Applications of CMOS Devices for the Diagnosis and Control of Infectious Diseases
Abstract
:1. Introduction
2. Optical Techniques
2.1. Luminescence Imaging for In-Vitro Diagnosis
2.2. Label-Free Imaging for In-Vitro Diagnosis
2.3. Contactless Monitoring of Diseases’ Symptoms
3. Electrochemical Techniques
3.1. Biological/Chemical Field-Effect Transistors (FETs)
3.2. Impedimetric, Capacitive and Conductometric Techniques
3.3. Other Electrochemical Techniques
4. Other Techniques
4.1. Magnetic Sensors
4.2. Mechanical Sensors
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Aytur, T.S. A CMOS Biosensor for Infectious Disease Detection; University of California: Berkeley, CA, USA, 2007. [Google Scholar]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Qi, Q.; Jing, Q.; Ao, S.; Zhang, Z.; Ding, M.; Wu, M.; Liu, K.; Wang, W.; Ling, Y. Electrical probing of COVID-19 spike protein receptor binding domain via a graphene field-effect transistor. arXiv 2020, arXiv:2003.12529. [Google Scholar]
- Cardean Transistors™ Made Available to Companies and Government Agencies Willing to Build Handheld Coronavirus Detection Devices. Available online: https://www.technologynetworks.com/diagnostics/product-news/cardean-transistors-made-available-to-companies-and-government-agencies-willing-to-build-handheld-332759 (accessed on 31 March 2020).
- Diagnostics with Molecular Scissors – Is This also Possible for on-site COVID-19 Tests? Available online: https://www.gesundheitsindustrie-bw.de/en/article/news/Diagnostics-with-molecular-scissors-is-this-also-possible-for-on-site-COVID-19-tests (accessed on 3 April 2020).
- Hajian, R.; Balderston, S.; Tran, T.; DeBoer, T.; Etienne, J.; Sandhu, M.; Wauford, N.A.; Chung, J.-Y.; Nokes, J.; Athaiya, M. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 2019, 3, 427–437. [Google Scholar] [CrossRef]
- Bruch, R.; Baaske, J.; Chatelle, C.; Meirich, M.; Madlener, S.; Weber, W.; Dincer, C.; Urban, G.A. CRISPR/Cas13a-powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics. Adv. Mater. 2019, 31, 1905311. [Google Scholar] [CrossRef] [Green Version]
- Mahari, S.; Roberts, A.; Shahdeo, D.; Gandhi, S. eCovSens-Ultrasensitive Novel In-House Built Printed Circuit Board Based Electrochemical Device for Rapid Detection of nCovid-19 antigen, a spike protein domain 1 of SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano 2020, 14, 5268–5277. [Google Scholar] [CrossRef] [Green Version]
- A Biosensor for the COVID-19 Virus. Available online: https://tectales.com/wearables-sensors/a-biosensor-for-the-covid-19-virus.html (accessed on 24 April 2020).
- The HEMEMICS Platform Empowers Healthcare Providers to Rapidly Identify Infectious Disease Pathogens in 60 Seconds or Less and Generate Real-time Alerts to an Outbreak. Available online: https://hememics.com/ (accessed on 16 April 2020).
- HEMEMICS Biotechnologies, Inc. Receives HHS Support to Develop Rapid Antigen, Antibody Diagnostic to Identify COVID-19 Infected Americans. Available online: https://www.newswise.com/coronavirus/hememics-biotechnologies-inc-receives-hhs-support-to-develop-rapid-antigen-antibody-diagnostic-to-identify-covid-19-infected-americans/?article_id=730032 (accessed on 16 April 2020).
- Roswell Biotechnologies and Imec to Develop First Molecular Electronics Biosensor Chips for Infectious Disease Surveillance, Precision Medicine and DNA Storage. Available online: https://www.roswellbiotech.com/wp-content/uploads/2020/05/roswell-biotechnologies-and-imec-to-develop-first-molecular-electronics-biosensor-chips-for-infectious-disease.pdf (accessed on 5 May 2020).
- Moran, A.; Beavis, K.G.; Matushek, S.M.; Ciaglia, C.; Francois, N.; Tesic, V.; Love, N. The detection of SARS-CoV-2 using the cepheid xpert xpress SARS-CoV-2 and Roche cobas SARS-CoV-2 assays. J. Clin. Microbiol. 2020. [Google Scholar] [CrossRef] [Green Version]
- 4 Fast-track Covid-19 Diagnostic Tests. Available online: https://www.eetimes.eu/4-fast-track-covid-19-diagnostic-tests/ (accessed on 31 March 2020).
- Jeong, H.; Rogers, J.A.; Xu, S. Continuous on-body sensing for the COVID-19 pandemic: Gaps and opportunities. Sci. Adv. 2020, 6, eabd4794. [Google Scholar] [CrossRef]
- Monitoring COVID-19 from Hospital to Home: First Wearable Device Continuously Tracks Key Symptoms. Available online: https://news.northwestern.edu/stories/2020/04/monitoring-covid-19-from-hospital-to-home-first-wearable-device-continuously-tracks-key-symptoms/ (accessed on 4 May 2020).
