1. Introduction
The yeast
Saccharomyces cerevisiae has been commonly used as a model organism for aging studies in eukaryotic cells [
1,
2,
3,
4,
5]. A common measure used in aging studies is the Replicative Life Span (RLS), which is the measure of how long a cell lives while still having the ability to replicate itself [
1]. Traditionally, these measurements were done by using the microdissection technique [
6,
7,
8,
9,
10,
11]. In this technique, cells are grown in agar pads and, whenever they replicate, daughter cells are removed with the help of micromanipulators. This technique has several drawbacks: First, it is laborious and time consuming. Second, experiments must be either performed 24/7 or stopped at night while refrigerating the cells under observation, which may bias the interpretation of the results since temperature fluctuations might affect aging. Third, cells may be affected physically by the use of the micromanipulators.
To address these problems, many novel microfluidic devices have been designed for conducting RLS measurements [
12,
13,
14,
15,
16,
17,
18,
19,
20,
21,
22]. These devices, commonly referred to as Mother Machines, work by trapping cell in a way that they have access to fresh media but are not moving with the flow. Additionally, they are designed in such a way that as the cell replicates the daughter cells are taken away by the flow. Such microfluidic devices work better for RLS measurements than micromanipulators but have two major drawbacks: First, the trapping method subjects cells to mechanical pressure, which could bias the results, as we do not know how pressure affects the aging process in unicellular organisms. It has been demonstrated that mechanical pressure affects physiological processes such as growth rate [
23,
24] and diffusion rates of intracellular components [
25,
26]. Second, all the published devices require microfabrication techniques with resolution under 5 µm. This means that custom masks are required, which makes the fabrication process expensive, and thus out of reach for some labs.
To address these issues, we present a microfluidic device that traps cells without subjecting them to high mechanical pressures. To accomplish low-pressure trapping, we take advantage of the slipstreaming effect. This happens when a fluid flows around a barrier and a pressure gradient is created in the back of the barrier, generating a force that points against the direction of the flow. The device is designed in such a way that a gradient zone of the dimensions of a yeast cell is created, allowing the mother cells to be trapped while daughter cells are born outside this area. This means that the mother cells can be kept trapped indefinitely while the daughter cells are rapidly taken away by the flow. This device thus allows measurements of populations of cells at the single-cell level over long periods. Some examples are RLS measurements, distributions of replication times, division strategies, etc. Our device has the added advantage of being fabricated with low-cost techniques since we do not use custom masks but a UV light projector (SF-100 Micropatterning) with 5-µm resolution instead.
In this paper, we present the device and the protocol for its use, as well as a sketch of the optimization used to design it. We present basic gene expression measurements that allow us to conclude that, in the pressure ranges estimated to be used in existing devices, the gene expression pattern is likely to be altered. Finally, we show an example of its use in obtaining the distribution of replication times for a population and an RLS curve and comparing it with the results from previous devices.
2. Materials and Methods
2.1. Master Design and Fabrication
We used an SF100 Micropatterning UV projector (ScoTech, Renfrewshire, Scotland, UK) instead of a high-resolution printed mask. This projector requires a virtual mask in bmp (bitmap) format, so we designed it pixel by pixel using Illustrator and Paint. One pixel in the design is equivalent to 5 µm by 5 µm in the physical chip. In a cleanroom, we spun SC 1827 Photoresist at 2000 RPM for 1 min and then soft baked it at 115 °C for 50 s. We then exposed it for 50 s to UV light using the projector and developed the master for 1 min before hard baking it at 120 °C for 1 min.
2.2. Polydimethylsiloxane (PDMS) Chips Fabrication
Curing agent was mixed with pre-polymer for PDMS in a proportion of 1:10 and cured in an oven at 65 °C. The PDMS was then peeled off the master, and inlet/outlet channels were punched with a 0.75-mm biopsy punch. Chips were then cut and sonicated in isopropanol for 30–40 min, and then blown, dried and heated at 65 °C for 4 h. Glass coverslips were cleaned in 1 M KOH for 30 min, and then sonicated in milliQ water for 10 min. After that, the coverslips were blow-dried and heated at 65 °C for 10 min. The PDMS chips were cleaned with magic tape and treated with oxygen plasma for 1 min for bonding and hot bonded for 1 min at 150 °C.
2.3. Chip Dimensions
The device fits on a 22 mm × 22 mm coverslip. Each channel contains an array of 255 trapping units, with 140 μm between units in the direction of the flow and 115 μm between lines of units. Each unit consists of three PDMS pillars of 5-μm height that extend from the ceiling to the glass floor.
2.4. Media Preparation
We grew the cells in either Synthetic Dropout (SD) or Synthetic Complete (SC) medium. Every liter of SD medium contained 6700 mg of YNB (Yeast Nitrogen Base), 750 mg of DO supplement, 20 g of D-glucose, 20 mg of uracil and 20 mg of histidine. Every liter of SC medium contained 6700 mg of YNB, 2000 mg of DO supplement, 20 g of D-glucose, 80 mg of D-L tryptophan, 80 mg of D histidine and 80 mg of D-uracil.
