Recent Advances in Liposome-Based Molecular Robots
Abstract
1. Introduction
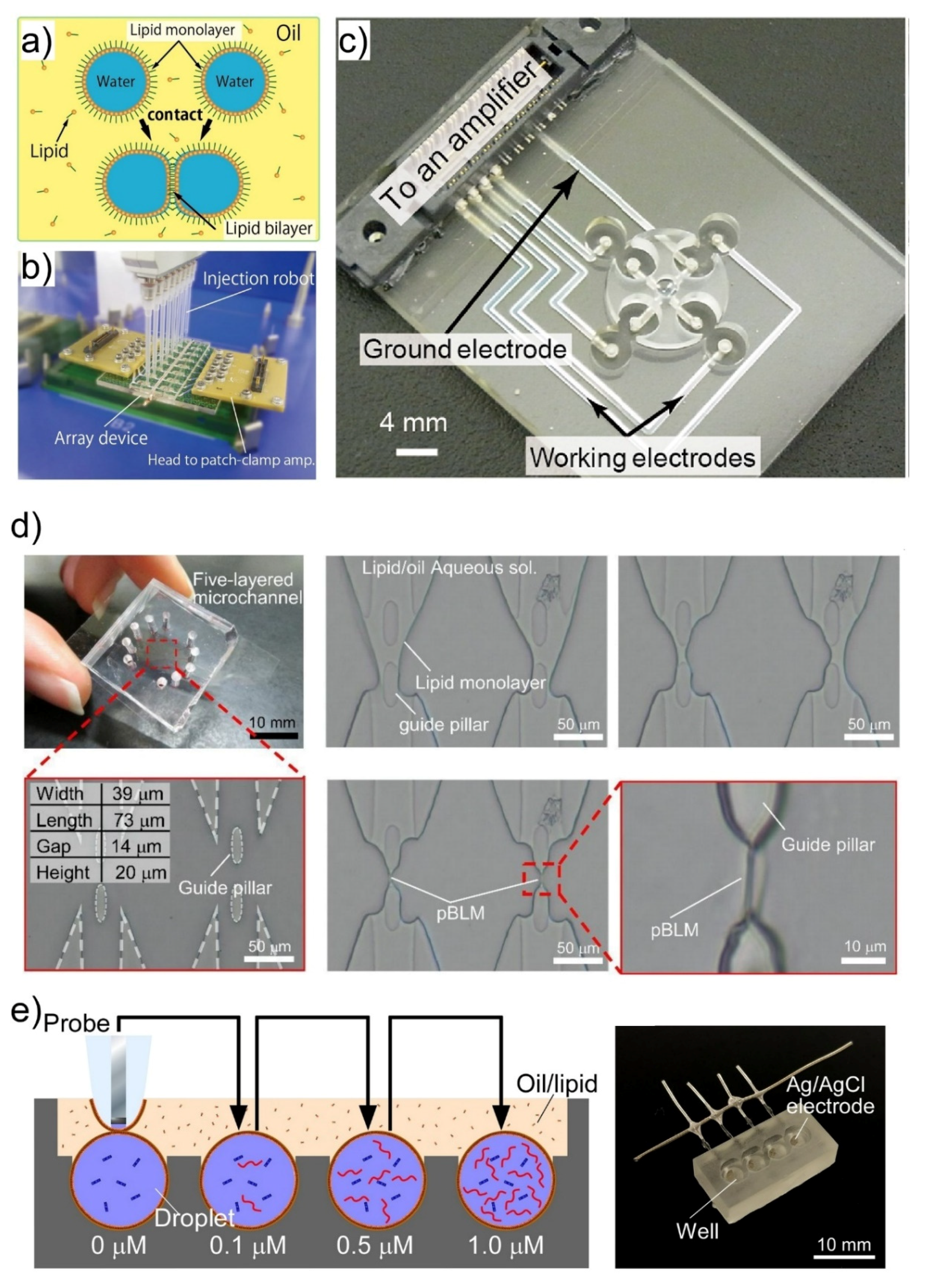
2. Giant Unilamellar Vesicles (GUVs) Generation Using Microfluidic Devices

3. Functions of Liposomal Membranes
3.1. Functionalization of Lipid Membranes
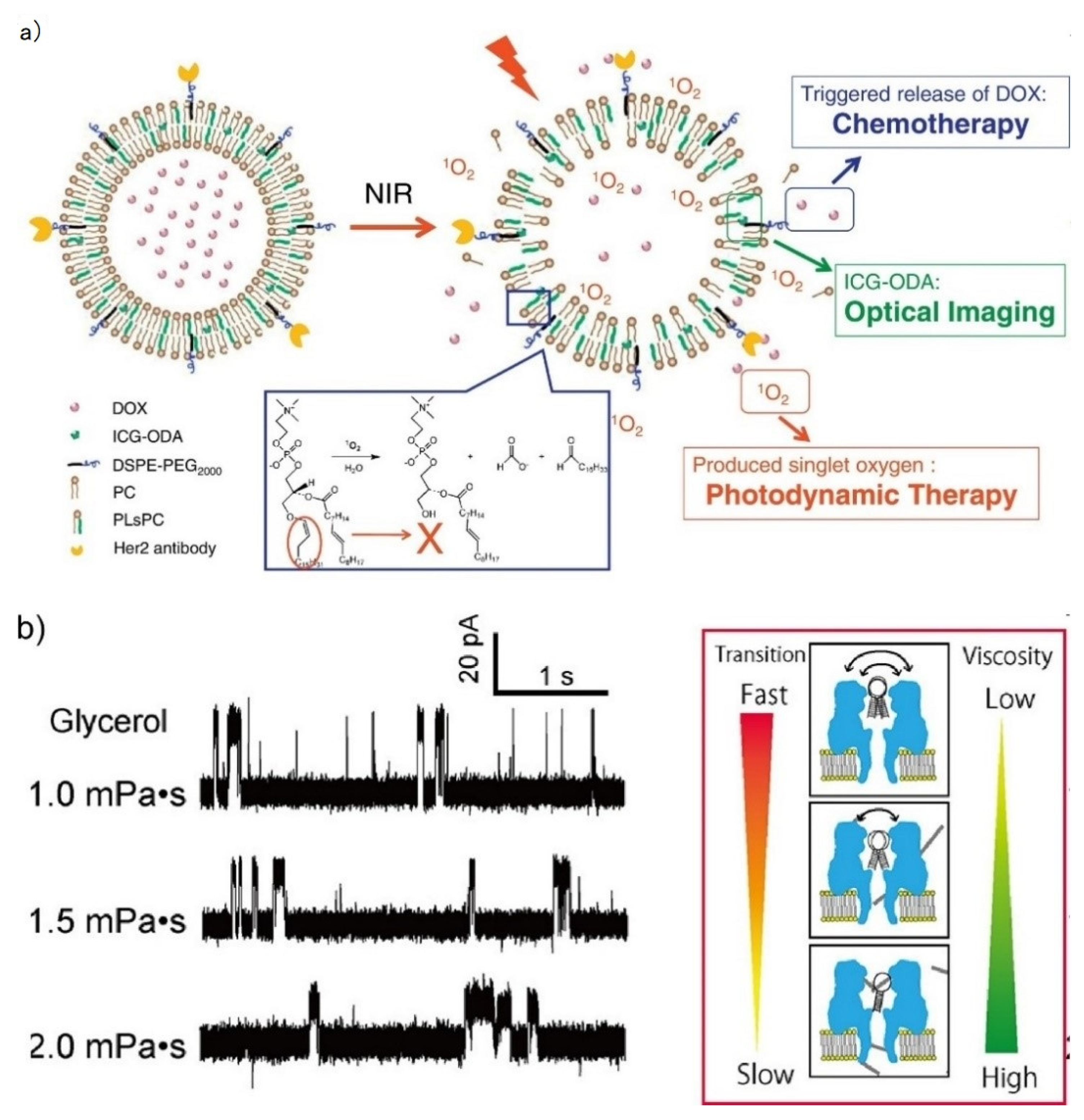
3.1.1. Chemical Sensing
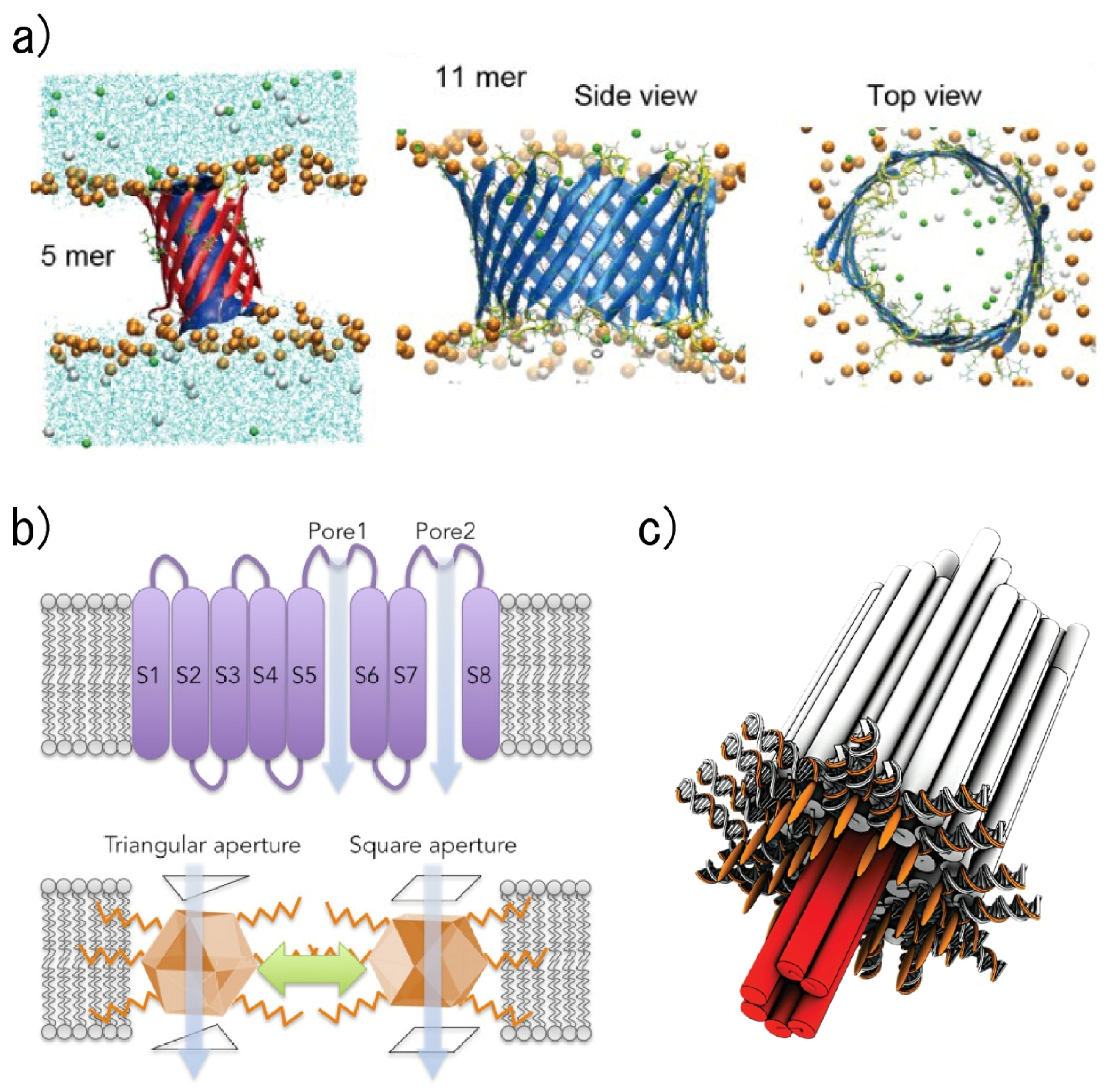
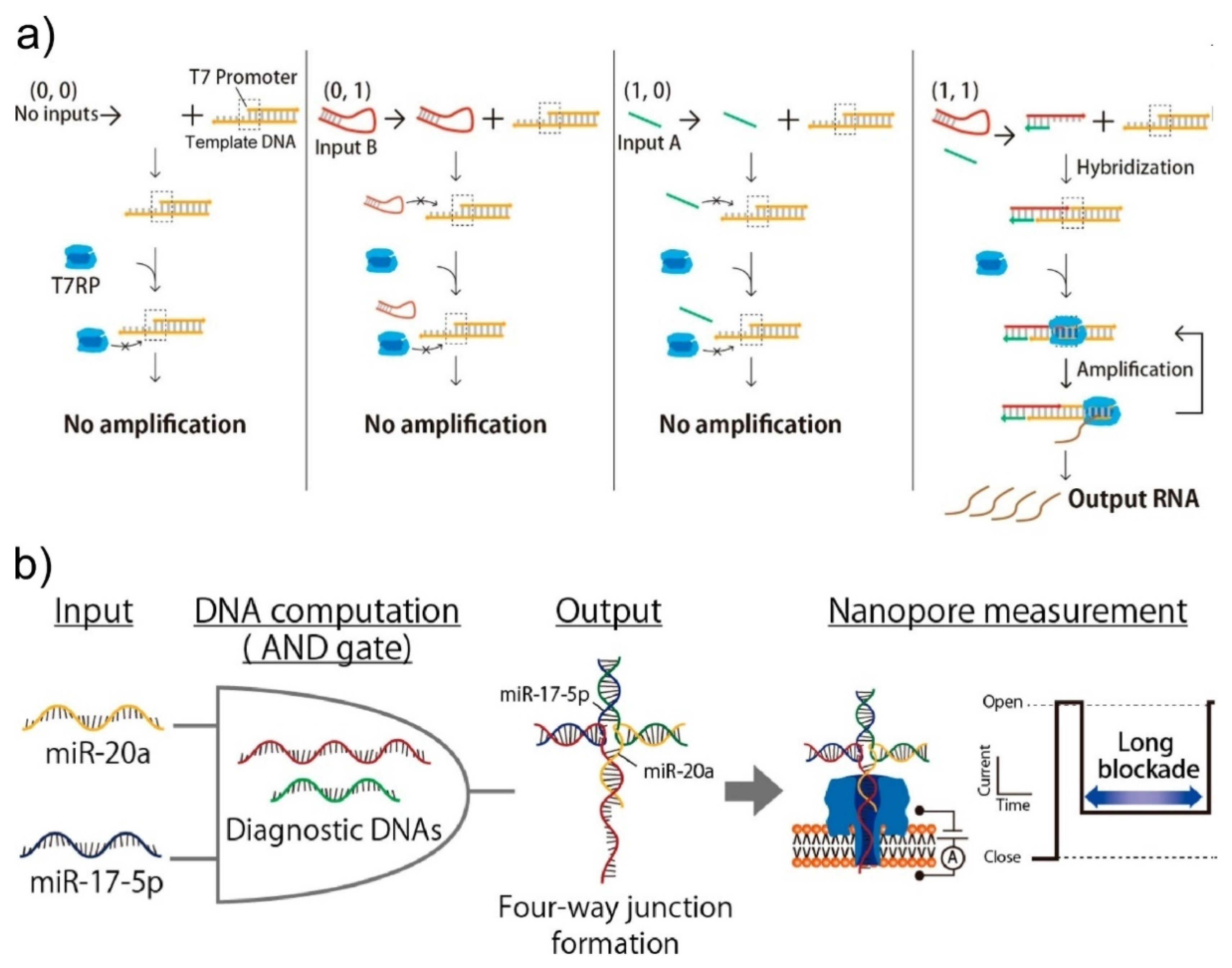
3.1.2. External Condition Sensing
3.2. Use of Semi-Permeability of Lipid Membranes
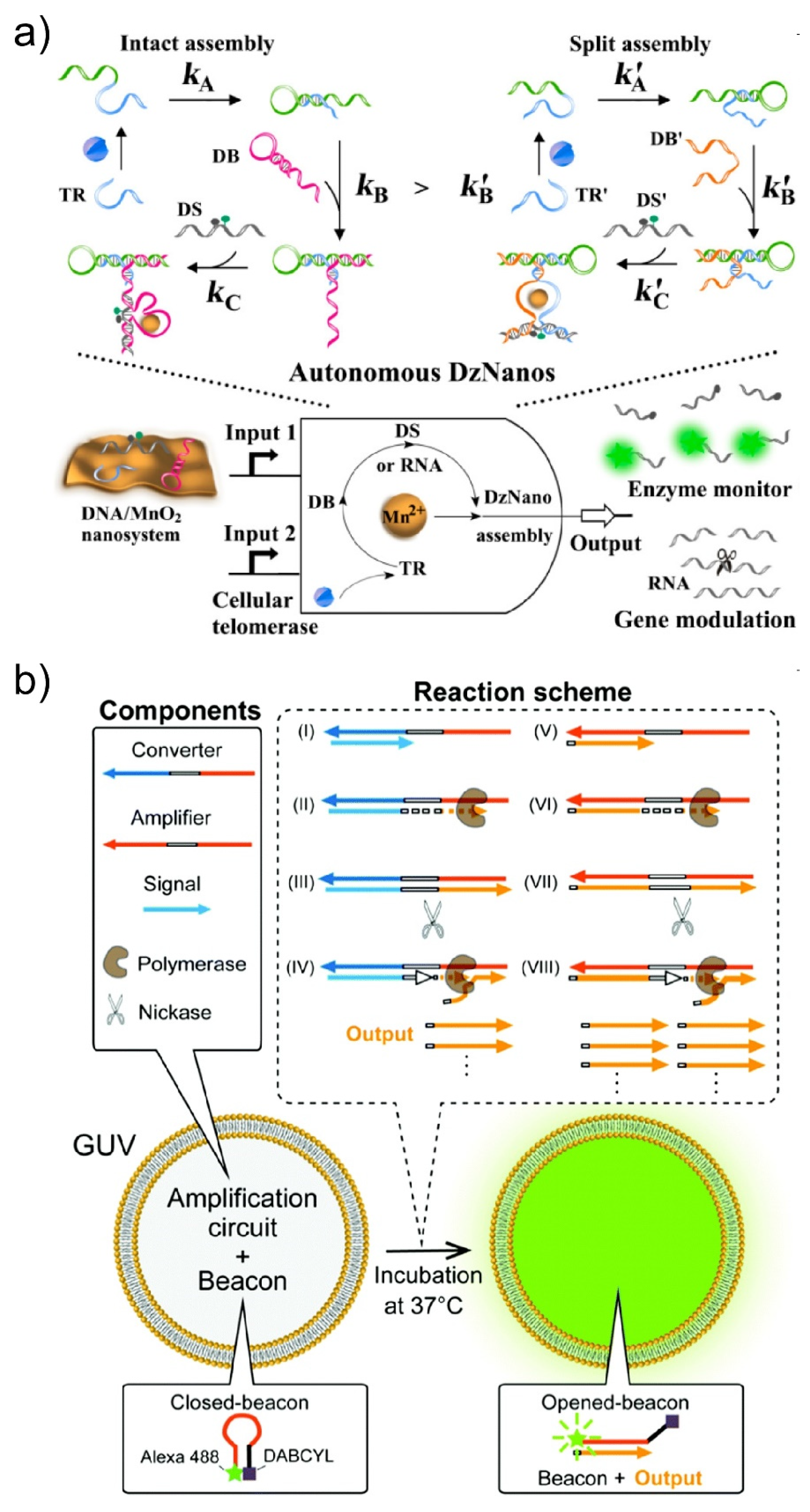
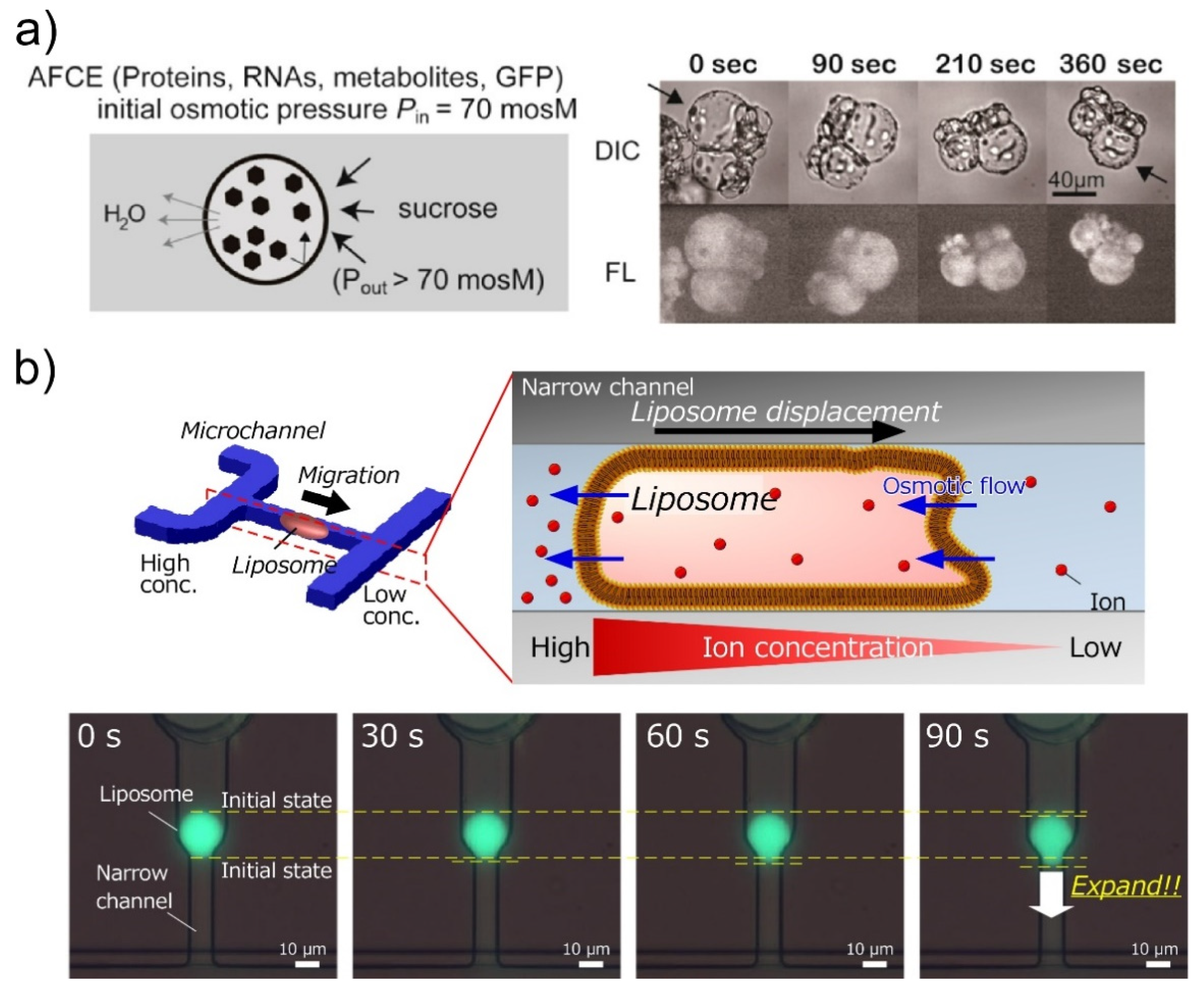
4. Encapsulation in Giant Unilamellar Vesicles
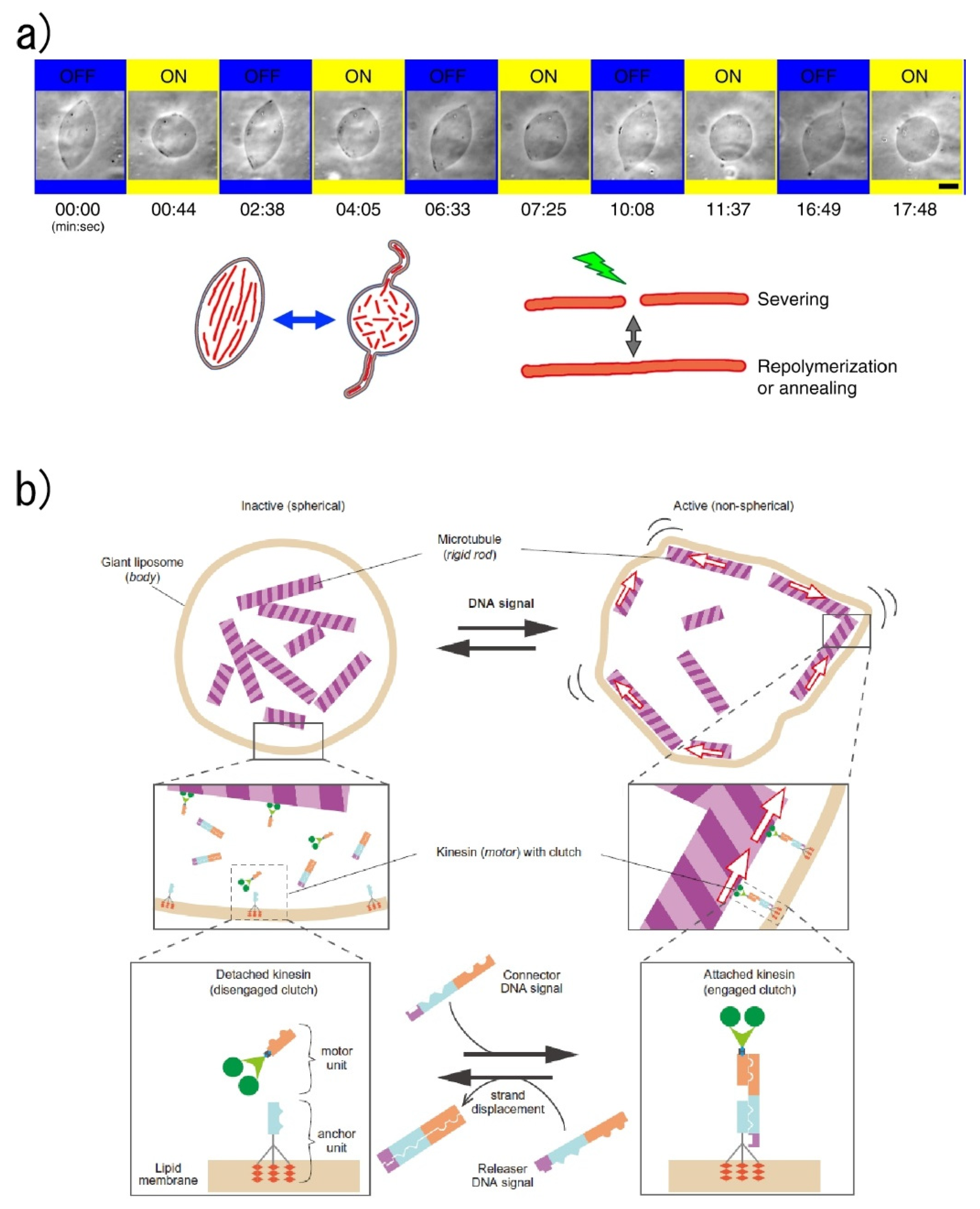
4.1. Actuators of Molecular Robots
4.2. Intelligence of Molecular Robots
5. Conclusions and Future Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Xu, C.; Hu, S.; Chen, X. Artificial cells: From basic science to applications. Mater. Today 2016, 19, 516–532. [Google Scholar] [CrossRef]
- Buddingh’, B.C.; van Hest, J.C.M. Artificial Cells: Synthetic Compartments with Life-like Functionality and Adaptivity. Acc. Chem. Res. 2017, 50, 769–777. [Google Scholar] [CrossRef]
- Simonsson, L.; Kurczy, M.E.; Trouillon, R.; Hook, F.; Cans, A.-S. A functioning artificial secretory cell. Sci. Rep. 2012, 2, 824. [Google Scholar] [CrossRef]
- Shin, J.; Noireaux, V. An E. coli Cell-Free Expression Toolbox: Application to Synthetic Gene Circuits and Artificial Cells. ACS Synth. Biol. 2012, 1, 29–41. [Google Scholar] [CrossRef]
- Pols, T.; Sikkema, H.R.; Gaastra, B.F.; Frallicciardi, J.; Śmigiel, W.M.; Singh, S.; Poolman, B. A synthetic metabolic network for physicochemical homeostasis. Nat. Commun. 2019, 10, 4239. [Google Scholar] [CrossRef]
- Choi, H.-J.; Montemagno, C.D. Artificial Organelle: ATP Synthesis from Cellular Mimetic Polymersomes. Nano Lett. 2005, 5, 2538–2542. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.F.; Szostak, J.W. Coupled Growth and Division of Model Protocell Membranes. J. Am. Chem. Soc. 2009, 131, 5705–5713. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, K.; Tamura, M.; Shohda, K.; Toyota, T.; Suzuki, K.; Sugawara, T. Self-reproduction of supramolecular giant vesicles combined with the amplification of encapsulated DNA. Nat. Chem. 2011, 3, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Peters, R.J.R.W.; Nijemeisland, M.; van Hest, J.C.M. Reversibly Triggered Protein-Ligand Assemblies in Giant Vesicles. Angew. Chem. Int. Ed. 2015, 54, 9614–9617. [Google Scholar] [CrossRef]
- Sun, S.; Li, M.; Dong, F.; Wang, S.; Tian, L.; Mann, S. Chemical Signaling and Functional Activation in Colloidosome-Based Protocells. Small 2016, 12, 1920–19273. [Google Scholar] [CrossRef]
- Adamala, K.P.; Martin-Alarcon, D.A.; Guthrie-Honea, K.R.; Boyden, E.S. Engineering genetic circuit interactions within and between synthetic minimal cells. Nat. Chem. 2017, 9, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Lin, X.; Si, T.; He, Q. Recent Progress on Bioinspired Self-Propelled Micro/Nanomotors via Controlled Molecular Self-Assembly. Small 2016, 12, 3080–3093. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Contreras-Llano, L.E.; Morris, E.; Mao, M.; Tan, C. Minimizing Context Dependency of Gene Networks Using Artificial Cells. ACS Appl. Mater. Interfaces 2018, 10, 30137–30146. [Google Scholar] [CrossRef] [PubMed]
