Drug Toxicity Evaluation Based on Organ-on-a-chip Technology: A Review
Abstract
1. Introduction
2. Biomarkers for Drug Toxicity Testing
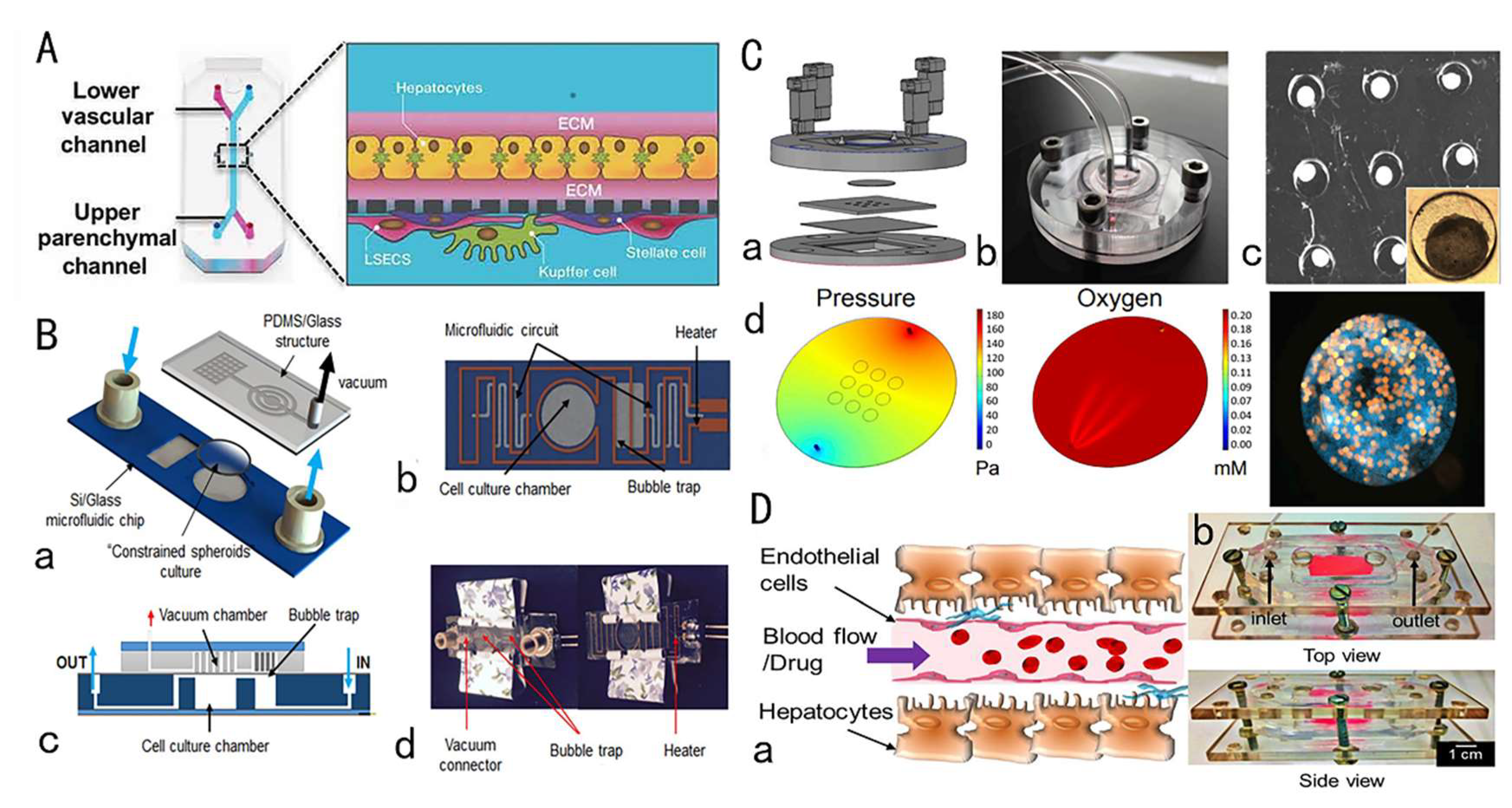
3. Drug Hepatotoxicity Testing by Liver-on-a-Chip
3.1. Overview of Liver and Hepatotoxicity
3.2. Research Progress of Liver Chips for Drug Toxicity Testing
3.3. Brief Summary
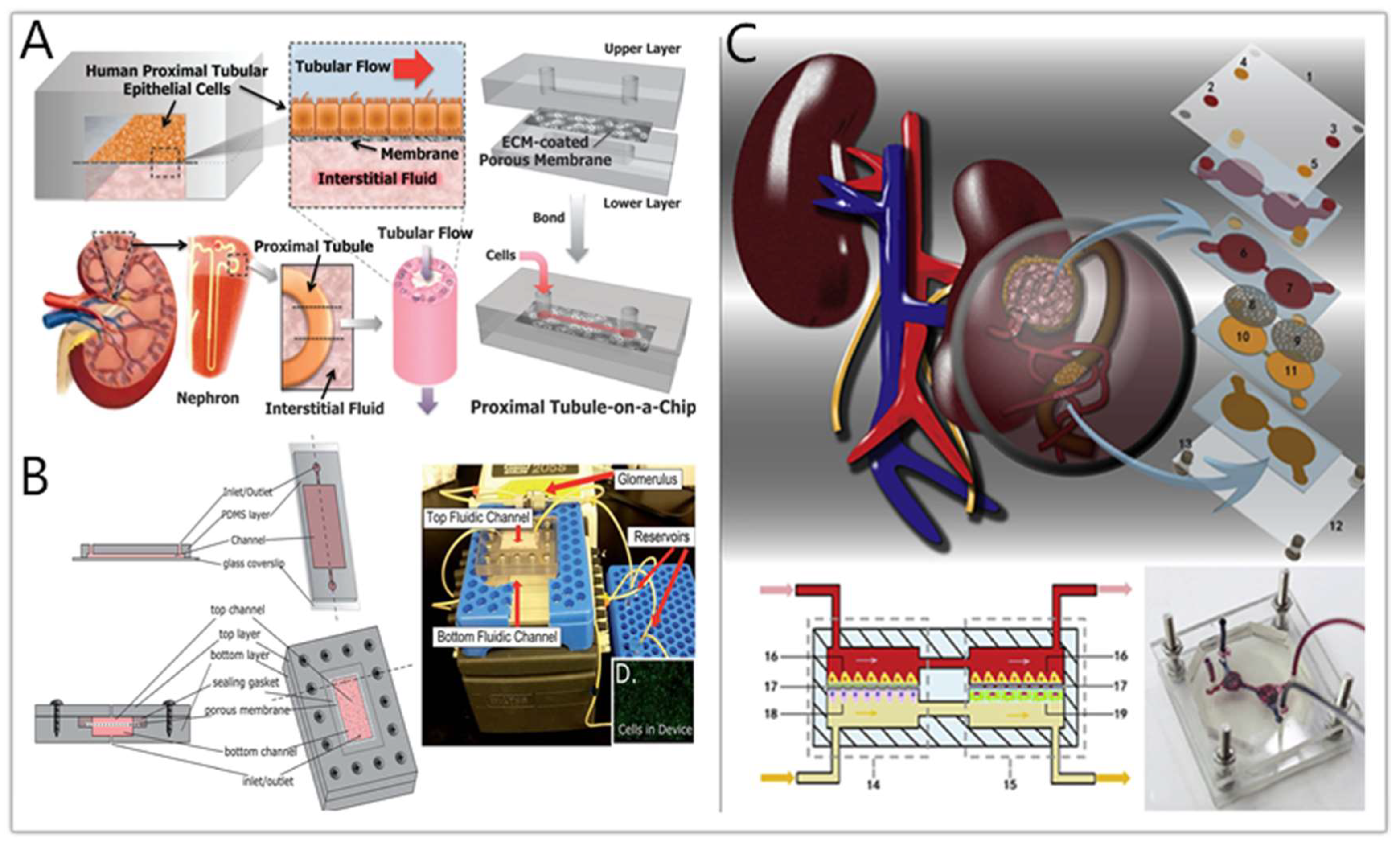
4. Drug Nephrotoxicity Testing by Kidney-on-a-Chip
4.1. Overview of Drug-Induced Nephrotoxicity
4.2. Kidney-on-a-Chip for Drug Nephrotoxicity Testing
4.3. Brief Summary
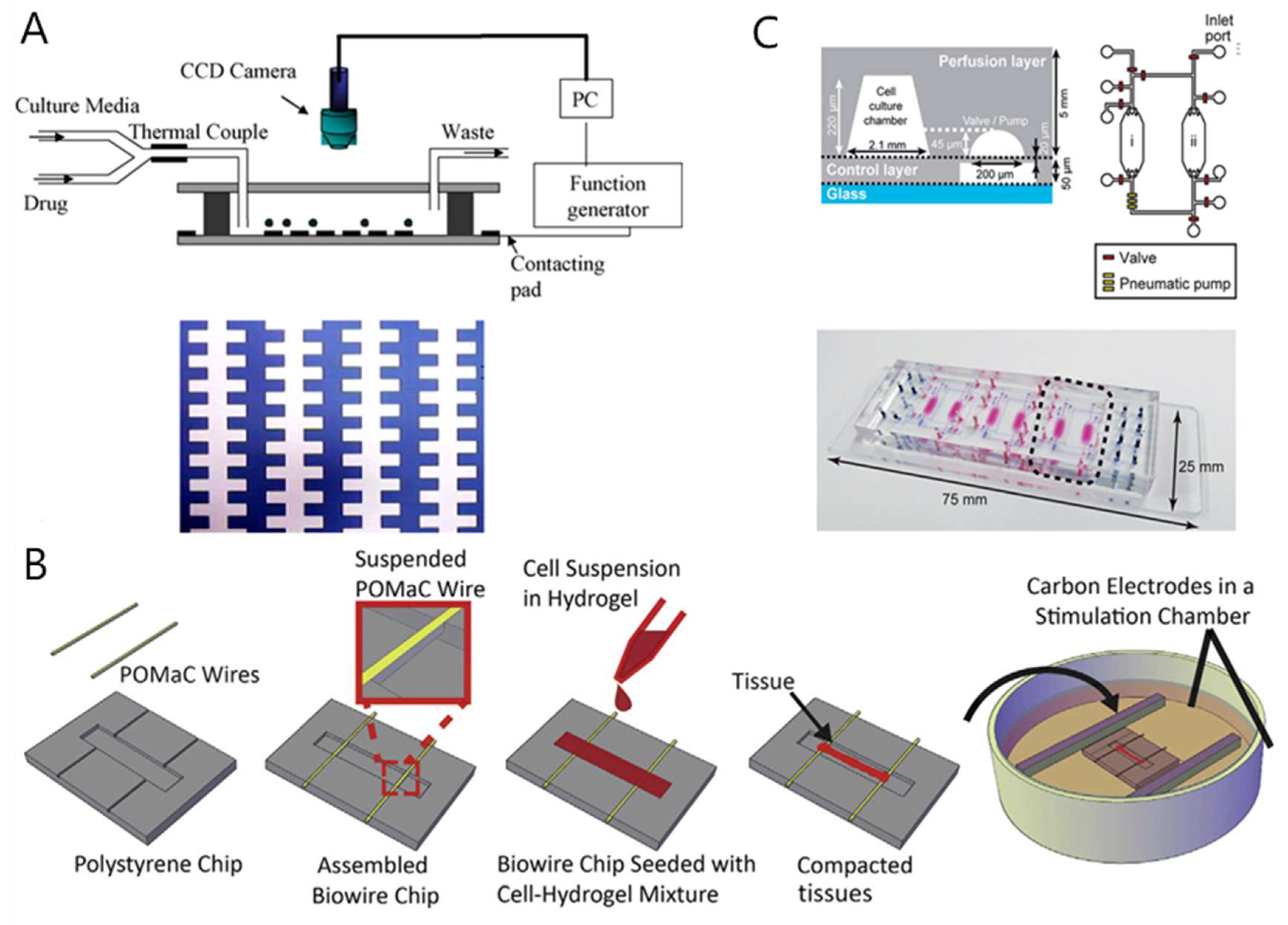
5. Drug Cardiotoxicity Testing by Heart-on-a-Chip
5.1. Overview of Drug-Induced Cardiotoxicity
5.2. Heart-on-a-Chip for Drug-Induced Cardiotoxicity Testing
5.3. Brief Summary
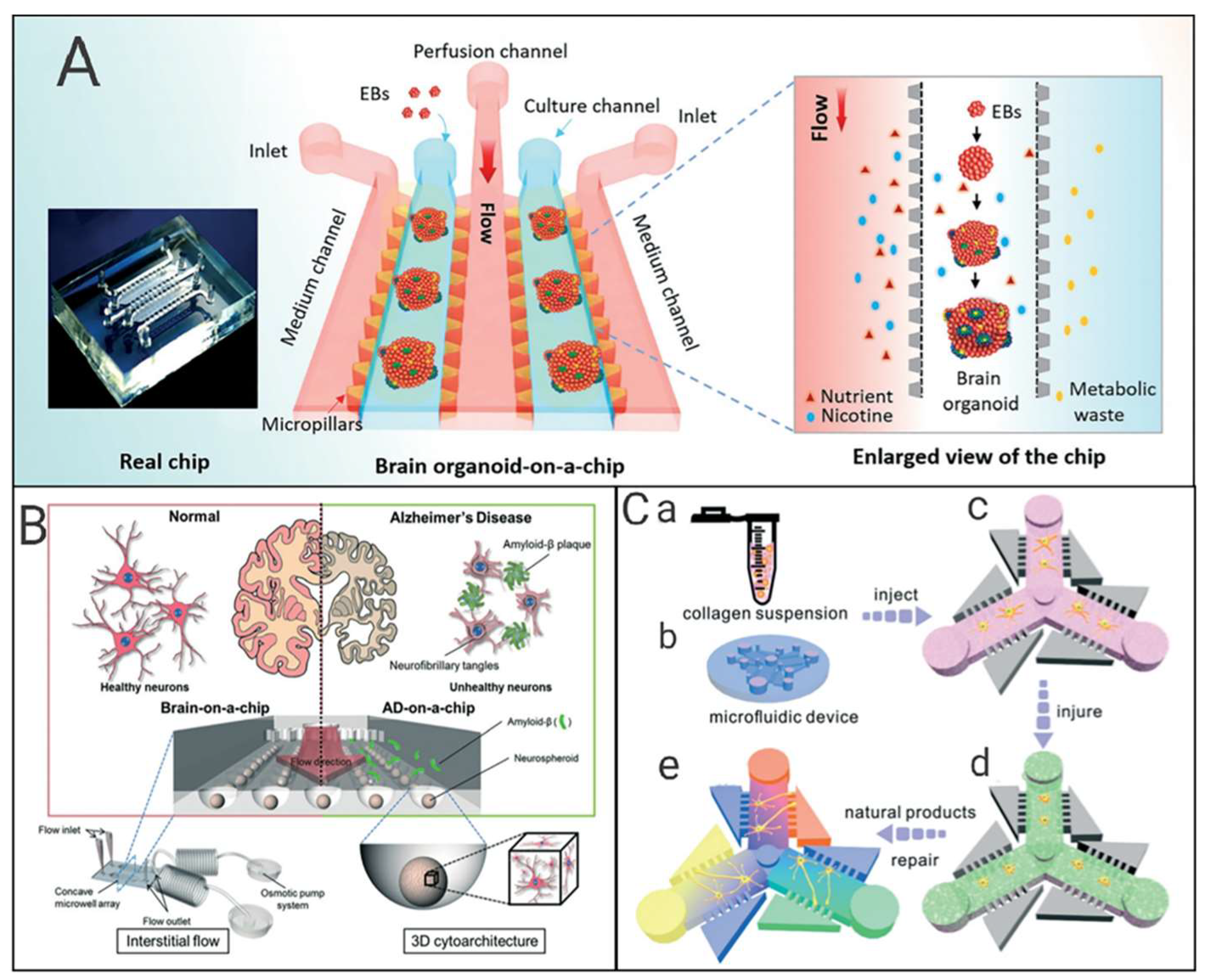
6. Drug Neurotoxicity Testing by Nerve-on-a-Chip
6.1. Research Progress of Neural Chips
6.2. Application of Neural Chips in Toxicity Detection
6.3. Neural Chips and Neurodegenerative Diseases
6.4. Brief Summary
7. Drug Toxicity Evaluation by Other Organ-on-Chips
7.1. Gut-on-a-Chip
7.2. Lung-on-a-Chip
7.3. Blood–brain Barrier (BBB)-on-a-Chip
7.4. Brief Summary
8. Drug Toxicity Evaluation by Multi-Organ-on-a-Chip
8.1. Liver-Kidney-on-Chip
8.2. Other Multi-Organs-on-Chips
8.3. Brief Summary
9. Conclusions and Future Perspective
Author Contributions
Funding
Conflicts of Interest
References
- Barré-Sinoussi, F.O.; Montagutelli, X. Animal models are essential to biological research: Issues and perspectives. Future Sci. OA 2015, 1, FSO63. [Google Scholar] [CrossRef] [PubMed]
- Yeo, L.Y.; Hsueh-Chia, C.; Peggy, P.Y.C.; James, R.F. Microfluidic devices for bioapplications. Small 2011, 1, 12–48. [Google Scholar] [CrossRef] [PubMed]
- Volpatti, L.R.; Yetisen, A.K. Commercialization of microfluidic devices. Trends Biotechnol. 2014, 7, 347–350. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Q.; Ning, R.; Ma, Y.; Lin, J. Recent developments in microfluidic chip for In vitro cell-based research. Chin. J. Anal. Chem. 2016, 4, 522–532. [Google Scholar] [CrossRef]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 8, 760–772. [Google Scholar] [CrossRef]
- Zhang, B.; Korolj, A.; Lai, B.F.L.; Radisic, M. Advances in organ-on-a-chip engineering. Nat. Rev. Mater. 2018, 8, 257–278. [Google Scholar] [CrossRef]
- Hendriks, D.F.G.; Fredriksson Puigvert, L.; Messner, S.; Mortiz, W.; Ingelman-Sundberg, M. Hepatic 3D spheroid models for the detection and study of compounds with cholestatic liability. Sci. Rep. 2016, 6, 35434. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, X.; Li, X.; Zhang, Y.; Hou, T.; Wei, L.; Qu, L.; Shi, L.; Liu, Y.; Zou, L.; et al. Anti-gastric cancer activity in three-dimensional tumor spheroids of bufadienolides. Sci. Rep. 2016, 6, 24772. [Google Scholar] [CrossRef]
- Zhuang, P.; Sun, A.X.; An, J.; Chua, C.K.; Chew, S.Y. 3D neural tissue models: From spheroids to bioprinting. Biomaterials 2018, 154, 113–133. [Google Scholar] [CrossRef]
- Fatehullah, A.; Tan, S.H.; Barker, N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016, 3, 246–254. [Google Scholar] [CrossRef]
- Park, S.E.; Georgescu, A.; Huh, D. Organoids-on-a-chip. Science 2019, 364, 960–965. [Google Scholar] [CrossRef] [PubMed]
- Caplin, J.D.; Granados, N.G.; James, M.R.; Montazami, R.; Hashemi, N. Microfluidic organ-on-a-chip technology for advancement of drug development and toxicology. Adv. Healthc. Mater. 2015, 10, 1426–1450. [Google Scholar] [CrossRef] [PubMed]
- Knowlton, S.; Tasoglu, S. A bioprinted liver-on-a-chip for drug screening applications. Trends Biotechnol. 2016, 9, 681–682. [Google Scholar] [CrossRef] [PubMed]
- Faria, J.; Ahmed, S.; Gerritsen, K.G.F.; Mihaila, S.M.; Masereeuw, R. Kidney-based In vitro models for drug-induced toxicity testing. Arch. Toxicol. 2019, 93, 3397–3418. [Google Scholar] [CrossRef] [PubMed]
- Conant, G.; Lai, B.F.L.; Lu, R.X.Z.; Korolj, A.; Wang, E.Y.; Radisic, M. High-content assessment of cardiac function using heart-on-a-chip devices as drug screening model. Stem Cell Rev. Rep. 2017, 3, 335–346. [Google Scholar] [CrossRef] [PubMed]
- van de Wijdeven, R.; Ramstad, O.H.; Bauer, U.S.; Halaas, O.; Sandvig, A.; Sandvig, I. Structuring a multi-nodal neural network In vitro within a novel design microfluidic chip. Biomed. Microdevices 2018, 1, 9. [Google Scholar] [CrossRef] [PubMed]
- Rezaei Kolahchi, A.; Khadem Mohtaram, N.; Pezeshgi Modarres, H.; Mohammadi, M.; Geraili, A.; Jafari, P.; Akbari, M.; Sanati-Nezhad, A. Microfluidic-based multi-organ platforms for drug discovery. Micromachines 2016, 9, 162. [Google Scholar] [CrossRef]
- Zheng, F.; Fu, F.; Cheng, Y.; Wang, C.; Zhao, Y.; Gu, Z. Organ-on-a-chip systems: Microengineering to biomimic living systems. Small 2016, 17, 2253–2282. [Google Scholar] [CrossRef]
- Hamilton, L.A.; Collins-Yoder, A.; Collins, R.E. Drug-induced liver injury. AACN Adv. Crit. Care 2016, 4, 430–440. [Google Scholar] [CrossRef]