2.5. Cell Growth and Insertion in Device
We recovered cells from −80 °C storage and grew them in SD Medium (2% Glucose) for 24 h. We diluted 20 μL in 5 mL fresh medium and grew the cells at 30 °C and 245 RPM for 12 h to get them back to an exponential phase (0.05–0.1 OD600). We then took media with these cells and centrifuged it at 5000 RPM for 5 min and then resuspended in 1/5 of the original volume. The cells were then manually inserted into the chip in the direction opposite to the normal flow before turning on the syringe pump and starting the experiment.
2.6. Data Acquisition and Analysis
Growth experiments were performed on a Zeiss inverted microscope controlled with Micromanager. We used 40× (air), 60× (air) and 100× (immersion) objectives. Images were acquired using a Photometrics CoolSnap Camera EZ CCD Camera with 1392 × 1040. Media was delivered using a syringe pump (Harvard Apparatus) at a rate of 3.6 mL/h. Expression experiments were performed on a Nikon Ti Eclipse inverted microscope with a 100× (immersion) objective.
2.7. Growth Conditions for Measuring Genetic Expression under Pressure
Cells were grown in SD medium supplemented with amino acids (uracil and histidine) and glucose. Cells were grown overnight (24 h) on a plate. We took 20 μL from the overnight culture and diluted it in 10 mL fresh media, and then let it grow for 12 h.
3. Results
3.1. Fabrication of the Microfluidic Device
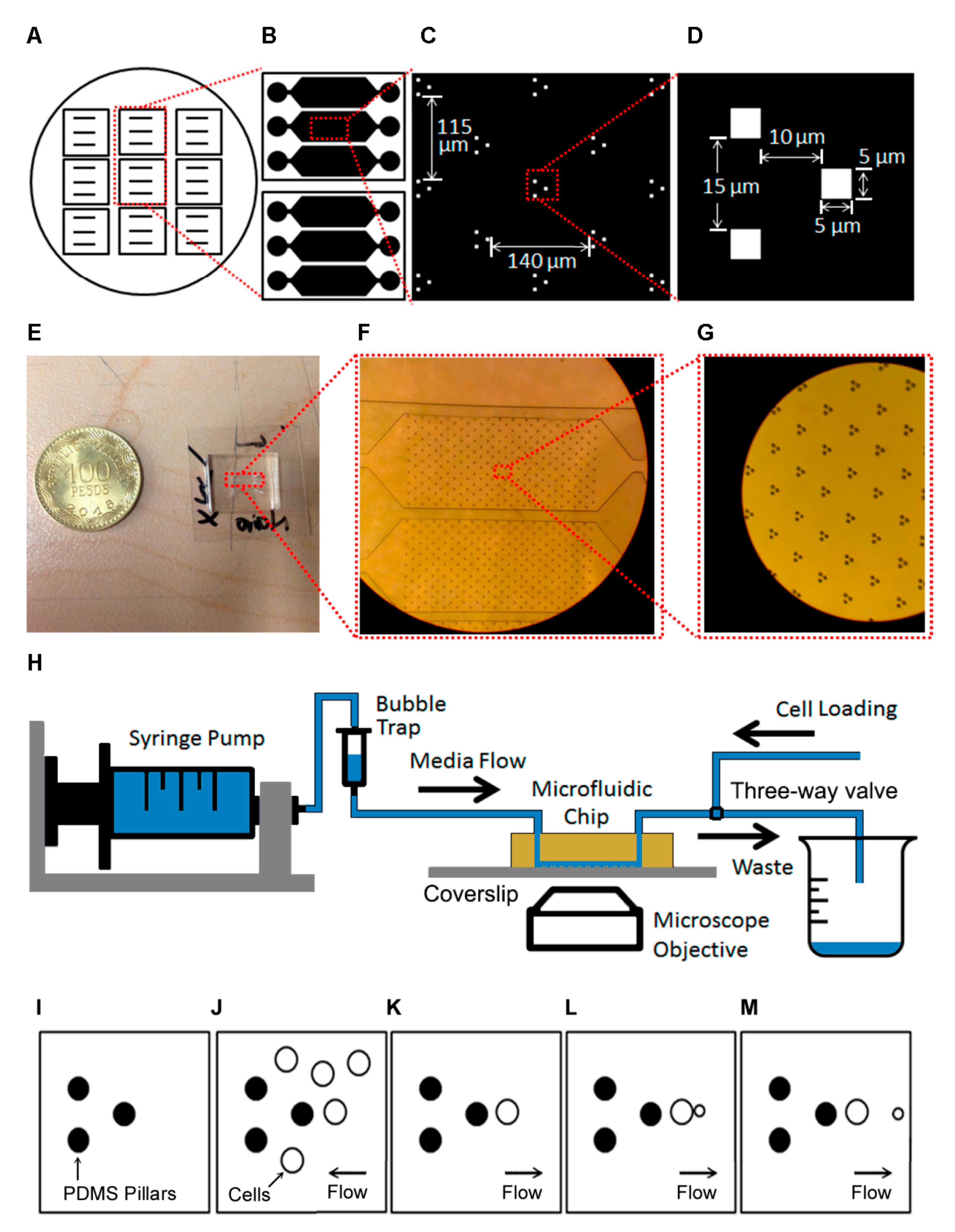
We designed and fabricated a novel microfluidic platform (
Figure 1A–H) that can be used to measure single cell dynamics in a population of yeast cells. A silicon wafer contains masters for nine devices (
Figure 1A). The microfluidic device at the core of the platform has three separate channels that permit the running of experiments with different media or strains independently (
Figure 1B). Each channel contains an array of 255 trapping units, with 140 μm between units in the direction of the flow and 115 μm between lines of units (
Figure 1C). Each unit consists of three PDMS pillars of 5-μm height that extend from the ceiling to the glass floor (
Figure 1D). Cells are trapped to the right of the rightmost pillar while the other two pillars are used for flow focusing (
Figure 1D). The z-dimension was chosen such that only one cell fits the channel but is not pressed by the roof of the channel. The device fits in a 22 mm × 22 mm coverslip (
Figure 1E).