- Wong, C.K.; Ho, D.T.Y.; Tam, A.R.; Zhou, M.; Lau, Y.M.; Tang, M.O.Y.; Tong, R.C.F.; Rajput, K.S.; Chen, G.; Chan, S.C. Artificial intelligence mobile health platform for early detection of COVID-19 in quarantine subjects using a wearable biosensor: protocol for a randomised controlled trial. BMJ Open 2020, 10, e038555. [Google Scholar] [CrossRef]
- Zhao, H.; Liu, F.; Xie, W.; Zhou, T.-C.; OuYang, J.; Jin, L.; Li, H.; Zhao, C.-Y.; Zhang, L.; Wei, J. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sens. Actuators B Chem. 2020, 128899. [Google Scholar]
- Mao, K.; Zhang, H.; Yang, Z. The potential of an integrated biosensor system with mobile health and wastewater-based epidemiology (iBMW) for the prevention, surveillance, monitoring and intervention of the COVID-19 pandemic. Biosens. Bioelectron. 2020, 112617. [Google Scholar] [CrossRef] [PubMed]
- Chandra, P. Miniaturized label-free smartphone assisted electrochemical sensing approach for personalized COVID-19 diagnosis. Sens. Int. 2020, 1, 100019. [Google Scholar] [CrossRef]
- Alvarez, A. Introduction to BiCMOS. In BiCMOS Technology and Applications; Springer: New York, NY, USA, 1990; pp. 1–20. [Google Scholar]
- Nishi, Y. Comparison of new technologies for VLSI: Possibilities and limitations. Microelectron. J. 1981, 12, 5–14. [Google Scholar] [CrossRef]
- Gamo, K.; Nakazato, K.; Niitsu, K. A current-integration-based CMOS amperometric sensor with 1024× 1024 bacteria-sized microelectrode array for high-sensitivity bacteria counting. IEICE Trans. Electron. 2017, 100, 602–606. [Google Scholar] [CrossRef]
- Jeon, J.-W.; Kim, J.-H.; Lee, J.-M.; Lee, W.-H.; Lee, D.-Y.; Paek, S.-H. Rapid immuno-analytical system physically integrated with lens-free CMOS image sensor for food-borne pathogens. Biosens. Bioelectron. 2014, 52, 384–390. [Google Scholar] [CrossRef]
- Ghafar-Zadeh, E.; Sawan, M.; Chodavarapu, V.P.; Hosseini-Nia, T. Bacteria growth monitoring through a differential CMOS capacitive sensor. IEEE Trans. Biomed. Circuits Syst. 2010, 4, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Devadhasan, J.P.; Lee, D.Y.; Kim, S. Real-time DNA amplification and detection system based on a CMOS image sensor. Anal. Sci. 2016, 32, 653–658. [Google Scholar] [CrossRef] [Green Version]
- Norian, H.; Field, R.M.; Kymissis, I.; Shepard, K.L. An integrated CMOS quantitative-polymerase-chain-reaction lab-on-chip for point-of-care diagnostics. Lab Chip 2014, 14, 4076–4084. [Google Scholar] [CrossRef]
- Van Dorst, B.; Brivio, M.; Van Der Sar, E.; Blom, M.; Reuvekamp, S.; Tanzi, S.; Groenhuis, R.; Adojutelegan, A.; Lous, E.-J.; Frederix, F. Integration of an optical CMOS sensor with a microfluidic channel allows a sensitive readout for biological assays in point-of-care tests. Biosens. Bioelectron. 2016, 78, 126–131. [Google Scholar] [CrossRef]
- Sun, G.; Nakayama, Y.; Dagdanpurev, S.; Abe, S.; Nishimura, H.; Kirimoto, T.; Matsui, T. Remote sensing of multiple vital signs using a CMOS camera-equipped infrared thermography system and its clinical application in rapidly screening patients with suspected infectious diseases. Int. J. Infect. Dis. 2017, 55, 113–117. [Google Scholar] [CrossRef] [Green Version]