- Dwidar, M.; Seike, Y.; Kobori, S.; Whitaker, C.; Matsuura, T.; Yokobayashi, Y. Programmable Artificial Cells Using Histamine-Responsive Synthetic Riboswitch. J. Am. Chem. Soc. 2019, 141, 11103–11114. [Google Scholar] [CrossRef] [PubMed]
- Murata, S.; Konagaya, A.; Kobayashi, S.; Saito, H.; Hagiya, M. Molecular Robotics: A New Paradigm for Artifacts. New Gener. Comput. 2013, 31, 27–45. [Google Scholar] [CrossRef]
- Hagiya, M.; Konagaya, A.; Kobayashi, S.; Saito, H.; Murata, S. Molecular Robots with Sensors and Intelligence. Acc. Chem. Res. 2014, 47, 1681–1690. [Google Scholar] [CrossRef]
- Kawano, R. Synthetic Ion Channels and DNA Logic Gates as Components of Molecular Robots. ChemPhysChem 2018, 19, 359–366. [Google Scholar] [CrossRef]
- Masubuchi, T.; Endo, M.; Iizuka, R.; Iguchi, A.; Yoon, D.H.; Sekiguchi, T.; Qi, H.; Iinuma, R.; Miyazono, Y.; Shoji, S.; et al. Construction of integrated gene logic-chip. Nat. Nanotechnol. 2018, 13, 933–940. [Google Scholar]
- Willner, E.M.; Kamada, Y.; Suzuki, Y.; Emura, T.; Hidaka, K.; Dietz, H.; Sugiyama, H.; Endo, M. Single-Molecule Observation of the Photoregulated Conformational Dynamics of DNA Origami Nanoscissors. Angew. Chem. Int. Ed. 2017, 56, 15324–15328. [Google Scholar] [CrossRef]
- Inoue, D.; Nitta, T.; Kabir, A.M.R.; Sada, K.; Gong, J.P.; Konagaya, A.; Kakugo, A. Sensing surface mechanical deformation using active probes driven by motor proteins. Nat. Commun. 2016, 7, 12557. [Google Scholar] [CrossRef]
- Keya, J.J.; Suzuki, R.; Kabir, A.M.R.; Inoue, D.; Asanuma, H.; Sada, K.; Hess, H.; Kuzuya, A.; Kakugo, A. DNA-assisted swarm control in a biomolecular motor system. Nat. Commun. 2018, 9, 453. [Google Scholar] [CrossRef] [PubMed]
- Inaba, H.; Yamamoto, T.; Iwasaki, T.; Kabir, A.M.R.; Kakugo, A.; Sada, K.; Matsuura, K. Stabilization of microtubules by encapsulation of the GFP using a Tau-derived peptide. Chem. Commun. 2019, 55, 9072–9075. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, K.; Kabir, A.M.R.; Akamatsu, N.; Saito, A.; Ishikawa, S.; Matsuyama, T.; Ditzer, O.; Islam, M.S.; Ohya, Y.; Sada, K.; et al. Artificial Smooth Muscle Model Composed of Hierarchically Ordered Microtubule Asters Mediated by DNA Origami Nanostructures. Nano Lett. 2019, 19, 3933–3938. [Google Scholar] [CrossRef] [PubMed]
- Moscho, A.; Orwar, O.; Chiu, D.T.; Modi, B.P.; Zare, R.N. Rapid preparation of giant unilamellar vesicles. Proc. Natl. Acad. Sci. USA 1996, 93, 11443–11447. [Google Scholar] [CrossRef] [PubMed]
- Akashi, K.; Miyata, H.; Itoh, H.; Kinosita, K. Preparation of giant liposomes in physiological conditions and their characterization under an optical microscope. Biophys. J. 1996, 71, 3242–3250. [Google Scholar] [CrossRef]
- Kurokawa, C.; Fujiwara, K.; Morita, M.; Kawamata, I.; Kawagishi, Y.; Sakai, A.; Murayama, Y.; Nomura, S.M.; Murata, S.; Takinoue, M.; et al. DNA cytoskeleton for stabilizing artificial cells. Proc. Natl. Acad. Sci. USA 2017, 114, 7228–7233. [Google Scholar] [CrossRef]
- Tohgasaki, T.; Shitomi, Y.; Feng, Y.; Honna, S.; Emura, T.; Hidaka, K.; Sugiyama, H.; Endo, M. A Photocaged DNA Nanocapsule for Controlled Unlocking and Opening inside the Cell. Bioconjugate Chem. 2019, 30, 1860–1863. [Google Scholar] [CrossRef]
- Hagiya, M.; Wang, S.; Kawamata, I.; Murata, S.; Isokawa, T.; Peper, F.; Imai, K. On DNA-based gellular automata. In Proceedings of the Unconventional Computation and Natural Computation. 13th International Conference, UCNC 2014, Lecture Notes in Computer Science, London, ON, Canada, 14–18 July 2014; pp. 177–189. [Google Scholar]