Figure 1F,G shows zoom-ins of the channel and traps. The horizontal dimensions of the device, including unit separation, were determined after several in silico optimization rounds.
The system setup is sketched in
Figure 1H. First, the chip is set on the (inverted) microscope and fresh media is loaded into a syringe pump, which is connected to a bubble trap which in turn is connected to the chip inlet. The outlet is connected to a three-way valve, with exits connecting to the waste beaker and a tube used for initial loading of the cells. The cells are then loaded manually through that tube, making sure they do not reach the bubble trap. The three-way valve is then set to the waste beaker tube and flow from the syringe is started. The operation of the trap during this process is sketched in
Figure 1I–M.
3.2. Optimization of Experimental Procedure
We optimized the experimental procedure for loading and maintenance of the mother cells. To do this, we examined various options for the Optical Density at 600 nm (OD600 or simply OD) at which cells were inserted, the loading procedure and the flow rate for operation. We grew cells to an OD corresponding to peak exponential growth (0.1 OD600), after which we tried two different strategies: inserting cells directly into the chip and concentration by centrifugation before insertion. This was achieved by re-suspending cells in different fractions of the original volume. Concentrating the cells 5× gave the best results in number of trapped cells for various values of OD. Loading the cells in the same direction of the maintenance flow resulted in low trapping rates because the cells mostly flowed around the traps when flow was started, so we settled on loading the cells in the opposite direction before starting the syringe pump. This resulted in a trapping rate of approximately 70%, or about 180 cells per channel. The disadvantage of this method is that it is possible to introduce air bubbles when changing the direction of flow, so we had to introduce a bubble trap between the syringe pump and the chip to avoid this. Lastly, we varied the flow rate from 1 to 3.6 mL/h. The trapping rates ranged from 42% to 73% and the retention rates from 38% to 54%. We found that a low flow rate can result in the formation of microcolonies, and, although high flow can still result in the loss of some of the trapped cells during operation, overall trapping and retention rates increased with flow. We settled on a maximum flow of 3.6 mL/h because syringes need to be changed manually and higher flows would imply a more frequent change of the syringe and more media use, which was impractical.
3.3. Performance Test of the Setup
Using the parameters found in the optimization stage, we tested the chip by measuring cell retention. We grew cells overnight (24 h) in minimal media with 2% Glucose (
w/
v) diluted 20 μL in 5 mL of fresh media and grew it for 12 h. This gave us an OD600 of 0.1–0.2. We observed a very clear exponential phase for times between 12 and 17 h. We centrifuged the cells for 5 min at 5000 RPM and re-suspended in 1/5 of the original volume. We loaded the cells, started the flow and measured the percentage of retained cells. We observed retention rates between 25% and 55%. These retention rates compare well with equivalent systems: while in some they are not reported [
12,
14,
20], in others they are below 30% [
16,
21] and the best ones are between 40% and 60% [
21]. If the retention rate is the main consideration, higher flows can be used at the cost of more media use and more frequent syringe changes.
3.4. Finite Elements Simulation of the Slipstreaming Effect
To design the device, we modeled the slipstreaming effect by numerically solving the Navier–Stokes equations using the finite element method [
27] in COMSOL Multiphysics, using the MEMS and Microfluidics modules for incompressible fluid flow. We modeled the effect of fluid flow for different configurations of the pillars and settled on a symmetric three column design (
Figure 1D).