- Thulin, P.; Hornby, R.J.; Auli, M.; Nordahl, G.; Antoine, D.J.; Starkey Lewis, P.; Goldring, C.E.; Park, B.K.; Prats, N.; Glinghammar, B.; et al. A longitudinal assessment of miR-122 and GLDH as biomarkers of drug-induced liver injury in the rat. Biomarkers 2017, 5, 461–469. [Google Scholar] [CrossRef]
- Huebener, P.; Pradere, J.; Hernandez, C.; Gwak, G.; Caviglia, J.M.; Mu, X.; Loike, J.D.; Jenkins, R.E.; Antoine, D.J.; Schwabe, R.F. The HMGB1/RAGE axis triggers neutrophil-mediated injury amplification following necrosis. J. Clin. Investig. 2015, 2, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Church, R.J.; Watkins, P.B. The transformation in biomarker detection and management of drug-induced liver injury. Liver Int. 2017, 37, 1582–1590. [Google Scholar] [CrossRef]
- Mcgill, M.R.; Sharpe, M.R.; Williams, C.D.; Taha, M.; Jaeschke, H. The mechanism underlying acetaminophen-induced hepatotoxicity in humans and mice involves mitochondrial damage and nuclear DNA fragmentation. J. Clin. Investig. 2012, 4, 1574–1583. [Google Scholar] [CrossRef] [PubMed]
- Church, R.J.; Ublick, G.A.K.; Aubrecht, J.; Bonkovsky, H.L.; Chalasani, N.; Fontana, R.J.; Goepfert, J.C.; Hackman, F.; King, N.M.P.; Kirby, S. Candidate biomarkers for the diagnosis and prognosis of drug-induced liver injury: An international collaborative effort. Hepatology 2019, 69, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Desai, P.K.; Tseng, H.; Souza, G.R. Assembly of Hepatocyte Spheroids Using Magnetic 3D Cell Culture for CYP450 Inhibition/Induction. Int. J. Mol. Sci. 2017, 18, 1085. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, S.; Marzolf, B.; Troisch, P.; Brightman, A.; Hu, Z.; Hood, L.E.; Galas, D.J. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl. Acad. Sci. USA 2009, 11, 4402–4407. [Google Scholar] [CrossRef]
- Mikus, M.; Drobin, K.; Gry, M.; Bachmann, J.; Lindberg, J.; Yimer, G.; Aklillu, E.; Makonnen, E.; Aderaye, G.; Roach, J. Elevated levels of circulating CDH5 and FABP1 in association with human drug-induced liver injury. Liver Int. 2017, 37, 132–140. [Google Scholar] [CrossRef]
- Swan, S.K. The search continues—An ideal marker of GFR. Clin. Chem. 1997, 6, 913–914. [Google Scholar] [CrossRef]
- Rewa, O.; Bagshaw, S.M. Acute kidney injury—Epidemiology, outcomes and economics. Nat. Rev. Nephrol. 2014, 10, 193–207. [Google Scholar] [CrossRef]
- Vlasakova, K.; Erdos, Z.; Troth, S.P.; McNulty, K.; Chapeau-Campredon, V.; Mokrzycki, N.; Muniappa, N.; Gu, Y.Z.; Holder, D.; Bailey, W.J. Evaluation of the relative performance of 12 urinary biomarkers for renal safety across 22 rat sensitivity and specificity studies. Toxicol. Sci. Off. J. Soc. Toxicol. 2014, 1, 3–20. [Google Scholar] [CrossRef]
- George, B.; Joy, M.S.; Aleksunes, L.M. Urinary protein biomarkers of kidney injury in patients receiving cisplatin chemotherapy. Exp. Biol. Med. 2018, 243, 272–282. [Google Scholar] [CrossRef]
- Katagiri, D.; Hamasaki, Y.; Doi, K.; Negishi, K.; Noiri, E. Interstitial renal fibrosis due to multiple cisplatin treatments is ameliorated by semicarbazide-sensitive amine oxidase inhibition. Kidney Int. 2015, 2, 374–385. [Google Scholar] [CrossRef]
- Liu, X.; Guan, Y.; Xu, S.; Li, Q.; Sun, Y.; Han, R.; Jiang, C. Early predictors of acute kidney injury: A narrative review. Kidney Blood Press. Res. 2016, 5, 680–700. [Google Scholar] [CrossRef]
- Pianta, T.J.; Pickering, J.W.; Succar, L.; Chin, M.; Davidson, T.; Buckley, N.A.; Mohamed, F.; Endre, Z.H. Dexamethasone modifies cystatin c-based diagnosis of acute kidney injury during cisplatin-based chemotherapy. Kidney Blood Press Res. 2017, 1, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.X.; Blaskovich, M.A.; Cooper, M.A. Cell- and biomarker-based assays for predicting nephrotoxicity. Expert Opin. Drug Metab. Toxicol. 2014, 12, 1621–1635. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.Y.; Yang, H.Y.; Chien, H.P.; Tseng, M.H.; Huang, J.L. Urinary clusterin—A novel urinary biomarker associated with pediatric lupus renal histopathologic features and renal survival. Pediatr. Nephrol. 2018, 33, 1189–1198. [Google Scholar] [CrossRef]
- Chapman, A.B.; Devuyst, O.; Eckardt, K.U.; Gansevoort, R.T. Autosomal-dominant polycystic kidney disease (ADPKD): Executive summary from a kidney disease: Improving global outcomes (KDIGO) controversies conference. Kidney Int. 2015, 88, 17–27. [Google Scholar] [CrossRef]
- Odijk, M.; van der Meer, A.D.; Levner, D.; Kim, H.J.; van der Helm, M.W.; Segerink, L.I.; Frimat, J.; Hamilton, G.A.; Ingber, D.E.; van den Berg, A. Measuring direct current trans-epithelial electrical resistance in organ-on-a-chip microsystems. Lab Chip 2015, 3, 745–752. [Google Scholar] [CrossRef]