- Abe, K.; Kawamata, I.; Nomura, S.M.; Murata, S. Programmable reactions and diffusion using DNA for pattern formation in hydrogel medium. Mol. Syst. Des. Eng. 2019, 4, 639–643. [Google Scholar] [CrossRef]
- Bermudez, H.; Brannan, A.K.; Hammer, D.A.; Bates, F.S.; Discher, D.E. Molecular Weight Dependence of Polymersome Membrane Structure, Elasticity, and Stability. Macromolecules 2002, 35, 8203–8208. [Google Scholar] [CrossRef]
- Bangham, A.D.; Standish, M.M.; Watkins, J.C. Diffusion of univalent ions across the lamellae of swollen phospholipids. J. Mol. Biol. 1965, 13, 238–252. [Google Scholar] [CrossRef]
- Angelova, M.I.; Dimitrov, D.S. Liposome electroformation. Faraday Discuss. Chem. Soc. 1986, 81, 303–311. [Google Scholar] [CrossRef]
- Pautot, S.; Frisken, B.J.; Weitz, D.A. Production of Unilamellar Vesicles Using an Inverted Emulsion. Langmuir 2003, 19, 2870–2879. [Google Scholar] [CrossRef]
- Pautot, S.; Frisken, B.J.; Weitz, D.A. Engineering asymmetric vesicles. Proc. Natl. Acad. Sci. USA 2003, 100, 10718–10721. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.C.; Li, S.; Malmstadt, N. Microfluidic Fabrication of Asymmetric Giant Lipid Vesicles. ACS Appl. Mater. Interfaces 2011, 3, 1434–1440. [Google Scholar] [CrossRef]
- Matosevic, S.; Paegel, B.M. Stepwise Synthesis of Giant Unilamellar Vesicles on a Microfluidic Assembly Line. J. Am. Chem. Soc. 2011, 133, 2798–2800. [Google Scholar] [CrossRef] [PubMed]
- Karamdad, K.; Law, R.V.; Seddon, J.M.; Brooks, N.J.; Ces, O. Preparation and mechanical characterisation of giant unilamellar vesicles by a microfluidic method. Lab. Chip 2014, 15, 557–562. [Google Scholar] [CrossRef]
- Karamdad, K.; Law, R.V.; Seddon, J.M.; Brooks, N.J.; Ces, O. Studying the effects of asymmetry on the bending rigidity of lipid membranes formed by microfluidics. Chem. Commun. 2016, 52, 5277–5280. [Google Scholar] [CrossRef]
- Abkarian, M.; Loiseau, E.; Massiera, G. Continuous droplet interface crossing encapsulation (cDICE) for high throughput monodisperse vesicle design. Soft Matter. 2011, 7, 4610–4614. [Google Scholar] [CrossRef]
- Morita, M.; Onoe, H.; Yanagisawa, M.; Ito, H.; Ichikawa, M.; Fujiwara, K.; Saito, H.; Takinoue, M. Droplet-Shooting and Size-Filtration (DSSF) Method for Synthesis of Cell-Sized Liposomes with Controlled Lipid Compositions. ChemBioChem 2015, 16, 2029–2035. [Google Scholar] [CrossRef]
- Shum, H.C.; Lee, D.; Yoon, I.; Kodger, T.; Weitz, D.A. Double Emulsion Templated Monodisperse Phospholipid Vesicles. Langmuir 2008, 24, 7651–7653. [Google Scholar] [CrossRef]
- Arriaga, L.R.; Datta, S.S.; Kim, S.-H.; Amstad, E.; Kodger, T.E.; Monroy, F.; Weitz, D.A. Ultrathin Shell Double Emulsion Templated Giant Unilamellar Lipid Vesicles with Controlled Microdomain Formation. Small 2014, 10, 950–956. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, S.; Caspi, Y.; Meijering, A.E.C.; Dekker, C. Octanol-assisted liposome assembly on chip. Nat. Commun. 2016, 7, 10447. [Google Scholar] [CrossRef] [PubMed]
- Deng, N.-N.; Yelleswarapu, M.; Huck, W.T.S. Monodisperse Uni- and Multicompartment Liposomes. J. Am. Chem. Soc. 2016, 138, 7584–7591. [Google Scholar] [CrossRef] [PubMed]