- Zourob, M.; Elwary, S.; Turner, A.P. Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems; Springer Science & Business Media: New York, NY, USA, 2008. [Google Scholar]
- Lazcka, O.; Del Campo, F.J.; Munoz, F.X. Pathogen detection: A perspective of traditional methods and biosensors. Biosens. Bioelectron. 2007, 22, 1205–1217. [Google Scholar] [CrossRef] [PubMed]
- Nikkhoo, N.; Gulak, P.G.; Maxwell, K. Rapid detection of E. coli bacteria using potassium-sensitive FETs in CMOS. IEEE Trans. Biomed. Circuits Syst. 2013, 7, 621–630. [Google Scholar] [CrossRef]
- Preechakasedkit, P.; Pinwattana, K.; Dungchai, W.; Siangproh, W.; Chaicumpa, W.; Tongtawe, P.; Chailapakul, O. Development of a one-step immunochromatographic strip test using gold nanoparticles for the rapid detection of Salmonella typhi in human serum. Biosens. Bioelectron. 2012, 31, 562–566. [Google Scholar] [CrossRef] [PubMed]
- Zeinhom, M.M.A.; Wang, Y.; Song, Y.; Zhu, M.-J.; Lin, Y.; Du, D. A portable smart-phone device for rapid and sensitive detection of E. coli O157: H7 in Yoghurt and Egg. Biosens. Bioelectron. 2018, 99, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.; Barizuddin, S.; Darr, C.M.; Mathai, C.J.; Ball, A.; Minch, K.; Somoskovi, A.; Hamasur, B.; Connelly, J.T.; Weigl, B. Ultrasensitive detection of lipoarabinomannan with plasmonic grating biosensors in clinical samples of HIV negative patients with tuberculosis. PLoS ONE 2019, 14. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liu, X.; Dang, T.C.; Huang, X.; Feng, H.; Zhang, Q.; Yu, H. A High-Sensitivity Potentiometric 65-nm CMOS ISFET Sensor for Rapid E. coli Screening. IEEE Trans. Biomed. Circuits Syst. 2018, 12, 402–415. [Google Scholar] [CrossRef] [PubMed]
- Couniot, N.; Bol, D.; Poncelet, O.; Francis, L.A.; Flandre, D. A capacitance-to-frequency converter with on-chip passivated microelectrodes for bacteria detection in saline buffers up to 575 MHz. IEEE Trans. Circuits Syst. Ii: Express Briefs 2014, 62, 159–163. [Google Scholar] [CrossRef]
- Mujika, M.; Arana, S.; Castano, E.; Tijero, M.; Vilares, R.; Ruano-Lopez, J.; Cruz, A.; Sainz, L.; Berganza, J. Magnetoresistive immunosensor for the detection of Escherichia coli O157: H7 including a microfluidic network. Biosens. Bioelectron. 2009, 24, 1253–1258. [Google Scholar] [CrossRef]
- Huang, Y.-J.; Huang, C.-W.; Lin, T.-H.; Lin, C.-T.; Chen, L.-G.; Hsiao, P.-Y.; Wu, B.-R.; Hsueh, H.-T.; Kuo, B.-J.; Tsai, H.-H. A CMOS cantilever-based label-free DNA SoC with improved sensitivity for hepatitis B virus detection. IEEE Trans. Biomed. Circuits Syst. 2013, 7, 820–831. [Google Scholar] [CrossRef]
- Wang, T.; Kim, S.; An, J.H. A novel CMOS image sensor system for quantitative loop-mediated isothermal amplification assays to detect food-borne pathogens. J. Microbiol. Methods 2017, 133, 1–7. [Google Scholar] [CrossRef]
- Pérez, J.M.; Jofre, M.; Martínez, P.; Yáñez, M.; Catalan, V.; Pruneri, V. An image cytometer based on angular spatial frequency processing and its validation for rapid detection and quantification of waterborne microorganisms. Analyst 2015, 140, 7734–7741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishara, W.; Sikora, U.; Mudanyali, O.; Su, T.-W.; Yaglidere, O.; Luckhart, S.; Ozcan, A. Holographic pixel super-resolution in portable lensless on-chip microscopy using a fiber-optic array. Lab Chip 2011, 11, 1276–1279. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.; Keles, H.O.; Ozcan, A.; Khademhosseini, A.; Hæggstrom, E.; Kuritzkes, D.; Demirci, U. Integrating microfluidics and lensless imaging for point-of-care testing. Biosens. Bioelectron. 2009, 24, 3208–3214. [Google Scholar] [CrossRef] [PubMed]