- Saikumar, J.; Hoffmann, D.; Kim, T.M.; Gonzalez, V.R.; Zhang, Q.; Goering, P.L.; Brown, R.P.; Bijol, V.; Park, P.J.; Waikar, S.S. Expression, circulation, and excretion profile of microRNA-21, -155, and -18a following acute kidney injury. Toxicol. Sci. 2012, 2, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Ewer, M.S.; Ali, M.K.; Mackay, B.; Wallace, S.; Valdivieso, M.; Legha, S.S.; Benjamin, R.S.; Haynie, T.P. A comparison of cardiac biopsy grades and ejection fraction estimations in patients receiving adriamycin. J. Clin. Oncol. 1984, 2, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Wallace, K.B.; Hausner, E.; Herman, E.; Holt, G.D.; MacGregor, J.T.; Metz, A.L.; Murphy, E.; Rosenblum, I.Y.; Sistare, F.D.; York, M.J. Serum troponins as biomarkers of drug-induced cardiac toxicity. Toxicol. Pathol. 2004, 1, 106–121. [Google Scholar] [CrossRef] [PubMed]
- Causey, M.W.; Niten Singh, M.D. Clinical implications of B-type Natriuretic Peptide (BNP) and N-terminal proBNP (NT-proBNP) in the care of the vascular surgery patient. Semin. Vasc. Surg. 2015, 27, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Lyon, A.R. Role of biomarkers in prediction of cardiotoxicity during cancer treatment. Curr. Treat. Options Cardiovasc. Med. 2018, 20, 55. [Google Scholar] [CrossRef] [PubMed]
- Marta, O.P.O.; Gurda, D.; Fedoruk-Wyszomirska, A.; Smolarek, I.; Wyszko, E. Circulating microRNAs in cardiovascular diseases. Acta Biochim. Pol. 2016, 4, 725–729. [Google Scholar]
- Kujala, V.J.; Pasqualini, F.S.; Goss, J.A.; Nawroth, J.C.; Parker, K.K. Laminar ventricular myocardium on a microelectrode array-based chip. J. Mater. Chem. B 2016, 2, 3534–3543. [Google Scholar] [CrossRef]
- Agarwal, A.; Goss, J.A.; Cho, A.; McCain, M.L.; Parker, K.K. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip 2013, 18, 3599. [Google Scholar] [CrossRef]
- Maoz, B.M.; Herl, A.; Henry, O.Y.F.; Leineweber, W.; Yadid, M.; Doyle, J.; Mannix, R.; Kujala, V.; Fitzgerald, E.A.; Parker, K.K. Organs-on-chips with combined multi-electrode array and transepithelial electrical resistance measurement capabilities. Lab Chip 2017, 17, 2294–2302. [Google Scholar] [CrossRef]
- Higuchi, M.; Virginia, M.Y.L.; John, Q.T. Tau and axonopathy in neurodegenerative disorders. Neuromol. Med. 2002, 2, 131–150. [Google Scholar] [CrossRef]
- Siman, R.; Toraskar, N.; Dang, A.; McNeil, E.; McGarvey, M.; Plaum, J.; Maloney, E.; Grady, M.S. A panel of neuron-enriched proteins as markers for traumatic brain injury in humans. J. Neurotrauma 2009, 11, 1867–1877. [Google Scholar] [CrossRef]
- Martinez, B.; Peplow, P.V. MicroRNAs as diagnostic markers and therapeutic targets for traumatic brain injury. Neural Regen. Res. 2017, 12, 1749–1761. [Google Scholar]
- Lescuyer, P.; Allard, L.; Zimmermann-Ivol, C.G.; Burgess, J.A.; Hochstrasser, D.F. Identification of post-mortem cerebrospinal fluid proteins as potential biomarkers of ischemia and neurodegeneration. Proteomics 2004, 8, 2234–2241. [Google Scholar] [CrossRef] [PubMed]
- Salehpoor, F.; Meshkini, A.; Razmgiri, A.; Mahdkhah, A. Prognostic serum factors in patients with traumatic brain injury: A systematic review. Neurosurg. Q. 2016, 1, 19–36. [Google Scholar] [CrossRef]
- Oliver, J.; Abbas, K.; Lightfoot, J.; Baskin, K.; Collins, B.; Wier, D.; Bramhall, J.; Huang, J.; Puschett, J. Comparison of neurocognitive testing and the measurement of marinobufagenin in mild traumatic brain injury: A preliminary report. J. Exp. Neurosci. 2015, 9, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Carr, D.F.; Ayehunie, S.; Davies, A.; Duckworth, C.A.; French, S.; Hall, N.; Hussain, S.; Mellor, H.R.; Norris, A.; Park, B.K.; et al. Towards better models and mechanistic biomarkers for drug-induced gastrointestinal injury. Pharmacol. Ther. 2017, 172, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Liu, Y.; Duan, Y. Breath biomarkers in diagnosis of pulmonary diseases. Clin. Chim. Acta 2012, 21–22, 1770–1780. [Google Scholar] [CrossRef] [PubMed]
- Muller, T. The cellular stress response induced by aqueous extracts of cigarette smoke is critically dependent on the intracellular glutathione concentration. Carcinogenesis 1998, 5, 797–801. [Google Scholar] [CrossRef]
- Stolarek, R.; Bialasiewicz, P.; Krol, M.; Nowak, D. Breath analysis of hydrogen peroxide as a diagnostic tool. Clin. Chim. Acta 2010, 411, 1849–1861. [Google Scholar] [CrossRef]
- Phillips, M.; Cataneo, R.N.; Greenberg, J.; Grodman, R.; Gunawardena, R.; Naidu, A. Effect of oxygen on breath markers of oxidative stress. Eur. Respir. J. 2003, 1, 48–51. [Google Scholar] [CrossRef]
- Henry, O.Y.F.; Villenave, R.; Cronce, M.J.; Leineweber, W.D.; Benz, M.A.; Ingber, D.E. Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (TEER) measurements of human epithelial barrier function. Lab Chip 2017, 13, 2264–2271. [Google Scholar] [CrossRef]
- Hartung, T. Toxicology for the twenty-first century. Nature 2009, 7252, 208–212. [Google Scholar] [CrossRef]
- Han, W.; Wu, Q.; Zhang, X.; Duan, Z. Innovation for hepatotoxicity in vitro research models: A review. J. Appl. Toxicol. 2019, 1, 146–162. [Google Scholar] [CrossRef] [PubMed]