- Funakoshi, K.; Suzuki, H.; Takeuchi, S. Formation of Giant Lipid Vesiclelike Compartments from a Planar Lipid Membrane by a Pulsed Jet Flow. J. Am. Chem. Soc. 2007, 129, 12608–12609. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, K.; Kawano, R.; Osaki, T.; Akiyoshi, K.; Takeuchi, S. Cell-sized asymmetric lipid vesicles facilitate the investigation of asymmetric membranes. Nat. Chem. 2016, 8, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Funakoshi, K.; Suzuki, H.; Takeuchi, S. Lipid Bilayer Formation by Contacting Monolayers in a Microfluidic Device for Membrane Protein Analysis. Anal. Chem. 2006, 78, 8169–8174. [Google Scholar] [CrossRef] [PubMed]
- Kawano, R.; Tsuji, Y.; Sato, K.; Osaki, T.; Kamiya, K.; Hirano, M.; Ide, T.; Miki, N.; Takeuchi, S. Automated Parallel Recordings of Topologically Identified Single Ion Channels. Sci. Rep. 2013, 3, 1995. [Google Scholar] [CrossRef]
- Abate, A.R.; Hung, T.; Mary, P.; Agresti, J.J.; Weitz, D.A. High-throughput injection with microfluidics using picoinjectors. Proc. Natl. Acad. Sci. USA 2010, 107, 19163–19166. [Google Scholar] [CrossRef]
- Weiss, M.; Frohnmayer, J.P.; Benk, L.T.; Haller, B.; Janiesch, J.-W.; Heitkamp, T.; Börsch, M.; Lira, R.B.; Dimova, R.; Lipowsky, R.; et al. Sequential bottom-up assembly of mechanically stabilized synthetic cells by microfluidics. Nat. Mater. 2018, 17, 89–96. [Google Scholar] [CrossRef]
- Khan, S.; Li, M.; Muench, S.P.; Jeuken, L.J.C.; Beales, P.A. Durable proteo-hybrid vesicles for the extended functional lifetime of membrane proteins in bionanotechnology. Chem. Commun. 2016, 52, 11020–11023. [Google Scholar] [CrossRef]
- Thiele, J.; Abate, A.R.; Shum, H.C.; Bachtler, S.; Förster, S.; Weitz, D.A. Fabrication of Polymersomes using Double-Emulsion Templates in Glass-Coated Stamped Microfluidic Devices. Small 2010, 6, 1723–1727. [Google Scholar] [CrossRef] [PubMed]
- Shum, H.C.; Zhao, Y.; Kim, S.-H.; Weitz, D.A. Multicompartment Polymersomes from Double Emulsions. Angew. Chem. Int. Ed. 2011, 50, 1648–1651. [Google Scholar] [CrossRef] [PubMed]
- Morton, D.; Mortezaei, S.; Yemenicioglu, S.; Isaacman, M.J.; Nova, I.C.; Gundlach, J.H.; Theogarajan, L. Tailored polymeric membranes for Mycobacterium smegmatis porin A (MspA) based biosensors. J. Mater. Chem. B 2015, 3, 5080–5086. [Google Scholar] [CrossRef] [PubMed]
- Rofeh, J.; Schankweiler, S.; Morton, D.; Mortezaei, S.; Qiang, L.; Gundlach, J.; Fisher, J.; Theogarajan, L. Microfluidic block copolymer membrane arrays for nanopore DNA sequencing. Appl. Phys. Lett. 2019, 114, 213701. [Google Scholar] [CrossRef]
- Kleineberg, C.; Wölfer, C.; Abbasnia, A.; Pischel, D.; Bednarz, C.; Ivanov, I.; Heitkamp, T.; Börsch, M.; Sundmacher, K.; Vidaković-Koch, T. Light-Driven ATP Regeneration in Diblock/Grafted Hybrid Vesicles. ChemBioChem 2020. [Google Scholar] [CrossRef] [PubMed]
- Sakai, N.; Matile, S. Synthetic Ion Channels. Langmuir 2013, 29, 9031–9040. [Google Scholar] [CrossRef] [PubMed]
- Joh, N.H.; Wang, T.; Bhate, M.P.; Acharya, R.; Wu, Y.; Grabe, M.; Hong, M.; Grigoryan, G.; DeGrado, W.F. De novo design of a transmembrane Zn2+-transporting four-helix bundle. Science 2014, 346, 1520–1524. [Google Scholar] [CrossRef]
- Lu, P.; Min, D.; DiMaio, F.; Wei, K.Y.; Vahey, M.D.; Boyken, S.E.; Chen, Z.; Fallas, J.A.; Ueda, G.; Sheffler, W.; et al. Accurate computational design of multipass transmembrane proteins. Science 2018, 359, 1042–1046. [Google Scholar] [CrossRef]