- Viswanath, B.; Kristine, Y.M.; Kim, S. Recent trends in the development of complementary metal oxide semiconductor image sensors to detect foodborne bacterial pathogens. Trac Trends Anal. Chem. 2018, 98, 47–57. [Google Scholar] [CrossRef]
- Shin, D.; Athamanolap, P.; Chen, L.; Hardick, J.; Lewis, M.; Hsieh, Y.-H.; Rothman, R.; Gaydos, C.A.; Wang, T. Mobile nucleic acid amplification testing (mobiNAAT) for Chlamydia trachomatis screening in hospital emergency department settings. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Priye, A.; Ball, C.S.; Meagher, R.J. Colorimetric-luminance readout for quantitative analysis of fluorescence signals with a smartphone CMOS sensor. Anal. Chem. 2018, 90, 12385–12389. [Google Scholar] [CrossRef] [PubMed]
- Natesan, M.; Wu, S.-W.; Chen, C.-I.; Jensen, S.M.; Karlovac, N.; Dyas, B.K.; Mudanyali, O.; Ulrich, R.G. A smartphone-based rapid telemonitoring system for Ebola and Marburg disease surveillance. ACS Sens. 2018, 4, 61–68. [Google Scholar] [CrossRef] [Green Version]
- Seo, S.-M.; Kim, S.-W.; Jeon, J.-W.; Kim, J.-H.; Kim, H.-S.; Cho, J.-H.; Lee, W.-H.; Paek, S.-H. Food contamination monitoring via internet of things, exemplified by using pocket-sized immunosensor as terminal unit. Sens. Actuators B: Chem. 2016, 233, 148–156. [Google Scholar] [CrossRef]
- Tsouti, V.; Boutopoulos, C.; Zergioti, I.; Chatzandroulis, S. Capacitive microsystems for biological sensing. Biosens. Bioelectron. 2011, 27, 1–11. [Google Scholar] [CrossRef]
- Göröcs, Z.; Ozcan, A. On-chip biomedical imaging. IEEE Rev. Biomed. Eng. 2012, 6, 29–46. [Google Scholar] [CrossRef] [Green Version]
- Song, J.M.; Culha, M.; Kasili, P.M.; Griffin, G.D.; Vo-Dinh, T. A compact CMOS biochip immunosensor towards the detection of a single bacteria. Biosens. Bioelectron. 2005, 20, 2203–2209. [Google Scholar] [CrossRef] [PubMed]
- Ho, D.; Noor, M.O.; Krull, U.J.; Gulak, G.; Genov, R. CMOS spectrally-multiplexed Fret-on-a-Chip for DNA analysis. IEEE Trans. Biomed. Circuits Syst. 2013, 7, 643–654. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, K.; Zhang, J.; Qin, W.; Yan, X.; Shen, G.; Gao, G.; Pan, F.; Cui, D. Simultaneous quantitative detection of Helicobacter pylori based on a rapid and sensitive testing platform using quantum dots-labeled immunochromatiographic test strips. Nanoscale Res. Lett. 2016, 11, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baader, J.; Klapproth, H.; Bednar, S.; Brandstetter, T.; Rühe, J.; Lehmann, M.; Freund, I. Polysaccharide microarrays with a CMOS based signal detection unit. Biosens. Bioelectron. 2011, 26, 1839–1846. [Google Scholar] [CrossRef]
- Pilavaki, E.; Valente, V.; Demosthenous, A. CMOS Image Sensor for Lateral Flow Immunoassay Readers. IEEE Trans. Circuits Syst. Ii Express Briefs 2018, 65, 1405–1409. [Google Scholar] [CrossRef]