- Bhise, N.S.; Manoharan, V.; Massa, S.; Tamayol, A.; Ghaderi, M.; Miscuglio, M.; Lang, Q.; Shrike, Z.Y.; Shin, S.R.; Calzone, G.; et al. A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication 2016, 8, 14101. [Google Scholar] [CrossRef]
- Zuchowska, A.; Kwapiszewska, K.; Chudy, M.; Dybko, A.; Brzozka, Z. Studies of anticancer drug cytotoxicity based on long-term HepG2 spheroid culture in a microfluidic system. Electrophoresis 2017, 8, 1206–1216. [Google Scholar] [CrossRef] [PubMed]
- Kostadinova, R.; Boess, F.; Applegate, D.; Suter, L.; Weiser, T.; Singer, T.; Naughton, B.; Roth, A. A long-term three dimensional liver co-culture system for improved prediction of clinically relevant drug-induced hepatotoxicity. Toxicol. Appl. Pharm. 2013, 1, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Zhao, L.; Zhou, E.; Xu, J.; Shen, S.; Wang, J. On-chip construction of liver lobule-like microtissue and its application for adverse drug reaction assay. Anal. Chem. 2016, 3, 1719–1727. [Google Scholar] [CrossRef]
- Jang, K.; Otieno, M.A.; Ronxhi, J.; Lim, H.; Ewart, L.; Kodella, K.R.; Petropolis, D.B.; Kulkarni, G.; Rubins, J.E.; Conegliano, D.; et al. Reproducing human and cross-species drug toxicities using a liver-chip. Sci. Transl. Med. 2019, 517, x5516. [Google Scholar] [CrossRef]
- Yu, F.; Deng, R.; Hao Tong, W.; Huan, L.; Chan Way, N.; IslamBadhan, A.; Iliescu, C.; Yu, H. A perfusion incubator liver chip for 3D cell culture with application on chronic hepatotoxicity testing. Sci. Rep. 2017, 7, 14528. [Google Scholar] [CrossRef]
- Bavli, D.; Prill, S.; Ezra, E.; Levy, G.; Cohen, M.; Vinken, M.; Vanfleteren, J.; Jaeger, M.; Nahmias, Y. Real-time monitoring of metabolic function in liver-on-chip microdevices tracks the dynamics of mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA 2016, 16, E2231–E2240. [Google Scholar] [CrossRef]
- Massa, S.; Sakr, M.A.; Seo, J.; Bandaru, P.; Arneri, A.; Bersini, S.; Zare-Eelanjegh, E.; Jalilian, E.; Cha, B.; Antona, S.; et al. Bioprinted 3D vascularized tissue model for drug toxicity analysis. BiomicrofluIdics 2017, 4, 44109. [Google Scholar] [CrossRef]
- Deng, J.; Zhang, X.; Chen, Z.; Luo, Y.; Lu, Y.; Liu, T.; Wu, Z.; Jin, Y.; Zhao, W.; Lin, B. A cell lines derived microfluidic liver model for investigation of hepatotoxicity induced by drug-drug interaction. Biomicrofluidic 2019, 2, 24101. [Google Scholar] [CrossRef]
- Jang, M.; Neuzil, P.; Volk, T.; Manz, A.; Kleber, A. On-chip three-dimensional cell culture in phaseguides improves hepatocyte functionsin vitro. Biomicrofluidic 2015, 3, 34113. [Google Scholar] [CrossRef] [PubMed]
- Riahi, R.; Shaegh, S.A.M.; Ghaderi, M.; Zhang, Y.S.; Shin, S.R.; Aleman, J.; Massa, S.; Kim, D.; Dokmeci, M.R.; Khademhosseini, A. Automated microfluidic platform of bead-based electrochemical immunosensor integrated with bioreactor for continual monitoring of cell secreted biomarkers. Sci. Rep. 2016, 6, 24598. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.J.; Lee, D.W.; Shah, D.A.; Ku, B.; Jeon, S.Y.; Solanki, K.; Ryan, J.D.; Clark, D.S.; Dordick, J.S.; Lee, M. High-throughput and combinatorial gene expression on a chip for metabolism-induced toxicology screening. Nat. Commun. 2014, 5, 3739. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.G.; Funk, J.; Robbins, J.B.; Crogan-Grundy, C.; Presnell, S.C.; Singer, T.; Roth, A.B. Bioprinted 3D primary liver tissues allow assessment of organ-level response to clinical drug induced toxicity in vitro. PLoS ONE 2016, 7, e158674. [Google Scholar] [CrossRef]
- Prior, N.; Inacio, P.; Huch, M. Liver organoids: From basic research to therapeutic applications. Gut 2019, 68, 2228–2237. [Google Scholar] [CrossRef]
- Shen, T.; Liu, Y.; Shang, J.; Xie, Q.; Li, J.; Yan, M.; Xu, J.; Niu, J.; Liu, J.; Watkins, P.B.; et al. Incidence and Etiology of Drug-Induced Liver Injury in Mainland China. Gastroenterology 2019, 156, 2230–2241.e11. [Google Scholar] [CrossRef]
- Jang, K.; Mehr, A.P.; Hamilton, G.A.; McPartlin, L.A.; Chung, S.; Suh, K.; Ingber, D.E. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr. Biol. 2013, 9, 1119–1129. [Google Scholar] [CrossRef]
- Snouber, L.C.; Letourneur, F.; Chafey, P.; Broussard, C.; Monge, M.; Legallais, C.; Leclerc, E. Analysis of transcriptomic and proteomic profiles demonstrates improved madin-darby canine kidney cell function in a renal microfluidic biochip. Biotechnol. Prog. 2012, 2, 474–484. [Google Scholar] [CrossRef]
- Ferrell, N.; Ricci, K.B.; Groszek, J.; Marmerstein, J.T.; Fissell, W.H. Albumin handling by renal tubular epithelial cells in a microfluidic bioreactor. Biotechnol. Bioeng. 2012, 3, 797–803. [Google Scholar] [CrossRef]
- Kim, S.; LesherPerez, S.C.; Kim, B.C.C.; Yamanishi, C.; Labuz, J.M.; Leung, B.; Takayama, S. Pharmacokinetic profile that reduces nephrotoxicity of gentamicin in a perfused kidney-on-a-chip. Biofabrication 2016, 1, 15021. [Google Scholar] [CrossRef]