- Shimizu, K.; Mijiddorj, B.; Yoshida, S.; Akayama, S.; Hamada, Y.; Ohyama, A.; Usui, K.; Kawamura, I.; Kawano, R. De Novo Design of a Nanopore for DNA Detection Incorporating a β-hairpin Peptide. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Kaucher, M.S.; Harrell, W.A.; Davis, J.T. A Unimolecular G-Quadruplex that Functions as a Synthetic Transmembrane Na+ Transporter. J. Am. Chem. Soc. 2006, 128, 38–39. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, G.; Yamato, K.; Shen, Y.; Cheng, R.; Wei, X.; Bai, W.; Gao, Y.; Li, H.; Liu, Y.; et al. Self-assembling subnanometer pores with unusual mass-transport properties. Nat. Commun. 2012, 3, 949. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Kim, H.; Baek, K.; Kim, K. Synthetic Ion Channel Based on Metal-Organic Polyhedra. Angew. Chem. Int. Ed. 2008, 47, 5755–5757. [Google Scholar] [CrossRef] [PubMed]
- Kawano, R.; Horike, N.; Hijikata, Y.; Kondo, M.; Carné-Sánchez, A.; Larpent, P.; Ikemura, S.; Osaki, T.; Kamiya, K.; Kitagawa, S.; et al. Metal-Organic Cuboctahedra for Synthetic Ion Channels with Multiple Conductance States. Chem 2017, 2, 393–403. [Google Scholar] [CrossRef]
- Langecker, M.; Arnaut, V.; Martin, T.G.; List, J.; Renner, S.; Mayer, M.; Dietz, H.; Simmel, F.C. Synthetic Lipid Membrane Channels Formed by Designed DNA Nanostructures. Science 2012, 338, 932–936. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.R.; Stulz, E.; Howorka, S. Self-Assembled DNA Nanopores That Span Lipid Bilayers. Nano Lett. 2013, 13, 2351–2356. [Google Scholar] [CrossRef] [PubMed]
- Göpfrich, K.; Li, C.-Y.; Ricci, M.; Bhamidimarri, S.P.; Yoo, J.; Gyenes, B.; Ohmann, A.; Winterhalter, M.; Aksimentiev, A.; Keyser, U.F. Large-Conductance Transmembrane Porin Made from DNA Origami. ACS Nano 2016, 10, 8207–8214. [Google Scholar] [CrossRef]
- Krishnan, S.; Ziegler, D.; Arnaut, V.; Martin, T.G.; Kapsner, K.; Henneberg, K.; Bausch, A.R.; Dietz, H.; Simmel, F.C. Molecular transport through large-diameter DNA nanopores. Nat. Commun. 2016, 7, 12787. [Google Scholar] [CrossRef]
- Arnott, P.M.; Howorka, S. A Temperature-Gated Nanovalve Self-Assembled from DNA to Control Molecular Transport across Membranes. ACS Nano 2019, 13, 3334–3340. [Google Scholar] [CrossRef]
- Burns, J.R.; Seifert, A.; Fertig, N.; Howorka, S. A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nat. Nanotechnol. 2016, 11, 152–156. [Google Scholar] [CrossRef]
- Seifert, A.; Göpfrich, K.; Burns, J.R.; Fertig, N.; Keyser, U.F.; Howorka, S. Bilayer-Spanning DNA Nanopores with Voltage-Switching between Open and Closed State. ACS Nano 2015, 9, 1117–1126. [Google Scholar] [CrossRef]
- Kocabey, S.; Kempter, S.; List, J.; Xing, Y.; Bae, W.; Schiffels, D.; Shih, W.M.; Simmel, F.C.; Liedl, T. Membrane-Assisted Growth of DNA Origami Nanostructure Arrays. ACS Nano 2015, 9, 3530–3539. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Endo, M.; Sugiyama, H. Lipid-bilayer-assisted two-dimensional self-assembly of DNA origami nanostructures. Nat. Commun. 2015, 6, 8052. [Google Scholar] [CrossRef] [PubMed]
- Czogalla, A.; Kauert, D.J.; Franquelim, H.G.; Uzunova, V.; Zhang, Y.; Seidel, R.; Schwille, P. Amphipathic DNA Origami Nanoparticles to Scaffold and Deform Lipid Membrane Vesicles. Angew. Chem. Int. Ed. 2015, 54, 6501–6505. [Google Scholar] [CrossRef] [PubMed]
- Czogalla, A.; Petrov, E.P.; Kauert, D.J.; Uzunova, V.; Zhang, Y.; Seidel, R.; Schwille, P. Switchable domain partitioning and diffusion of DNA origami rods on membranes. Faraday Discuss. 2013, 161, 31–43. [Google Scholar] [CrossRef]