- Moon, S.; Manzur, F.; Manzur, T.; Klapperich, C.; Demirci, U. Escherichia coli counting using lens-free imaging for sepsis diagnosis. In Proceedings of Unmanned/Unattended Sensors and Sensor Networks; International Society for Optics and Photonics: Berlin, Germany, 2009; volume VI, p. 748011. [Google Scholar]
- Lee, J.; Kwak, Y.H.; Paek, S.-H.; Han, S.; Seo, S. CMOS image sensor-based ELISA detector using lens-free shadow imaging platform. Sens. Actuators B: Chem. 2014, 196, 511–517. [Google Scholar] [CrossRef]
- Ray, A.; Daloglu, M.U.; Ho, J.; Torres, A.; Mcleod, E.; Ozcan, A. Computational sensing of herpes simplex virus using a cost-effective on-chip microscope. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Devadhasan, J.P.; Kim, S. CMOS image sensor based HIV diagnosis: a smart system for point-of-care approach. Biochip J. 2013, 7, 258–266. [Google Scholar] [CrossRef]
- Devadhasan, J.P.; Kim, S. Label free quantitative immunoassay for Hepatitis B. J. Nanosci. Nanotechnol. 2015, 15, 85–92. [Google Scholar] [CrossRef]
- Tokel, O.; Yildiz, U.H.; Inci, F.; Durmus, N.G.; Ekiz, O.O.; Turker, B.; Cetin, C.; Rao, S.; Sridhar, K.; Natarajan, N. Portable microfluidic integrated plasmonic platform for pathogen detection. Sci. Rep. 2015, 5, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Soler, M.; Belushkin, A.; Cavallini, A.; Kebbi-Beghdadi, C.; Greub, G.; Altug, H. Multiplexed nanoplasmonic biosensor for one-step simultaneous detection of Chlamydia trachomatis and Neisseria gonorrhoeae in urine. Biosens. Bioelectron. 2017, 94, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Daaboul, G.G.; Freedman, D.S.; Scherr, S.M.; Carter, E.; Rosca, A.; Bernstein, D.; Mire, C.E.; Agans, K.N.; Hoenen, T.; Geisbert, T.W. Enhanced light microscopy visualization of virus particles from Zika virus to filamentous ebolaviruses. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Priego, P.; Martens, D.; Elamin, A.A.; Soetaert, P.; Van Roy, W.; Vos, R.; Anton, B.; Bockstaele, R.; Becker, H.; Singh, M. Label-free and real-time detection of tuberculosis in human urine samples using a nanophotonic point-of-care platform. ACS Sens. 2018, 3, 2079–2086. [Google Scholar] [CrossRef] [PubMed]
- Velasco-Garcia, M. Optical biosensors for probing at the cellular level: A review of recent progress and future prospects. In Proceedings of Seminars in Cell & Developmental Biology; Academic Press: San Diego, CA, USA, 2009; pp. 27–33. [Google Scholar]
- Lei, K.-M.; Mak, P.-I.; Law, M.-K.; Martins, R.P. CMOS biosensors for in vitro diagnosis–transducing mechanisms and applications. Lab Chip 2016, 16, 3664–3681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, B.; Cao, P.; Chevalier, A.; Ellington, A.; Hassibi, A. A CMOS fluorescent-based biosensor microarray. In Proceedings of the 2009 IEEE International Solid-State Circuits Conference-Digest of Technical Papers, San Francisco, CA, USA, 8–12 February 2009; pp. 436–437. [Google Scholar]
- Ta-chien, D.H.; Sorgenfrei, S.; Gong, P.; Levicky, R.; Shepard, K.L. A 0.18-µm CMOS Array Sensor for Integrated Time-Resolved Fluorescence Detection. IEEE J. Solid-State Circuits 2009, 44, 1644–1654. [Google Scholar]
- Hong, L.; McManus, S.; Yang, H.; Sengupta, K. A fully integrated CMOS fluorescence biosensor with on-chip nanophotonic filter. In Proceedings of the 2015 Symposium on VLSI Circuits (VLSI Circuits), Kyoto, Japan, 17–19 June 2015; pp. C206–C207. [Google Scholar]