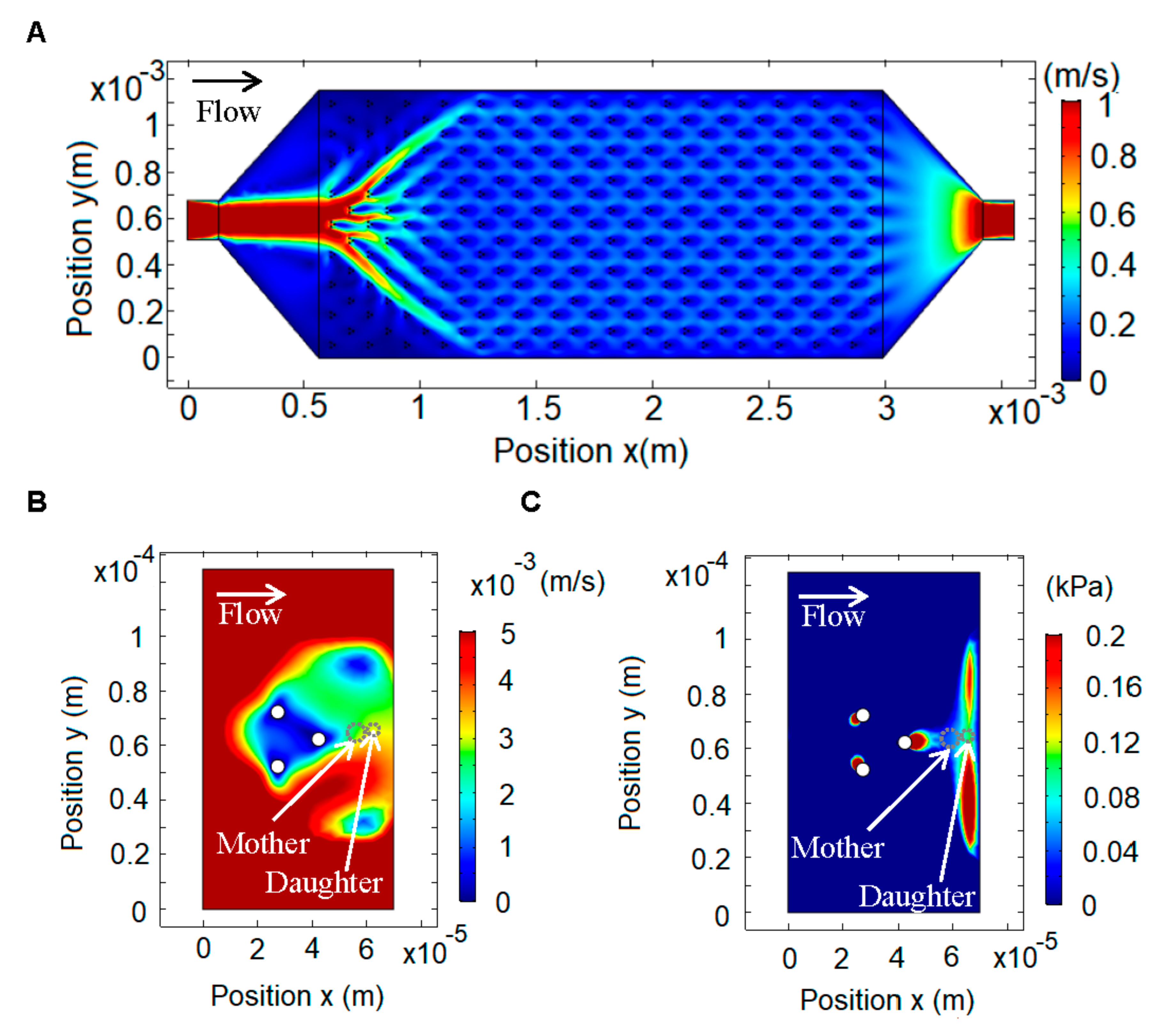
Figure 2 shows the simulation for a single channel with the inlet on the left and the outlet on the right. The average inlet velocity was calculated from the syringe pump flow (3.6 mL/h) and the transversal area of the chip, resulting in a velocity of 1.17 m/s. The boundary conditions were set as zero pressure on the outlet and zero velocity on the walls. The fluid properties were set using the density and viscosity of SD medium (density = 1000 kg/m
3, viscosity = 1 cp). We first modeled the whole channel (
Figure 2A) and from that simulation we obtained the boundary conditions and average velocity for each trap. This allowed us to do a second, zoomed-in simulation of each trap, from which we obtained the velocity and pressure profiles (
Figure 2B,C). This method allowed us to use a fine mesh for the traps, which would be computationally very costly if used for the whole chip.
The velocity profile shows that the lowest velocity is in the middle of the pillars, but there is also a low velocity zone to the right of the rightmost pillar (
Figure 1B). The pressure profile along a line bisecting the trap shows that in the zone where the mother cells are trapped there is a gradient, which results in a force towards the pillar (to the left), and in the area where the daughter cell is born there is a gradient, which results in a force (to the right) that carries the daughter cell away (
Figure 2C).