- Kasianowicz, J.J.; Brandin, E.; Branton, D.; Deamer, D.W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. USA 1996, 93, 13770–13773. [Google Scholar] [CrossRef]
- Akeson, M.; Branton, D.; Kasianowicz, J.J.; Brandin, E.; Deamer, D.W. Microsecond Time-Scale Discrimination Among Polycytidylic Acid, Polyadenylic Acid, and Polyuridylic Acid as Homopolymers or as Segments Within Single RNA Molecules. Biophys. J. 1999, 77, 3227–3233. [Google Scholar] [CrossRef]
- Gu, L.-Q.; Braha, O.; Conlan, S.; Cheley, S.; Bayley, H. Stochastic sensing of organic analytes by a pore-forming protein containing a molecular adapter. Nature 1999, 398, 686–690. [Google Scholar] [CrossRef]
- Meller, A.; Nivon, L.; Brandin, E.; Golovchenko, J.; Branton, D. Rapid nanopore discrimination between single polynucleotide molecules. Proc. Natl. Acad. Sci. USA 2000, 97, 1079–1084. [Google Scholar] [CrossRef]
- Ding, T.; Yang, J.; Pan, V.; Zhao, N.; Lu, Z.; Ke, Y.; Zhang, C. DNA nanotechnology assisted nanopore-based analysis. Nucleic Acids Res. 2020, 48, 2791–2806. [Google Scholar] [CrossRef]
- Song, L.; Hobaugh, M.R.; Shustak, C.; Cheley, S.; Bayley, H.; Gouaux, J.E. Structure of Staphylococcal α-Hemolysin, a Heptameric Transmembrane Pore. Science 1996, 274, 1859–1865. [Google Scholar] [CrossRef]
- Butler, T.Z.; Pavlenok, M.; Derrington, I.M.; Niederweis, M.; Gundlach, J.H. Single-molecule DNA detection with an engineered MspA protein nanopore. Proc. Natl. Acad. Sci. USA 2008, 105, 20647–20652. [Google Scholar] [CrossRef] [PubMed]
- Adleman, L.M. Molecular computation of solutions to combinatorial problems. Science 1994, 266, 1021–1024. [Google Scholar] [CrossRef] [PubMed]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Mueller, P.; Rudin, D.O.; Ti Tien, H.; Wescott, W.C. Reconstitution of Cell Membrane Structure in vitro and its Transformation into an Excitable System. Nature 1962, 194, 979–980. [Google Scholar] [CrossRef]
- Montal, M.; Mueller, P. Formation of Bimolecular Membranes from Lipid Monolayers and a Study of Their Electrical Properties. Proc. Natl. Acad. Sci. USA 1972, 69, 3561–3566. [Google Scholar] [CrossRef]
- Tsuji, Y.; Kawano, R.; Osaki, T.; Kamiya, K.; Miki, N.; Takeuchi, S. Droplet Split-and-Contact Method for High-Throughput Transmembrane Electrical Recording. Anal. Chem. 2013, 85, 10913–10919. [Google Scholar] [CrossRef]
- Tsuji, Y.; Kawano, R.; Osaki, T.; Kamiya, K.; Miki, N.; Takeuchi, S. Droplet-based lipid bilayer system integrated with microfluidic channels for solution exchange. Lab. Chip 2013, 13, 1476–1481. [Google Scholar] [CrossRef]
- Kawano, R.; Tsuji, Y.; Kamiya, K.; Kodama, T.; Osaki, T.; Miki, N.; Takeuchi, S. A Portable Lipid Bilayer System for Environmental Sensing with a Transmembrane Protein. PLoS ONE 2014, 9, e102427. [Google Scholar] [CrossRef]
- Shoji, K.; Kawano, R. Microfluidic Formation of Double-Stacked Planar Bilayer Lipid Membranes by Controlling the Water-Oil Interface. Micromachines 2018, 9, 253. [Google Scholar] [CrossRef]
- Shoji, K.; Kawano, R.; White, R.J. Spatially Resolved Chemical Detection with a Nanoneedle-Probe-Supported Biological Nanopore. ACS Nano 2019, 13, 2606–2614. [Google Scholar] [CrossRef]