- Erill, I.; Campoy, S.; Rus, J.; Fonseca, L.; Ivorra, A.; Navarro, Z.; Plaza, J.A.; Aguiló, J.; Barbé, J. Development of a CMOS-compatible PCR chip: comparison of design and system strategies. J. Micromech. Microeng. 2004, 14, 1558. [Google Scholar] [CrossRef] [Green Version]
- Toumazou, C.; Shepherd, L.M.; Reed, S.C.; Chen, G.I.; Patel, A.; Garner, D.M.; Wang, C.-J.A.; Ou, C.-P.; Amin-Desai, K.; Athanasiou, P. Simultaneous DNA amplification and detection using a pH-sensing semiconductor system. Nat. Methods 2013, 10, 641. [Google Scholar] [CrossRef]
- Sandeau, L.; Vuillaume, C.; Contié, S.; Grinenval, E.; Belloni, F.; Rigneault, H.; Owens, R.M.; Fournet, M.B. Large area CMOS bio-pixel array for compact high sensitive multiplex biosensing. Lab Chip 2015, 15, 877–881. [Google Scholar] [CrossRef]
- Arandian, A.; Bagheri, Z.; Ehtesabi, H.; Najafi Nobar, S.; Aminoroaya, N.; Samimi, A.; Latifi, H. Optical Imaging Approaches to Monitor Static and Dynamic Cell-on-Chip Platforms: A Tutorial Review. Small 2019, 15, 1900737. [Google Scholar] [CrossRef]
- Fan, X.; White, I.M.; Shopova, S.I.; Zhu, H.; Suter, J.D.; Sun, Y. Sensitive optical biosensors for unlabeled targets: A review. Anal. Chim. Acta 2008, 620, 8–26. [Google Scholar] [CrossRef]
- Damborský, P.; Švitel, J.; Katrlík, J. Optical biosensors. Essays Biochem. 2016, 60, 91–100. [Google Scholar] [PubMed] [Green Version]
- Garcia-Sucerquia, J.; Xu, W.; Jericho, S.K.; Klages, P.; Jericho, M.H.; Kreuzer, H.J. Digital in-line holographic microscopy. Appl. Opt. 2006, 45, 836–850. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Hakozaki, Y.; Suzuki, S.; Usui, T.; Kato, T.; Hasegawa, K.; Sugiyama, Y.; Sugamata, M.; Abe, S. A novel screening method for influenza patients using a newly developed non-contact screening system. J. Infect. 2010, 60, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.-A.; Lin, C.-H.; Yang, Y.-S.; Lu, M.S.-C. Ultrasensitive and label-free detection of pathogenic avian influenza DNA by using CMOS impedimetric sensors. Biosens. Bioelectron. 2012, 35, 456–460. [Google Scholar] [CrossRef]
- Couniot, N.; Francis, L.A.; Flandre, D. A 16×16 CMOS Capacitive Biosensor Array Towards Detection of Single Bacterial Cell. IEEE Trans. Biomed. Circuits Syst. 2016, 10, 364–374. [Google Scholar] [CrossRef]
- Malpartida-Cardenas, K.; Miscourides, N.; Rodriguez-Manzano, J.; Yu, L.-S.; Moser, N.; Baum, J.; Georgiou, P. Quantitative and rapid Plasmodium falciparum malaria diagnosis and artemisinin-resistance detection using a CMOS Lab-on-Chip platform. Biosens. Bioelectron. 2019, 145, 111678. [Google Scholar] [CrossRef]
- Rothberg, J.M.; Hinz, W.; Rearick, T.M.; Schultz, J.; Mileski, W.; Davey, M.; Leamon, J.H.; Johnson, K.; Milgrew, M.J.; Edwards, M. An integrated semiconductor device enabling non-optical genome sequencing. Nature 2011, 475, 348–352. [Google Scholar] [CrossRef]
- Huang, X.; Yu, H.; Liu, X.; Jiang, Y.; Yan, M.; Wu, D. A dual-mode large-arrayed CMOS ISFET sensor for accurate and high-throughput pH sensing in biomedical diagnosis. IEEE Trans. Biomed. Eng. 2015, 62, 2224–2233. [Google Scholar] [CrossRef]