3.5. Pressure Affects Cell Physiology
One of the main advantages of our device is the trapping of cells with low mechanical pressure, and we considered this to be an important factor because we found studies that show evidence of mechanical pressure affecting the growth rate [
23,
24]. We decided to do a quick check of whether gene expression was also affected by the pressure cells experience in a microfluidic device. We used a constitutive gene (LEU) tagged with GFP to measure gene expression in a population. After that, we grew the cells in three different conditions: under no pressure, under low pressure (69 kPa) and under higher pressure (103 kPa). These values were chosen based on the typical values used on the MACS (Microfluidic Activated Cells Screening) device, which is a microfluidic device used for pressing cells mechanically [
25,
26]. After 4 h (when most cells in the population have already had at least one daughter), we measured again the distribution of gene expression in the population and observed statistically significant changes which confirm that pressure can affect gene expression (
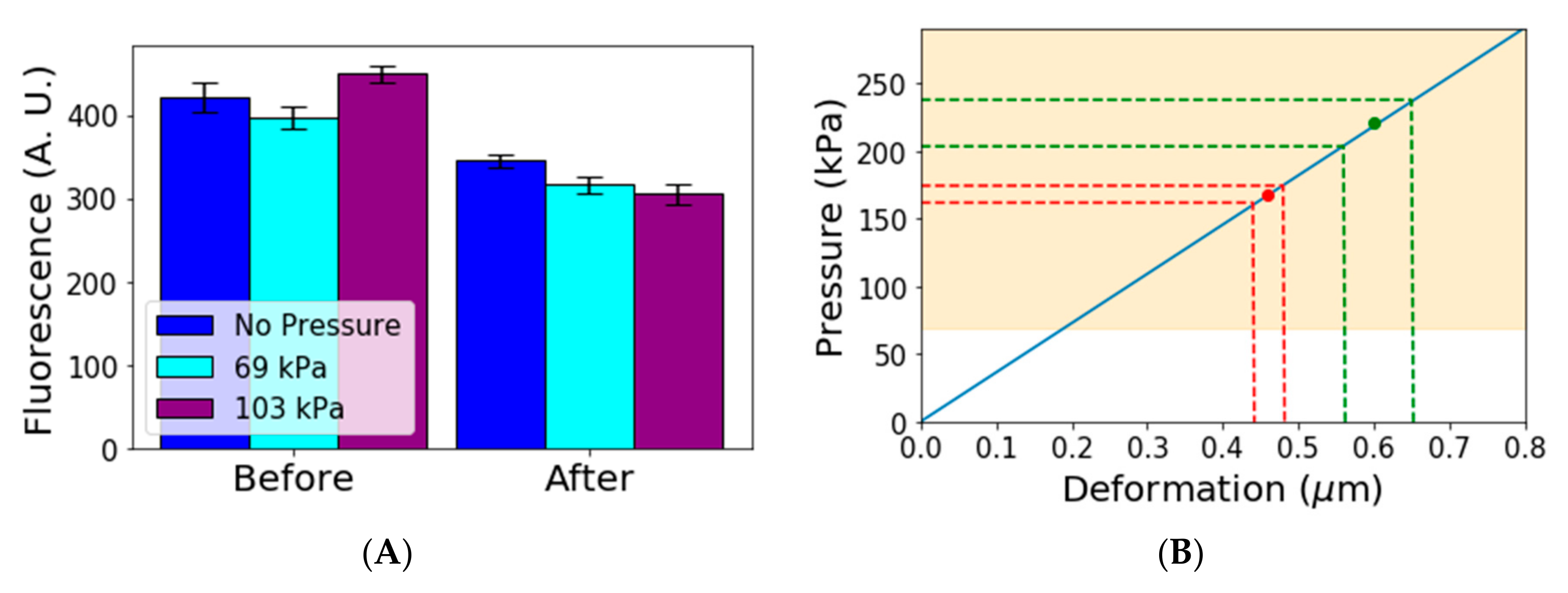
Figure 3A).
We performed an ANOVA statistical test for equal means for all of the three different groups. We found
p-value of less than 10
−4 when comparing the mean of gene expression under no pressure and at 69 or 103 kPa. This indicates that the means for cells under pressure and under no pressure can be assumed different, whereas the test for cells under 69 kPa and cells under 103 kPa gives a
p-value of 0.092, indicating these two means are not significantly different. Further experiments are required to determine whether there is a threshold pressure from which gene expression is affected or if it is changed continuously as pressure increases in a way similar to what has been found for growth rates under different pressures [
23,
24]. In any case, our simple experiment shows that gene expression is affected by mechanical pressure in the range used for cell trapping, which supports the claim that pressure in those experiments may affect the physiology of cells. If cell physiology is affected, studies of the aging process could also be affected.
To estimate the pressure exerted by the other Mother Machine devices, we looked at the deformation of the membrane in published images. We used a simple spring model using a surface modulus of 12 N/m [
28]. The model used was
where
is the stress,
is the Young’s Modulus and
is the deformation.
Figure 3B shows the results of the model applied to the available data. The continuous blue line shows the linear relation between deformation and pressure. The yellow area shows the region in which we observed pressure affects gene expression. The highlighted region starts at 69 kPa, but it is possible that gene expression is affected at even lower values. The red and green points show the average values estimated for two other devices found in the literature [
16,
21]. The dotted lines show the 95% confidence interval for the deformation measurement and corresponding pressure estimate in those devices. We observed that these devices subject cells to pressures that are high enough to affect gene expression.