- Sakolish, C.M.; Mahler, G.J. A novel microfluidic device to model the human proximal tubule and glomerulus. RSC Adv. 2017, 8, 4216–4225. [Google Scholar] [CrossRef]
- Sakolish, C.M.; Philip, B.; Mahler, G.J. A human proximal tubule-on-a-chip to study renal disease and toxicity. Biomicrofluidics 2019, 1, 14107. [Google Scholar] [CrossRef] [PubMed]
- Weber, E.J.; Lidberg, K.A.; Wang, L.; Bammler, T.K.; MacDonald, J.W.; Li, M.J.; Redhair, M.; Atkins, W.M.; Tran, C.; Hines, K.M.; et al. Human kidney on a chip assessment of polymyxin antibiotic nephrotoxicity. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tao, T.; Su, W.; Yu, H.; Yu, Y.; Qin, J. A disease model of diabetic nephropathy in a glomerulus-on-a-chip microdevice. Lab Chip 2017, 10, 1749–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jiang, L.; Tao, T.; Su, W.; Guo, Y.; Yu, H.; Qin, J. Assessment of cadmium-induced nephrotoxicity using a kidney-on-a-chip device. Toxicol. Res. 2017, 3, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; An, F.; Luo, Y.; Lu, Y.; Liu, T.; Zhao, W.; Lin, B. A nephron model for study of drug-induced acute kidney injury and assessment of drug-induced nephrotoxicity. Biomaterials 2018, 155, 41–53. [Google Scholar] [CrossRef]
- Brana, I.; Tabernero, J. Cardiotoxicity. Ann. Oncol. 2010, 7, i173–i179. [Google Scholar] [CrossRef]
- Chung, R.; Ghosh, A.K.; Banerjee, A. Cardiotoxicity: Precision medicine with imprecise definitions. Open Heart 2018, 2, e774. [Google Scholar] [CrossRef]
- Rajasingh, S.; Isai, D.G.; Samanta, S.; Zhou, Z.; Dawn, B.; Kinsey, W.H.; Czirok, A.; Rajasingh, J. Manipulation-free cultures of human iPSC-derived cardiomyocytes offer a novel screening method for cardiotoxicity. Acta Pharm. Sin. 2018, 10, 1590–1603. [Google Scholar] [CrossRef]
- Doherty, K.R.; Talbert, D.R.; Trusk, P.B.; Moran, D.M.; Shell, S.A.; Bacus, S. Structural and functional screening in human induced-pluripotent stem cell-derived cardiomyocytes accurately identifies cardiotoxicity of multiple drug types. Toxicol. Appl. Pharm. 2015, 1, 51–60. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, X. Electrical assisted patterning of cardiac myocytes with controlled macroscopic anisotropy using a microfluidic dielectrophoresis chip. Sens. Actuators A Phys. 2007, 1, 73–79. [Google Scholar] [CrossRef]
- Qian, F.; Huang, C.; Lin, Y.D.; Ivanovskaya, A.N.; O’Hara, T.J.; Booth, R.H.; Creek, C.J.; Enright, H.A.; Soscia, D.A.; Belle, A.M. Simultaneous electrical recording of cardiac electrophysiology and contraction on chip. Lab Chip 2017, 10, 10–1039. [Google Scholar] [CrossRef] [PubMed]
- Caluori, G.; Pribyl, J.; Pesl, M.; Jelinkova, S.; Rotrekl, V. Non-invasive electromechanical cell-based biosensors for improved investigation of 3D cardiac models. Biosens. Bioelectron. 2019, 124–125, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Marsano, A.; Conficconi, C.; Lemme, M.; Occhetta, P.; Gaudiello, E.; Votta, E.; Cerino, G.; Redaelli, A.; Rasponi, M. Beating heart on a chip: A novel microfluidic platform to generate functional 3D cardiac microtissues. Lab Chip 2016, 3, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Nunes, S.S.; Miklas, J.W.; Liu, J.; Aschar-Sobbi, R.; Xiao, Y.; Zhang, B.; Jiang, J.; Massé, S.; Gagliardi, M.; Hsieh, A.; et al. Biowire: A platform for maturation of human pluripotent stem cell—derived cardiomyocytes. Nat. Methods 2013, 8, 781–787. [Google Scholar] [CrossRef]
- Zhao, Y.; Rafatian, N.; Feric, N.T.; Cox, B.J.; Radisic, M. A platform for generation of chamber-specific cardiac tissues and disease modeling. Cell 2019, 176, 913–927.e18. [Google Scholar] [CrossRef]
- Ellis, B.W.; Acun, A.; Can, U.I.; Zorlutuna, P. Human iPSC-derived myocardium-on-chip with capillary-like flow for personalized medicine. Biomicrofluidics 2017, 2, 845–856. [Google Scholar] [CrossRef]
- Kamei, K.I.; Kato, Y.; Hirai, Y.; Ito, S.; Satoh, J.; Oka, A.; Tsuchiya, T.; Yong, C.; Tabata, O. Integrated heart/cancer on a chip to reproduce the side effects of anti-cancer drugs in vitro. RSC Adv. 2017, 58, 36777–36786. [Google Scholar] [CrossRef]
- Beulig, R.J.; Warias, R.; Heiland, J.J.; Ohla, S.; Zeitler, K.; Belder, D. A droplet-chip/mass spectrometry approach to study organic synthesis at nanoliter scale. Lab Chip 2017, 17, 1996–2002. [Google Scholar] [CrossRef]
- Maschmeyer, I.; Lorenz, A.K.; Schimek, K.; Hasenberg, T.; Ramme, A.P.; Hubner, J.; Lindner, M.; Drewell, C.; Bauer, S.; Thomas, A.; et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 2015, 12, 2688–2699. [Google Scholar] [CrossRef]
- Maher, M.; Wright, J.; Pine, J.; Tai, Y. A Microstructure for Interfacing with Neurons: The Neurochip; IEEE: Piscataway, NJ, USA, 1998; pp. 1698–1702. [Google Scholar]