- Singh, N.K.; Thungon, P.D.; Estrela, P.; Goswami, P. Development of an aptamer-based field effect transistor biosensor for quantitative detection of Plasmodium falciparum glutamate dehydrogenase in serum samples. Biosens. Bioelectron. 2019, 123, 30–35. [Google Scholar] [CrossRef]
- Nikkhoo, N.; Cumby, N.; Gulak, P.G.; Maxwell, K.L. Rapid Bacterial Detection via an All-Electronic CMOS Biosensor. PLoS ONE 2016, 11. [Google Scholar] [CrossRef]
- Lai, W.-A.; Lin, C.-H.; Yang, Y.-S.; Lu, M.S.-C. Ultrasensitive detection of avian influenza virus by using CMOS impedimetric sensor arrays. In Proceedings of the 2012 IEEE 25th International Conference on Micro Electro Mechanical Systems (MEMS), Paris, Franch, 29 January–2 February 2012; pp. 894–897. [Google Scholar]
- Hsu, C.-L.; Sun, A.; Zhao, Y.; Aronoff-Spencer, E.; Hall, D.A. A 16 × 20 electrochemical CMOS biosensor array with in-pixel averaging using polar modulation. In Proceedings of the 2018 IEEE Custom Integrated Circuits Conference (CICC), San Diego, CA, USA, 8–11 April 2017; pp. 1–4. [Google Scholar]
- Yao, L.; Lamarche, P.; Tawil, N.; Khan, R.; Aliakbar, A.M.; Hassan, M.H.; Chodavarapu, V.P.; Mandeville, R. CMOS conductometric system for growth monitoring and sensing of bacteria. IEEE Trans. Biomed. Circuits Syst. 2011, 5, 223–230. [Google Scholar] [CrossRef]
- Niitsu, K.; Ota, S.; Gamo, K.; Kondo, H.; Hori, M.; Nakazato, K. Development of microelectrode arrays using electroless plating for CMOS-based direct counting of bacterial and HeLa cells. IEEE Trans. Biomed. Circuits Syst. 2015, 9, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-W.; Huang, Y.-J.; Yen, P.-W.; Tsai, H.-H.; Liao, H.-H.; Juang, Y.-Z.; Lu, S.-S.; Lin, C.-T. A CMOS wireless biomolecular sensing system-on-chip based on polysilicon nanowire technology. Lab Chip 2013, 13, 4451–4459. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.C.; Alvarez-Fontecilla, E.; Venkatesh, A.; Aronoff-Spencer, E.; Hall, D.A. High-density redox amplified coulostatic discharge-based biosensor array. IEEE J. Solid-State Circuits 2018, 53, 2054–2064. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.; Alvarez-Fontecilla, E.; Venkatesh, A.; Aronoff-Spencer, E.; Hall, D.A. A 64 × 64 high-density redox amplified coulostatic discharge-based biosensor array in 180 nm CMOS. In Proceedings of the ESSCIRC 2017-43rd IEEE European Solid State Circuits Conference, Leuven, Belgium, 11–14 September 2017; pp. 368–371. [Google Scholar]
- Pourciel-Gouzy, M.; Sant, W.; Humenyuk, I.; Malaquin, L.; Dollat, X.; Temple-Boyer, P. Development of pH-ISFET sensors for the detection of bacterial activity. Sens. Actuators B Chem. 2004, 103, 247–251. [Google Scholar] [CrossRef]
- Bergveld, P. Development of an ion-sensitive solid-state device for neurophysiological measurements. IEEE Trans. Biomed. Eng. 1970, 70–71. [Google Scholar] [CrossRef]
- Bausells, J.; Carrabina, J.; Errachid, A.; Merlos, A. Ion-sensitive field-effect transistors fabricated in a commercial CMOS technology. Sens. Actuators B: Chem. 1999, 57, 56–62. [Google Scholar] [CrossRef] [Green Version]
- Ghafar-Zadeh, E. Wireless integrated biosensors for point-of-care diagnostic applications. Sensors 2015, 15, 3236–3261. [Google Scholar] [CrossRef] [Green Version]