3.6. Measurement Tests on Chip
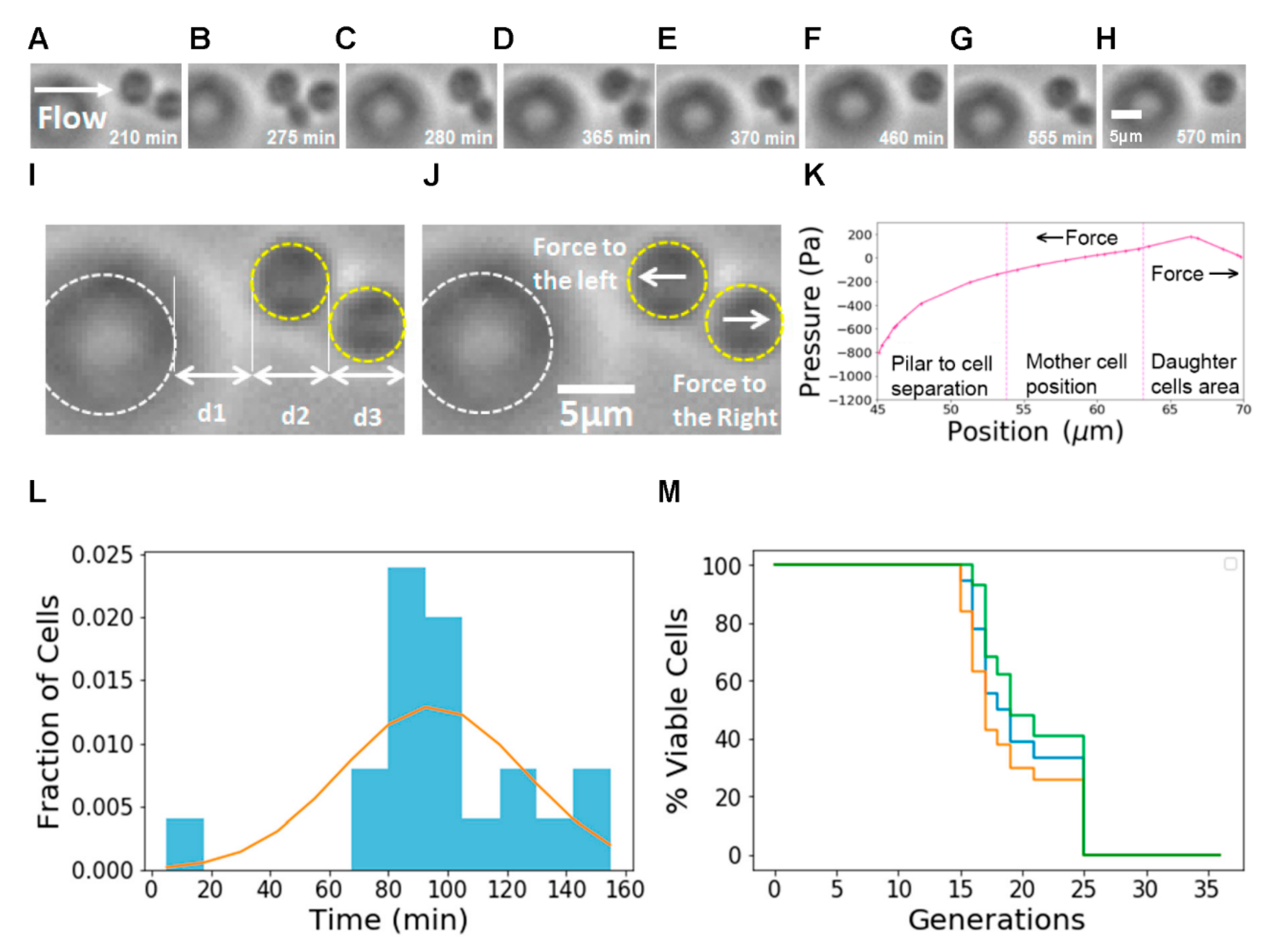
To test the usability of our design, we used our chip to measure yeast aging dynamics at the single-cell level. We were able to observe individual mother cells trapped during long periods of time and daughter cells as they grow and detach (
Figure 4A–H). To illustrate the trapping mechanism, we measured the distances from pillar to cell, mother cell size and daughter cell position and compared them with the distances and pressures obtained from the Multiphysics simulations (
Figure 4I–K). We can observe that, in the region where the mother cells are trapped, the pressure gradient generates a force to the left. In the region where the daughter cell is born and grows, the gradient generates a force to the right.
We measured doubling time distributions (
Figure 4L) and RLS (
Figure 4M) for BY4741 cells in SD medium and found a mean RLS of 19.2 generations, similar to measurements reported in other publications (
Table 1). The 95% confidence interval [
29] for the RLS is between 18.43 and 20.54 generations. This value matches the range obtained in other works: other authors [
16,
17,
21] reported average RLS times between 18 and 24 generations and 24.2 and 24.7, respectively. There are differences in the details between experiments, and the measured values of RLS times depend on the cell type and conditions used. Liu [
16] used S288C cells, while Jin [
17,
22] used BY4741 cells—similar to ours—but grown in a different medium (SC).
4. Discussion
We present a hydrodynamics based microfluidic “mother machine” device for observing cell dynamics by trapping mother cells and washing away daughter cells. The main advantage of this device is that it does not trap cells by pressing them mechanically as in other devices but uses hydrodynamic flow to trap the cells. In other devices, cells are subjected to external physical pressure of hundreds of kilopascals, whereas in our case the pressure difference that the cell is subjected to is of the order of hundreds of pascals. Our device has the added advantage that it can be fabricated with low-cost methods since it does not require masks with resolution under 5 µm. Other systems require a higher resolution because its physical features are used for the trapping directly, whereas in our case the trapping area is determined by the flow profile. Its only drawback is an imperfect retention rate, but since the device allows for imaging of multiple cells in one field of view this can be compensated by starting with a large number of trapped cells. We compared our device to previous designs and summarized the main features in
Table 1.