- Shoji, K.; Kawano, R.; White, R.J. Recessed Ag/AgCl Microelectrode-Supported Lipid Bilayer for Nanopore Sensing. Anal. Chem. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ohara, M.; Takinoue, M.; Kawano, R. Nanopore Logic Operation with DNA to RNA Transcription in a Droplet System. ACS Synth. Biol. 2017, 6, 1427–1432. [Google Scholar] [CrossRef] [PubMed]
- Hiratani, M.; Ohara, M.; Kawano, R. Amplification and Quantification of an Antisense Oligonucleotide from Target microRNA Using Programmable DNA and a Biological Nanopore. Anal. Chem. 2017, 89, 2312–2317. [Google Scholar] [CrossRef] [PubMed]
- Hiratani, M.; Kawano, R. DNA Logic Operation with Nanopore Decoding To Recognize MicroRNA Patterns in Small Cell Lung Cancer. Anal. Chem. 2018, 90, 8531–8537. [Google Scholar] [CrossRef] [PubMed]
- Yatvin, M.B.; Kreutz, W.; Horwitz, B.A.; Shinitzky, M. pH-sensitive liposomes: Possible clinical implications. Science 1980, 210, 1253–1255. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.-J.; Szoka, F.C. pH-Sensitive Liposomes. J. Liposome Res. 1994, 4, 361–395. [Google Scholar] [CrossRef]
- Sudimack, J.J.; Guo, W.; Tjarks, W.; Lee, R.J. A novel pH-sensitive liposome formulation containing oleyl alcohol. Biochim. Biophys. Acta Biomembr. 2002, 1564, 31–37. [Google Scholar] [CrossRef][Green Version]
- Yavlovich, A.; Smith, B.; Gupta, K.; Blumenthal, R.; Puri, A. Light-sensitive Lipid-based Nanoparticles for Drug Delivery: Design Principles and Future Considerations for Biological Applications. Mol. Membr. Biol. 2010, 27, 364–381. [Google Scholar] [CrossRef]
- Trier, S.; Henriksen, J.R.; Andresen, T.L. Membrane fusion of pH-sensitive liposomes—A quantitative study using giant unilamellar vesicles. Soft Matter. 2011, 7, 9027–9034. [Google Scholar] [CrossRef]
- Li, Q.; Li, W.; Di, H.; Luo, L.; Zhu, C.; Yang, J.; Yin, X.; Yin, H.; Gao, J.; Du, Y.; et al. A photosensitive liposome with NIR light triggered doxorubicin release as a combined photodynamic-chemo therapy system. J. Control. Release 2018, 277, 114–125. [Google Scholar] [CrossRef]
- Jia, H.; Litschel, T.; Heymann, M.; Eto, H.; Franquelim, H.G.; Schwille, P. Shaping Giant Membrane Vesicles in 3D-Printed Protein Hydrogel Cages. Small 2020, 16, 1906259. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, M.; Shoji, K.; Takai, N.; Kawano, R. Biological Nanopore Probe: Probing of Viscous Solutions in a Confined Nanospace. J. Phys. Chem. B 2020, 124, 2410–2416. [Google Scholar] [CrossRef] [PubMed]
- DeGuzman, V.S.; Lee, C.C.; Deamer, D.W.; Vercoutere, W.A. Sequence-dependent gating of an ion channel by DNA hairpin molecules. Nucleic Acids Res. 2006, 34, 6425–6437. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fujiwara, K.; Yanagisawa, M. Generation of Giant Unilamellar Liposomes Containing Biomacromolecules at Physiological Intracellular Concentrations using Hypertonic Conditions. ACS Synth. Biol. 2014, 3, 870–874. [Google Scholar] [CrossRef]
- Andes-Koback, M.; Keating, C.D. Complete Budding and Asymmetric Division of Primitive Model Cells To Produce Daughter Vesicles with Different Interior and Membrane Compositions. J. Am. Chem. Soc. 2011, 133, 9545–9555. [Google Scholar] [CrossRef]
- Shoji, K.; Kawano, R. Osmotic-engine-driven liposomes in microfluidic channels. Lab. Chip 2019, 19, 3472–3480. [Google Scholar] [CrossRef]
- Stroka, K.M.; Jiang, H.; Chen, S.-H.; Tong, Z.; Wirtz, D.; Sun, S.X.; Konstantopoulos, K. Water Permeation Drives Tumor Cell Migration in Confined Microenvironments. Cell 2014, 157, 611–623. [Google Scholar] [CrossRef]