- Service, R.F. Neurons and silicon get intimate. Science 1999, 5414, 578–579. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.W.; Cui, F.Z.; Chen, L.N.; Zhai, Y.; Xu, Q.Y. Improvement of neural cell adherence to silicon surface by hydroxyl ion implantation. Surf. Coat. Technol. 2000, 1, 355–359. [Google Scholar] [CrossRef]
- Liu, Q.; Cai, H.; Xu, Y.; Li, Y.; Li, R.; Wang, P. Olfactory cell-based biosensor: A first step towards a neurochip of bioelectronic nose. Biosens. Bioelectron. 2006, 2, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.; Giraldo-Vela, J.P.; Nathamgari, S.S.; McGuire, T.; McNaughton, R.L.; Kessler, J.A.; Espinosa, H.D. Microfluidic device for stem cell differentiation and localized electroporation of postmitotic neurons. Lab Chip 2014, 23, 4486–4495. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, B.Z.; Lehmann, M.; Gutbier, S.; Nembo, E.; Noel, S.; Smirnova, L.; Forsby, A.; Hescheler, J.; Avci, H.X. Thomas Hartung. In vitro acute and developmental neurotoxicity screening an overview of cellural platforms and high-throughput technical possibilities. Arch. Toxicol. 2017, 91, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.C.; Yang, Z. Study on the changes of the electrical activity of neural networks by nanomaterials based on the microelectrode array neural chip. Tianjin Med. J. 2011, 39, 135. [Google Scholar]
- Nierode, G.J.; Perea, B.C.; McFarland, S.K.; Pascoal, J.F.; Clark, D.S.; Schaffer, D.V.; Dordick, J.S. High-throughput toxicity and phenotypic screening of 3D human neural progenitor cell cultures on a microarray chip platform. Stem Cell Rep. 2016, 5, 970–982. [Google Scholar] [CrossRef]
- Kafi, M.A.; Cho, H.Y.; Choi, J.W. Neural cell chip based electrochemical detection of nanotoxicity. Nanomaterials 2015, 3, 1181–1199. [Google Scholar] [CrossRef]
- Qu, Z.; Lv, J.J.; Zang, S.; Geng, X.C.; Li, B.; Zhao, D.M. Study on neurotoxicity model of SD rat neural stem cell evaluation drug. China Pharm. Aff. 2018, 32, 1079–1087. [Google Scholar]
- Zheng, X.N. Study on Cytotoxicity of Quantum Dots based on Cell Chip Platform. Master’s Thesis, Fudan University, Shanghai, China, 2010. [Google Scholar]
- Wang, Y.; Wang, L.; Zhu, Y.; Qin, J. Human brain organoid-on-a-chip to model prenatal nicotine exposure. Lab Chip 2018, 6, 851–860. [Google Scholar] [CrossRef]
- Hardy, J.; Selkoe, D.J. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 2002, 5580, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, B.K.; Jeong, G.S.; Hyun, J.K.; Lee, C.J.; Lee, S. Three-dimensional brain-on-a-chip with an interstitial level of flow and its application as an In vitro model of Alzheimer’s disease. Lab Chip 2015, 1, 141–150. [Google Scholar] [CrossRef]
- Tang, Y.; Qiu, Q.; Zhang, F.; Xie, M.; Huang, W. Quantifying orientational regeneration of injured neurons by natural product concentration gradients in a 3D microfluidic device. Lab Chip 2018, 6, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Pabst, R. The anatomical basis for the immune function of the gut. Anat. Embryol. 1987, 2, 135–144. [Google Scholar] [CrossRef] [PubMed]
- McConnell, E.L.; Fadda, H.M.; Basit, A.W. Gut instincts: Explorations in intestinal physiology and drug delivery. Int. J. Pharm. 2008, 2, 213–226. [Google Scholar] [CrossRef]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012, 12, 2165. [Google Scholar] [CrossRef]
- Kim, H.J.; Ingber, D.E. Gut-on-a-chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. Quant. Biosci. Nano Macro 2013, 9, 1130. [Google Scholar] [CrossRef]
- Kim, H.J.; Li, H.; Collins, J.J.; Ingber, D.E. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. USA 2016, 1, E7–E15. [Google Scholar] [CrossRef]
- Guo, Y.; Li, Z.; Su, W.; Wang, L.; Zhu, Y.; Qin, J. A biomimetic human gut-on-a-chip for modeling drug metabolism in intestine. Artif. Organs 2018, 42, 1196–1205. [Google Scholar] [CrossRef]
- Beaurivage, C.; Naumovska, E.; Chang, Y.; Elstak, E.; Nicolas, A.; Wouters, H.; van Moolenbroek, G.; Lanz, H.; Trietsch, S.; Joore, J.; et al. Development of a gut-on-a-chip model for high throughput disease modeling and drug discovery. Int. J. Mol. Sci. 2019, 22, 5661. [Google Scholar] [CrossRef]
- De Haan, P.; Ianovska, M.A.; Mathwig, K.; van Lieshout, G.; Triantis, V.; Bouwmeester, H.; Verpoorte, E. Digestion-on-a-chip: A continuous-flow modular microsystem recreating enzymatic digestion in the gastrointestinal tract. Lab Chip 2019, 9, 1599–1609. [Google Scholar] [CrossRef] [PubMed]
- Delaunois, L.M. Mechanisms in pulmonary toxicology. Clin. Chest Med. 2004, 1, 1–14. [Google Scholar] [CrossRef]
- Zhang, M.; Xu, C.; Jiang, L.; Qin, J. A 3D human lung-on-a-chip model for nanotoxicity testing. Toxicol. Res. 2018, 7, 1048–1060. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting organ-level lung functions on a chip. Science 2010, 5986, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Leslie, D.C.; Matthews, B.D.; Fraser, J.P.; Jurek, S.; Hamilton, G.A.; Thorneloe, K.S.; McAlexander, M.A.; Ingber, D.E. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci. Transl. Med. 2012, 159, 159ra147. [Google Scholar] [CrossRef]