Long-term expression studies or experiments that measure response to changes in media are hard to do in the traditional setup of microcolonies on slides because neighboring cells change the microenvironment in a matter of minutes, and using micromanipulators to separate the cells is extremely time consuming. As with other Mother Machine systems, the removal of daughter cells in our setup allows for the measurement of long-term behavior of cells in stable media conditions, and using multiple input syringes would allow experiments on the response to changing media in individual cells over time. This device is thus adequate for many types of dynamic measurements, such as aging, replication times, response to media changes, drug screening applications and, in general, any experiment where there is interest in observing populations at the single-cell level for extended periods. Since the cells are trapped within 5 μm of a standard coverslip, all standard microscopy techniques, including fluorescence measurements, can be performed with this setup. As an example of use we demonstrated the capabilities of the device by measuring distributions of replications times and Replicative Life Span curves and obtained similar results to previous studies. Although we demonstrated the use of this device with yeast cells, it should be suitable for other cells with diameters ranging 5–10 µm since the trapping effect does not depend on the cell properties. Adjusting the size of the trapping area is in principle possible, although doing so would require moving back to standard photolithography techniques, negating the low-cost advantage of our design.
By removing the bias from pressure induced changes in the metrics of interest, and reducing the technological requirements for creating the chips, we expect that this design will help established labs make cleaner measurements and resource-limited labs to have access to Mother Machine microfluidic devices, thereby advancing fields such as aging and cell growth.
Author Contributions
D.C.D. and J.M.P. designed the experiments; D.C.D. and I.M.A. performed the FEM simulations; C.A.H. fabricated the masters; D.C.D., E.S. and R.A. performed the pressure experiments; D.C.D., E.S., D.A.F. and F.E.R. performed the time lapse experiments; D.C.D., I.M.A., R.A., D.A.F., F.E.R. and J.M.P. wrote the paper; D.C.D. and J.M.P. acquired funding; and J.M.P. supervised the project. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by UNIVERSIDAD DE LOS ANDES, FACULTAD DE CIENCIAS, Convocatoria para Proyectos de Investigación.
Acknowledgments
The authors would like to thank the Biophysics Group at Universidad de los Andes for useful discussions of the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mortimer, R.K.; Johnston, J.R. Life span of individual yeast cells. Nature 1959, 183, 1751–1752. [Google Scholar] [CrossRef]
- Kaeberlein, M.; Powers, R.W., 3rd; Steffen, K.; Westman, E.; Hu, D.; Dang, N.; Kerr, E.O.; Kirkland, K.T.; Fields, S.; Kennedy, B.K. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 2005, 310, 1193–1196. [Google Scholar] [CrossRef]
- Steinkraus, K.; Kaeberlein, M.K. Replicative aging in yeast: The means to the end. Annu. Rev. Cell Dev. Biol. 2008, 24, 29–54. [Google Scholar] [CrossRef]
- Longo, V.; Gralla, E.; Valentine, J. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J. Biol. Chem. 1996, 271, 12275–12280. [Google Scholar] [CrossRef]
- Fabrizio, P.; Longo, V. The chronological life span of Saccharomyces cerevisiae. Aging Cell 2003, 2, 73–81. [Google Scholar] [CrossRef]
- Mei, S.C.; Brenner, C. Calorie restriction-mediated replicative lifespan extension in yeast is non-cell autonomous. PLoS Biol 2015, 13, e1002048. [Google Scholar] [CrossRef]
- Fehrmann, S.; Paoletti, C.; Goulev, Y.; Ungureanu, A.; Aduilaniu, H.; Charvin, G. Aging Yeast Cells Undergo a Sharp Entry into Senescence Unrelated to the Loss of Mitochondrial Membrane Potential. Cell Rep. 2013, 5, 1589–1599. [Google Scholar] [CrossRef] [PubMed]
- Huberts, D.H.; Gonzalez, J.; Lee, S.S.; Litsios, A.; Hubmann, G.; Wit, E.C.; Heinemann, M. Calorie restriction does not elicit a robust extension of replicative lifespan in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2014, 111, 11727–11731. [Google Scholar] [CrossRef] [PubMed]
- Meitinger, F.; Khmelinskii, A.; Morlot, S.; Kurtulmus, B.; Palani, S.; Andres-Pons, A.; Hub, B.; Knop, M.; Charvin, G.; Pereira, G. A memory system of negative polarity cues prevents replicative aging. Cell 2014, 159, 1056–1069. [Google Scholar] [CrossRef]