- Honda, M.; Takiguchi, K.; Ishikawa, S.; Hotani, H. Morphogenesis of liposomes encapsulating actin depends on the type of actin-crosslinking11Edited by M. F. Moody. J. Mol. Biol. 1999, 287, 293–300. [Google Scholar] [CrossRef]
- Hotani, H.; Nomura, F.; Suzuki, Y. Giant liposomes: From membrane dynamics to cell morphogenesis. Curr. Opin. Colloid Interface Sci. 1999, 4, 358–368. [Google Scholar] [CrossRef]
- Hayashi, M.; Nishiyama, M.; Kazayama, Y.; Toyota, T.; Harada, Y.; Takiguchi, K. Reversible Morphological Control of Tubulin-Encapsulating Giant Liposomes by Hydrostatic Pressure. Langmuir 2016, 32, 3794–3802. [Google Scholar] [CrossRef]
- Tanaka, S.; Takiguchi, K.; Hayashi, M. Repetitive stretching of giant liposomes utilizing the nematic alignment of confined actin. Commun. Phys. 2018, 1, 1–10. [Google Scholar] [CrossRef]
- Sato, Y.; Hiratsuka, Y.; Kawamata, I.; Murata, S.; Nomura, S.M. Micrometer-sized molecular robot changes its shape in response to signal molecules. Sci. Robot. 2017, 2, eaal3735. [Google Scholar] [CrossRef]
- Toyota, T.; Sugiyama, H.; Hiroi, S.; Ito, H.; Kitahata, H. Chemically artificial rovers based on self-propelled droplets in micrometer-scale environment. Curr. Opin. Colloid Interface Sci. 2020, 49, 60–68. [Google Scholar] [CrossRef]
- Inaba, H.; Uemura, A.; Morishita, K.; Kohiki, T.; Shigenaga, A.; Otaka, A.; Matsuura, K. Light-induced propulsion of a giant liposome driven by peptide nanofibre growth. Sci. Rep. 2018, 8, 6243. [Google Scholar] [CrossRef]
- Benenson, Y. Biocomputers: From test tubes to live cells. Mol. BioSyst. 2009, 5, 675–685. [Google Scholar] [CrossRef]
- Ness, J.V.; Ness, L.K.V.; Galas, D.J. Isothermal reactions for the amplification of oligonucleotides. Proc. Natl. Acad. Sci. USA 2003, 100, 4504–4509. [Google Scholar] [CrossRef]
- Rinaudo, K.; Bleris, L.; Maddamsetti, R.; Subramanian, S.; Weiss, R.; Benenson, Y. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat. Biotechnol. 2007, 25, 795–801. [Google Scholar] [CrossRef]
- Chen, F.; Bai, M.; Cao, K.; Zhao, Y.; Cao, X.; Wei, J.; Wu, N.; Li, J.; Wang, L.; Fan, C.; et al. Programming Enzyme-Initiated Autonomous DNAzyme Nanodevices in Living Cells. ACS Nano 2017, 11, 11908–11914. [Google Scholar] [CrossRef]
- Komiya, K.; Komori, M.; Noda, C.; Kobayashi, S.; Yoshimura, T.; Yamamura, M. Leak-free million-fold DNA amplification with locked nucleic acid and targeted hybridization in one pot. Org. Biomol. Chem. 2019, 17, 5708–5713. [Google Scholar] [CrossRef]
- Sato, Y.; Komiya, K.; Kawamata, I.; Murata, S.; Nomura, S.M. Isothermal amplification of specific DNA molecules inside giant unilamellar vesicles. Chem. Commun. 2019, 55, 9084–9087. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shoji, K.; Kawano, R. Recent Advances in Liposome-Based Molecular Robots. Micromachines 2020, 11, 788. https://doi.org/10.3390/mi11090788
Shoji K, Kawano R. Recent Advances in Liposome-Based Molecular Robots. Micromachines. 2020; 11(9):788. https://doi.org/10.3390/mi11090788
Chicago/Turabian StyleShoji, Kan, and Ryuji Kawano. 2020. "Recent Advances in Liposome-Based Molecular Robots" Micromachines 11, no. 9: 788. https://doi.org/10.3390/mi11090788
APA StyleShoji, K., & Kawano, R. (2020). Recent Advances in Liposome-Based Molecular Robots. Micromachines, 11(9), 788. https://doi.org/10.3390/mi11090788