- Yang, X.; Li, K.; Zhang, X.; Liu, C.; Guo, B.; Wen, W.; Gao, X. Nanofiber membrane supported lung-on-a-chip microdevice for anti-cancer drug testing. Lab Chip 2018, 18, 486–495. [Google Scholar] [CrossRef]
- Felder, M.; Trueeb, B.; Stucki, A.O.; Borcard, S.; Stucki, J.D.; Schnyder, B.; Geiser, T.; Guenat, O.T. Impaired wound healing of alveolar lung epithelial cells in a breathing lung-on-a-chip. Front. Bioeng. Biotechnol. 2019, 7, 3. [Google Scholar] [CrossRef]
- Palmiotti, C.A.; Prasad, S.; Naik, P.; Abul, K.M.D.; Sajja, R.K.; Achyuta, A.H.; Cucullo, L. In vitro cerebrovascular modeling in the 21st century: Current and prospective technologies. Pharm. Res. 2014, 12, 3229–3250. [Google Scholar] [CrossRef]
- Nicolazzo, J.A.; Charman, S.A.; Charman, W.N. Methods to assess drug permeability across the blood-brain barrier. J. Pharm. Pharm. 2006, 3, 281–293. [Google Scholar] [CrossRef]
- Nakagawa, S.; Deli, M.A.; Kawaguchi, H.; Shimizudani, T.; Shimono, T.; Kittel, Á.; Tanaka, K.; Niwa, M. A new blood—Brain barrier model using primary rat brain endothelial cells, pericytes and astrocytes. Neurochem. Int. 2009, 3–4, 253–263. [Google Scholar] [CrossRef]
- Booth, R.; Kim, H. Characterization of a microfluidic In vitro model of the blood-brain barrier (muBBB). Lab Chip 2012, 10, 1784–1792. [Google Scholar] [CrossRef] [PubMed]
- Vatine, G.; Barrile, R.; Workman, M.; Chen, Z.; Eyk, J.; Svendsen, C. Human iPSC-derived blood-brain barrier chips enable disease modeling and personalized medicine applications. Cell Stem Cell 2019, 24, 995–1005.e6. [Google Scholar] [CrossRef] [PubMed]
- Wufuer, M.; Lee, G.; Hur, W.; Jeon, B.; Kim, B.J.; Choi, T.H.; Lee, S. Skin-on-a-chip model simulating inflammation, edema and drug-based treatment. Sci. Rep. 2016, 6, 37471. [Google Scholar] [CrossRef] [PubMed]
- Menon, N.V.; Tay, H.M.; Wee, S.N.; Li, K.H.H.; Hou, H.W. Micro-engineered perfusable 3D vasculatures for cardiovascular diseases. Lab Chip 2017, 17, 2960–2968. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.; Ma, X.; Luo, Y.; Fang, S.; Xie, Z.; Li, X.; Qi, D.; Zhang, F.; Kong, J.; Li, J.; et al. Application of a microfluidic-based perivascular tumor model for testing drug sensitivity in head and neck cancers and toxicity in endothelium. RSC Adv. 2016, 35, 29598–29607. [Google Scholar] [CrossRef]
- Ronaldson-Bouchard, K.; Vunjak-Novakovic, G. Organs-on-a-chip: A fast track for engineered human tissues in drug development. Cell Stem Cell 2018, 3, 310–324. [Google Scholar] [CrossRef]
- Chang, S.; Weber, E.J.; Sidorenko, V.S.; Chapron, A.; Yeung, C.K.; Gao, C.; Mao, Q.; Shen, D.; Wang, J.; Rosenquist, T.A.; et al. Human liver-kidney model elucidates the mechanisms of aristolochic acid nephrotoxicity. JCI Insight 2017, 22. [Google Scholar] [CrossRef]
- Theobald, J.; Ghanem, A.; Wallisch, P.; Banaeiyan, A.A.; Andrade-Navarro, M.A.; Taskova, K.; Haltmeier, M.; Kurtz, A.; Becker, H.; Reuter, S.; et al. Liver-kidney-on-chip to study toxicity of drug metabolites. ACS Biomater. Sci. Eng. 2018, 1, 78–89. [Google Scholar] [CrossRef]
- Theobald, J.; Abu El Maaty, M.A.; Kusterer, N.; Wetterauer, B.; Wink, M.; Cheng, X.; Wölfl, S. In vitro metabolic activation of vitamin D3 by using a multi-compartment microfluidic liver-kidney organ on chip platform. Sci. Rep. 2019, 9, 4616. [Google Scholar] [CrossRef]
- Sung, J.H.; Shuler, M.L. A micro cell culture analog (µCCA) with 3-D hydrogel culture of multiple cell lines to assess metabolism-dependent cytotoxicity of anti-cancer drugs. Lab Chip 2009, 10, 1385. [Google Scholar] [CrossRef]
- Oleaga, C.; Bernabini, C.; Smith, A.S.T.; Srinivasan, B.; Jackson, M.; McLamb, W.; Platt, V.; Bridges, R.; Cai, Y.; Santhanam, N.; et al. Multi-organ toxicity demonstration in a functional human In vitro system composed of four organs. Sci. Rep. 2016, 6, 20030. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Guo, Y.; Yu, Y.; Xu, C.; Xu, H.; Qin, J. Assessment of metabolism-dependent drug efficacy and toxicity on a multilayer organs-on-a-chip. Integr. Biol. 2016, 10, 1022–1029. [Google Scholar] [CrossRef] [PubMed]
- McAleer, C.W.; Pointon, A.; Long, C.J.; Brighton, R.L.; Wilkin, B.D.; Bridges, L.R.; Narasimhan Sriram, N.; Fabre, K.; McDougall, R.; Muse, V.P.; et al. On the potential of In vitro organ-chip models to define temporal pharmacokinetic-pharmacodynamic relationships. Sci. Rep. 2019, 9, 9619. [Google Scholar] [CrossRef] [PubMed]
- Ishida, S. Organs-on-a-chip: Current applications and consideration points for In vitro ADME-Tox studies. Drug Metab Pharm. 2018, 1, 49–54. [Google Scholar] [CrossRef]
- Pamies, D.; Price, A.; Chesne, C.; Coecke, S.; Dinnyes, A.; Eskes, C.; Grillari, R.; Hartung, T.; Leist, M.; Daneshian, M. Advanced good cell culture practice for human primary, stem cell-derived and organoid models as well as microphysiological systems. ALTEX 2018, 35, 353–378. [Google Scholar] [CrossRef]

