; 12(1):97. https://doi.org/10.3390/mi12010097
Chicago/Turabian StyleKim, Ga-Yeong, Jaejung Son, Jong-In Han, and Je-Kyun Park. 2021. "Inertial Microfluidics-Based Separation of Microalgae Using a Contraction–Expansion Array Microchannel" Micromachines 12, no. 1: 97. https://doi.org/10.3390/mi12010097
APA StyleKim, G.-Y., Son, J., Han, J.-I., & Park, J.-K. (2021). Inertial Microfluidics-Based Separation of Microalgae Using a Contraction–Expansion Array Microchannel. Micromachines, 12(1), 97. https://doi.org/10.3390/mi12010097