- Müller, I.; Zimmermann, M.; Becker, D.; Flömer, M. Calendar life span versus budding life span of Saccharomyces cerevisiae. Mech. Ageing Dev. 1980, 12, 47–52. [Google Scholar] [CrossRef]
- Xie, Z.; Jay, K.A.; Smith, D.L.; Zhang, Y.; Liu, Z.; Zheng, J.; Tian, R.; Li, H.; Blackburn, E.H. Early telomerase inactivation accelerates aging independently of telomere length. Cell 2015, 160, 928–939. [Google Scholar] [CrossRef] [PubMed]
- Ryley, J.; Pereira-Smith, O. Microfluidics device for single cell gene expression analysis in Saccharomyces cerevisiae. Yeast 2006, 23, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.S.; Avalos Vizcarra, I.; Huberts, D.H.; Lee, L.P.; Heinemann, M. Whole lifespan microscopic observation of budding yeast aging through a microfluidic dissection platform. Proc. Natl. Acad. Sci. USA 2012, 109, 4916–4920. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Luo, C.; Zou, K.; Xie, Z.; Brandman, O.; Ouyang, Q. Single cell analysis of yeast replicative aging using a new generation of microfluidic device. PLoS ONE 2012, 7, e48275. [Google Scholar] [CrossRef]
- Chen, K.L.; Crane, M.M.; Kaeberlein, M. Microfluidic technologies for yeast replicative lifespan studies. Mech. Ageing Dev. 2017, 161, 262–269. [Google Scholar] [CrossRef]
- Liu, P.; Young, T.Z.; Acar, M. Yeast replicator a high-throuput multiplexed microfluidics platform for automated measurements of single-cell aging. Cell Rep. 2015, 13, 634–644. [Google Scholar] [CrossRef]
- Jin, M.; Li, Y.; O’Laughlin, R.; Bittihn, P.; Pillus, L.; Tsimring, L.S.; Hasty, J.; Hao, N. Divergent Aging of Isogenic Yeast Cells Revealed through Single-Cell Phenotypic Dynamics. Cell Syst. 2019, 8, 242–253. [Google Scholar] [CrossRef]
- Jo, M.C.; Liu, W.; Gu, L.; Dang, W.; Qin, L. High-throughput analysis of yeast replicative aging using a microfluidic system. Proc. Natl. Acad. Sci. USA 2015, 112, 9364–9369. [Google Scholar] [CrossRef]
- Xie, Z.; Zhang, Y.; Zou, K.; Brandman, O.; Ouyang, Q.; Li, H. Molecular phenotyping of aging in single yeast cells using a novel microfluidic device. Aging Cell 2012, 11, 599–606. [Google Scholar] [CrossRef]
- Lee, P.; Helman, N.C.; Lim, W.A.; Hung, P.J. A microfluidic system for dynamic yeast cell imaging. BioTechniques 2008, 44, 91–95. [Google Scholar] [CrossRef]
- Crane, M.M.; Clark, I.B.; Bakker, E.; Smith, S.; Swain, P.S. A Microfluidic System for Studying Ageing and Dynamic Single-Cell Responses in Budding Yeast. PLoS ONE 2014, 9, e100042. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jin, M.; O’Laughlind, R.; Bittihn, P.; Tsimring, L.S.; Pillus, L.; Hasty, J.; Haoa, N. Multigenerational silencing dynamics control cell aging. Proc. Natl. Acad. Sci. USA 2017, 114, 11253–11258. [Google Scholar] [CrossRef]
- Delarue, M.; Poterewicz, G.; Hoxha, O.; Choi, J.; Yoo, W.; Kauser, J.; Holt, L.; Hallatschek, O. SCWISh network is essential for survival under mechanical pressure. Proc. Natl. Acad. Sci. USA 2017, 114, 13465–13470. [Google Scholar] [CrossRef] [PubMed]
- Holt, L.J.; Hallatschek, O.; Delarue, M. Chapter 12—Mechano-chemostats to study the effects of compressive stress on yeast. Methods Cell Biol. 2018, 147, 215–231. [Google Scholar] [PubMed]
- Okumus, B.; Landgraf, D.; Lai, G.C.; Bakshi, S.; Arias-Castro, J.C.; Yildiz, S.; Huh, D.; Fernandez-Lopez, R.; Peterson, C.N.; Toprak, E.; et al. Mechanical slowing-down of cytoplasmic diffusion allows in vivo counting of proteins in individual cells. Nat. Commun. 2016, 7, 11641. [Google Scholar] [CrossRef] [PubMed]
- Okumus, B.; Charles, J.; Baker, C.J.; Arias-Castro, J.C.; Lai, G.C.; Leoncini, E.; Bakshi, S.; Luro, S.; Landgraf, D.; Paulsson, J. Microfluidics-Assisted Cell Screening (MACS): An automated platform for single-cell microscopy of suspension cultures. Nat Protoc. 2018, 13, 170–194. [Google Scholar] [CrossRef] [PubMed]
- Scherer, P.O.J. Computational Physics, 2nd ed.; Springer: Heidelberg, Germany, 2013. [Google Scholar]
- Alexander, E.S.; Zhang, Z.; Colin, R.T.; Kennith, E.M.; Anton, P.J.M. The mechanical properties of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2000, 97, 9871–9874. [Google Scholar]
- Kleinbaum, D.G.; Klein, M. Survival Analysis: A Self-Learning Text, 3rd ed.; Statistics for Biology and Health: 2012; Springer: New York, NY, USA, 2012. [Google Scholar] [CrossRef]
| Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).