| Toxicity | Biomarker | Reference | Specification | Whether Utilized with Organ Chips |
|---|---|---|---|---|
| Hepatotoxicity | ALT (alanine aminotransferase) AST (aspartate aminotransferase) | [19] | Diagnostic marker of liver damage | √ |
| ALP (alkalinephosphatase) | [19] | Diagnostic marker of cholestatic injury | √ | |
| GLDH (glutamate dehydrogenase) | [20] | Early detection biomarker | × | |
| HMGB-1 (high-mobility group box 1) | [21] | Early detection biomarker | × | |
| K18 (keratin-18) | [22] | Early detection biomarker | √ | |
| OCT (ornithine carbamoyltransferase) | [23] | Early detection biomarker | × | |
| GST-α (glutathione S-transferase α) | [24] | Early detection biomarker | √ | |
| CYP (cytochrome P450) | [25] | Metabolic ability biomarke | √ | |
| miRNA-122, miRNA-192 | [26] | Genomic markers | √ | |
| CDH-5 (cadherin-5) | [27] | Proteomics biomarker | √ | |
| FABP1 (fatty acid binding protein 1) | [27] | Proteomics biomarker | × | |
| nephrotoxicity | GFR (glomerular filtration rate) | [28] | Diagnostic marker of renal function | √ |
| SCr levels and urine output | [29] | Diagnostic marker of AKI (acute kidney injury) | √ | |
| KIM-1 (kidney injury molecule-1) | [30] | Early detection biomarker | √ | |
| NAG (N-acetyl-β-glucosaminidase) | [30] | Early detection biomarker | √ | |
| NGAL (neutrophil gelatinase-associated lipocalin) | [31] | Early detection biomarker | √ | |
| L-FABP (liver type fatty acid binding protein) | [32] | Early detection biomarker | × | |
| MCP-1 (monocyte chemotactic peptide-1) | [33] | Early detection biomarker | × | |
| CYs C (cystatin C) | [34] | Early detection biomarker | √ | |
| OPN (osteopontin) | [35] | Early detection biomarker | × | |
| CLU (clusterin) | [36] | Early detection biomarker | × | |
| TFF3 (trefoil factor 3) | [37] | Early detection biomarker | × | |
| TEER (transendothelial resistance) | [38] | Biomarker of barrier functions | √ | |
| miRNAs | [39] | Genomic markers | √ | |
| cardiotoxicity | LVDP (left ventricular formation pressure) LVSP (left ventricular systolic pressure) | [40] | Diagnostic marker of myocardial injury | √ |
| TnI (troponin I) TnT (troponin T) | [41] | Early detection biomarker | × | |
| BNP (brain natriuretic peptide) NT-proBNP | [42] | Early detection biomarker | × | |
| MPO (myeloperoxidase) | [43] | Biomarker of oxidative stress | × | |
| miR-146a, miR-1, miR-133, miR-208, miR-499 | [44] | Genomic markers | √ | |
| beating frequency, systolic stress, field potential | [45,46] | Mechanical markers | √ | |
| TEER | [47] | Biomarker of barrier functions | √ | |
| neurotoxicity | plasma P-Tau (phosphorylated-Tau) T-Tau (total Tau) | [48] | Diagnostic marker of central nervous system (CNS) injury | × |
| NF-H (neurofilaments heavy subunit) | [49] | Diagnostic marker of axonal injury | × | |
| miR-425-p, miR-21, miR-93, miR-191, miR-499,miR-328, miR-362-3p, miR-451, miR-486a | [50] | Genomic markers | √ | |
| H-FABP (heart fatty acid binding protein) | [51] | Diagnostic marker of CNS injury | × | |
| SP, sCD40L, TIMP-1, MDA, CK-18 | [52] | Early detection biomarkers | √ | |
| MBG (marinobufagenin) | [53] | Biomarker of neuro-inflammation | × | |
| other toxicities | Ghrelin | [54] | Biomarker of stomach/small intestine injury | × |
| DAO (diamine oxidase) Citrulline | [54] | Biomarker of small intestine injury | × | |
| CD64, C-reactive protein | [54] | Biomarker of small/large intestine | √ | |
| NOS isoenzymes | [55] | Breath biomarkers | √ | |
| HO (heme oxygenase) | [56] | Biomarker of upper respiratory tract viral infections | √ | |
| CYP (cytochrome P450) H2O2 | [57] | Biomarker of pulmonary diseases | √ | |
| Breath methylated hydrocarbons | [58] | Lipid peroxidation markers | × | |
| TEER | [59] | Biomarker of barrier functions | √ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cong, Y.; Han, X.; Wang, Y.; Chen, Z.; Lu, Y.; Liu, T.; Wu, Z.; Jin, Y.; Luo, Y.; Zhang, X. Drug Toxicity Evaluation Based on Organ-on-a-chip Technology: A Review. Micromachines 2020, 11, 381. https://doi.org/10.3390/mi11040381
Cong Y, Han X, Wang Y, Chen Z, Lu Y, Liu T, Wu Z, Jin Y, Luo Y, Zhang X. Drug Toxicity Evaluation Based on Organ-on-a-chip Technology: A Review. Micromachines. 2020; 11(4):381. https://doi.org/10.3390/mi11040381
Chicago/Turabian StyleCong, Ye, Xiahe Han, Youping Wang, Zongzheng Chen, Yao Lu, Tingjiao Liu, Zhengzhi Wu, Yu Jin, Yong Luo, and Xiuli Zhang. 2020. "Drug Toxicity Evaluation Based on Organ-on-a-chip Technology: A Review" Micromachines 11, no. 4: 381. https://doi.org/10.3390/mi11040381
APA StyleCong, Y., Han, X., Wang, Y., Chen, Z., Lu, Y., Liu, T., Wu, Z., Jin, Y., Luo, Y., & Zhang, X. (2020). Drug Toxicity Evaluation Based on Organ-on-a-chip Technology: A Review. Micromachines, 11(4), 381. https://doi.org/10.3390/mi11040381