- Couniot, N.; Vanzieleghem, T.; Rasson, J.; Van Overstraeten-Schlögel, N.; Poncelet, O.; Mahillon, J.; Francis, L.; Flandre, D. Lytic enzymes as selectivity means for label-free, microfluidic and impedimetric detection of whole-cell bacteria using ALD-Al2O3 passivated microelectrodes. Biosens. Bioelectron. 2015, 67, 154–161. [Google Scholar] [CrossRef]
- Yao, L.; Hajj-Hassan, M.; Ghafar-Zadeh, E.; Shabani, A.; Chodavarapu, V.; Zourob, M. CMOS capactive sensor system for bacteria detection using phage organisms. In Proceedings of the Canadian Conference on Electrical and Computer Engineering CCECE, Niagara Falls, ON, Canada, 4–7 May 2008; pp. 000877–000880. [Google Scholar]
- Gamo, K.; Nakazato, K.; Niitsu, K. Design, theoretical analysis, and experimental verification of a CMOS current integrator with 1.2 × 2.05 µm2 microelectrode array for high-sensitivity bacterial counting. Jpn. J. Appl. Phys. 2016, 56, 01AH01. [Google Scholar] [CrossRef]
- Ambhorkar, P.; Wang, Z.; Ko, H.; Lee, S.; Koo, K.-i.; Kim, K.; Cho, D.-i.D. Nanowire-based biosensors: From growth to applications. Micromachines 2018, 9, 679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tigli, O.; Bivona, L.; Berg, P.; Zaghloul, M.E. Fabrication and characterization of a surface-acoustic-wave biosensor in CMOS technology for cancer biomarker detection. IEEE Trans. Biomed. Circuits Syst. 2009, 4, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Li, K.-W.; Yen, Y.-K. Gentamicin drug monitoring for peritonitis patients by using a CMOS-BioMEMS-based microcantilever sensor. Biosens. Bioelectron. 2019, 130, 420–426. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y. CMOS Magnetic Cell Manipulator and CMOS NMR Biomolecular Sensor; Harvard University: Cambridge, MA, USA, 2008. [Google Scholar]
- Aytur, T.; Foley, J.; Anwar, M.; Boser, B.; Harris, E.; Beatty, P.R. A novel magnetic bead bioassay platform using a microchip-based sensor for infectious disease diagnosis. J. Immunol. Methods 2006, 314, 21–29. [Google Scholar] [CrossRef]
- Pai, A.; Khachaturian, A.; Chapman, S.; Hu, A.; Wang, H.; Hajimiri, A. A handheld magnetic sensing platform for antigen and nucleic acid detection. Analyst 2014, 139, 1403–1411. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Chen, Y.; Hassibi, A.; Scherer, A.; Hajimiri, A. A frequency-shift CMOS magnetic biosensor array with single-bead sensitivity and no external magnet. In Proceedings of the Solid-State Circuits Conference-Digest of Technical Papers, San Francisco, CA, USA, 8–12 February 2009; pp. 438–439. [Google Scholar]
- Chen, C.-H.; Hwang, R.-Z.; Huang, L.-S.; Lin, S.-M.; Chen, H.-C.; Yang, Y.-C.; Lin, Y.-T.; Yu, S.-A.; Lin, Y.-S.; Wang, Y.-H. A wireless bio-MEMS sensor for C-reactive protein detection based on nanomechanics. IEEE Trans. Biomed. Eng. 2008, 56, 462–470. [Google Scholar] [CrossRef]
- Huang, C.-W.; Hsueh, H.-T.; Huang, Y.-J.; Liao, H.-H.; Tsai, H.-H.; Juang, Y.-Z.; Lin, T.-H.; Lu, S.-S.; Lin, C.-T. A fully integrated wireless CMOS microcantilever lab chip for detection of DNA from Hepatitis B virus (HBV). Sens. Actuators B Chem. 2013, 181, 867–873. [Google Scholar] [CrossRef]
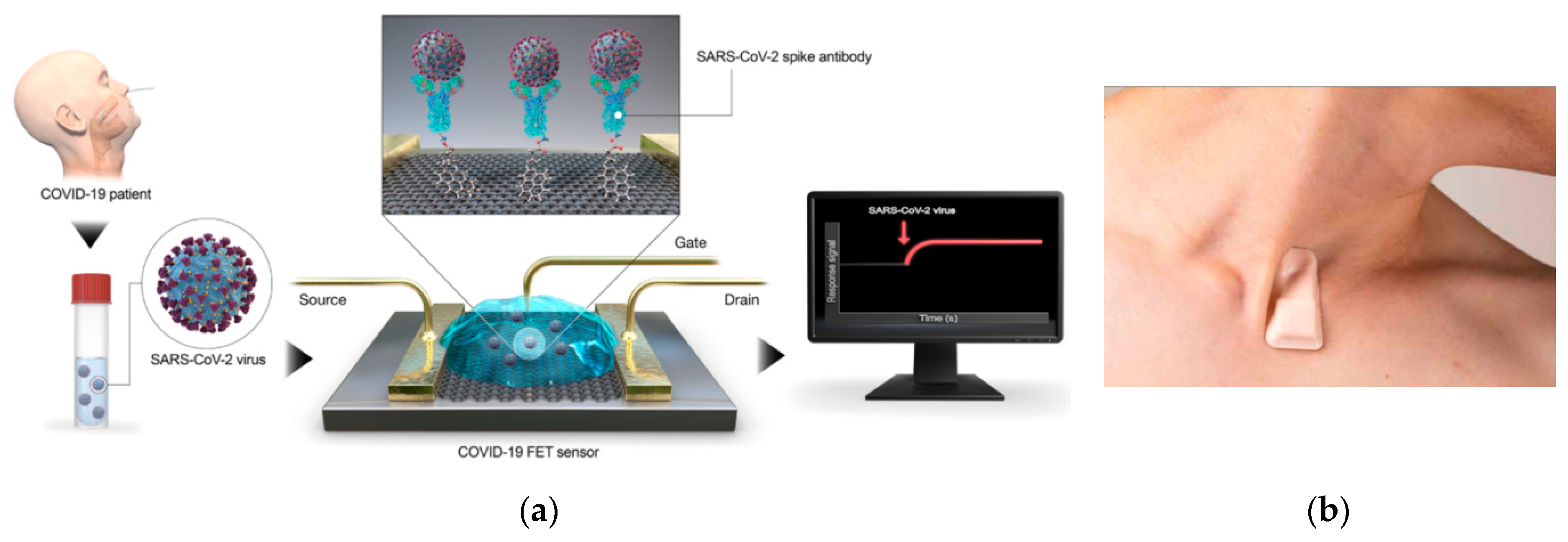
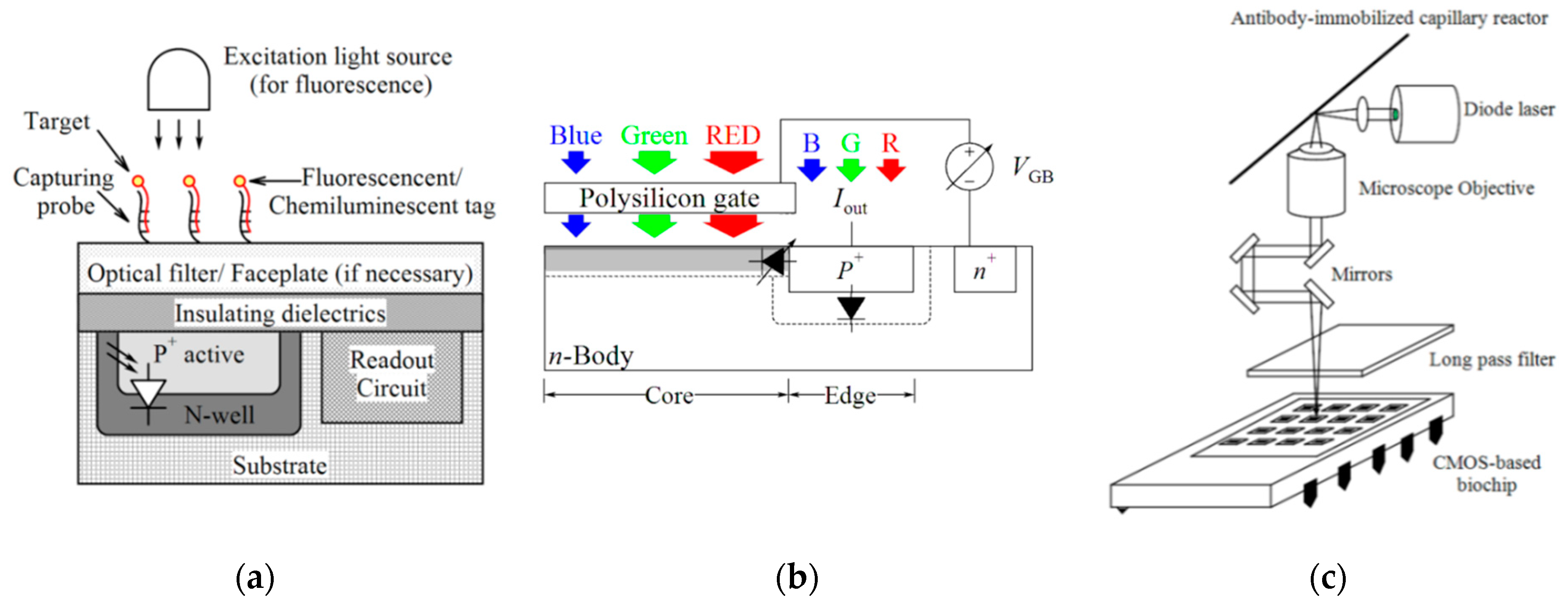
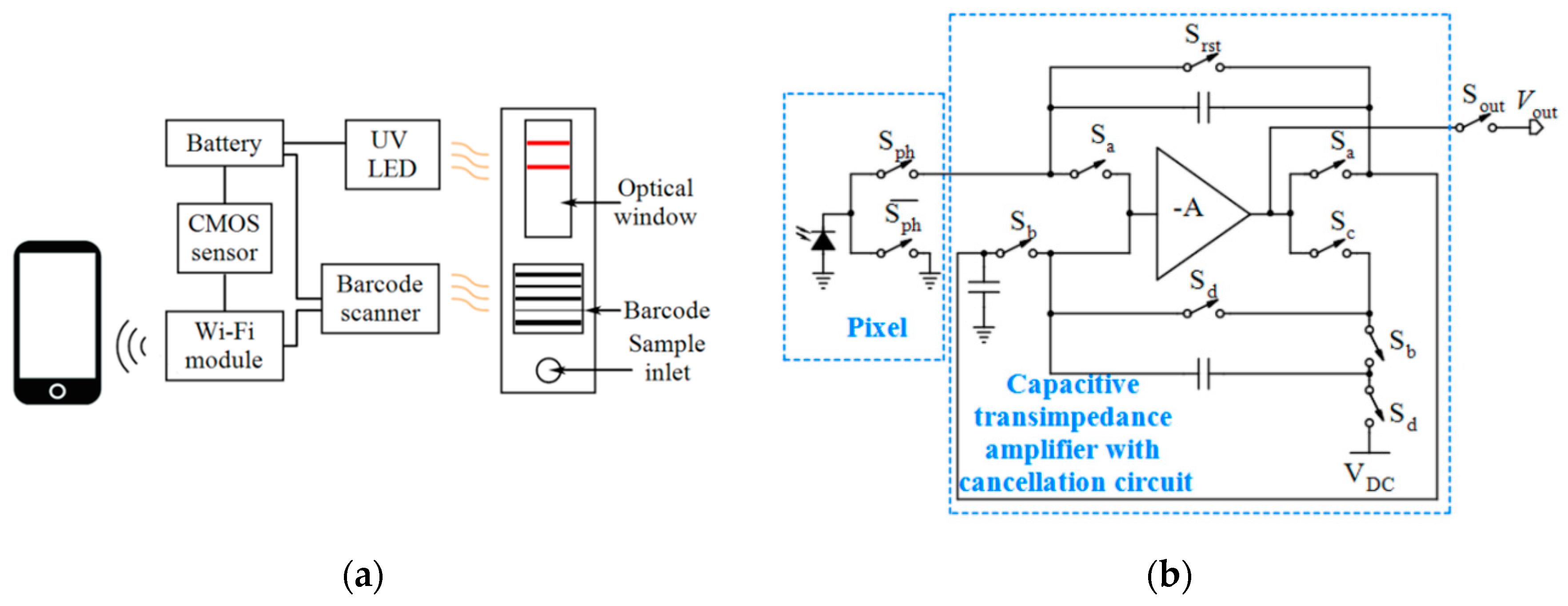
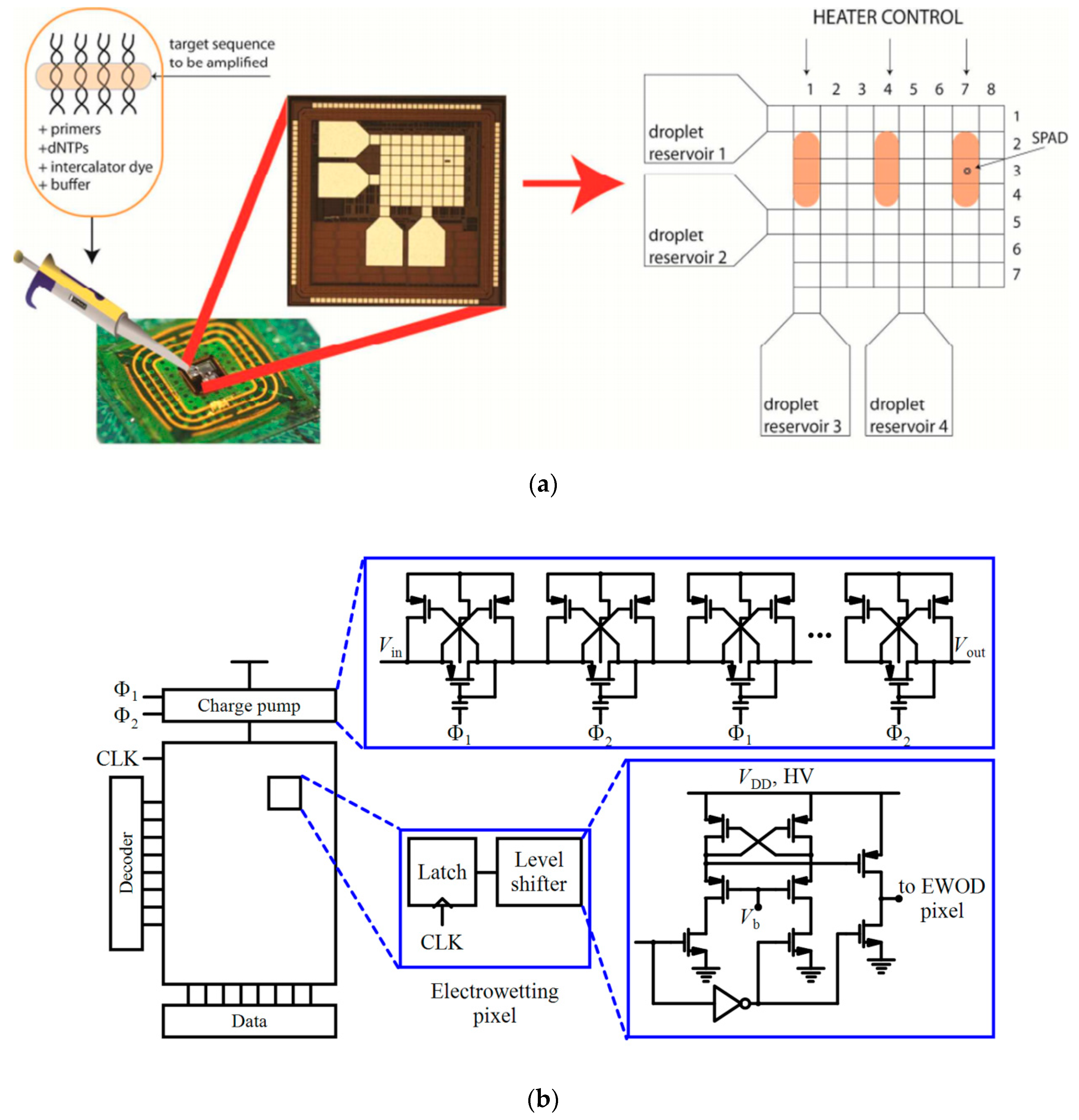
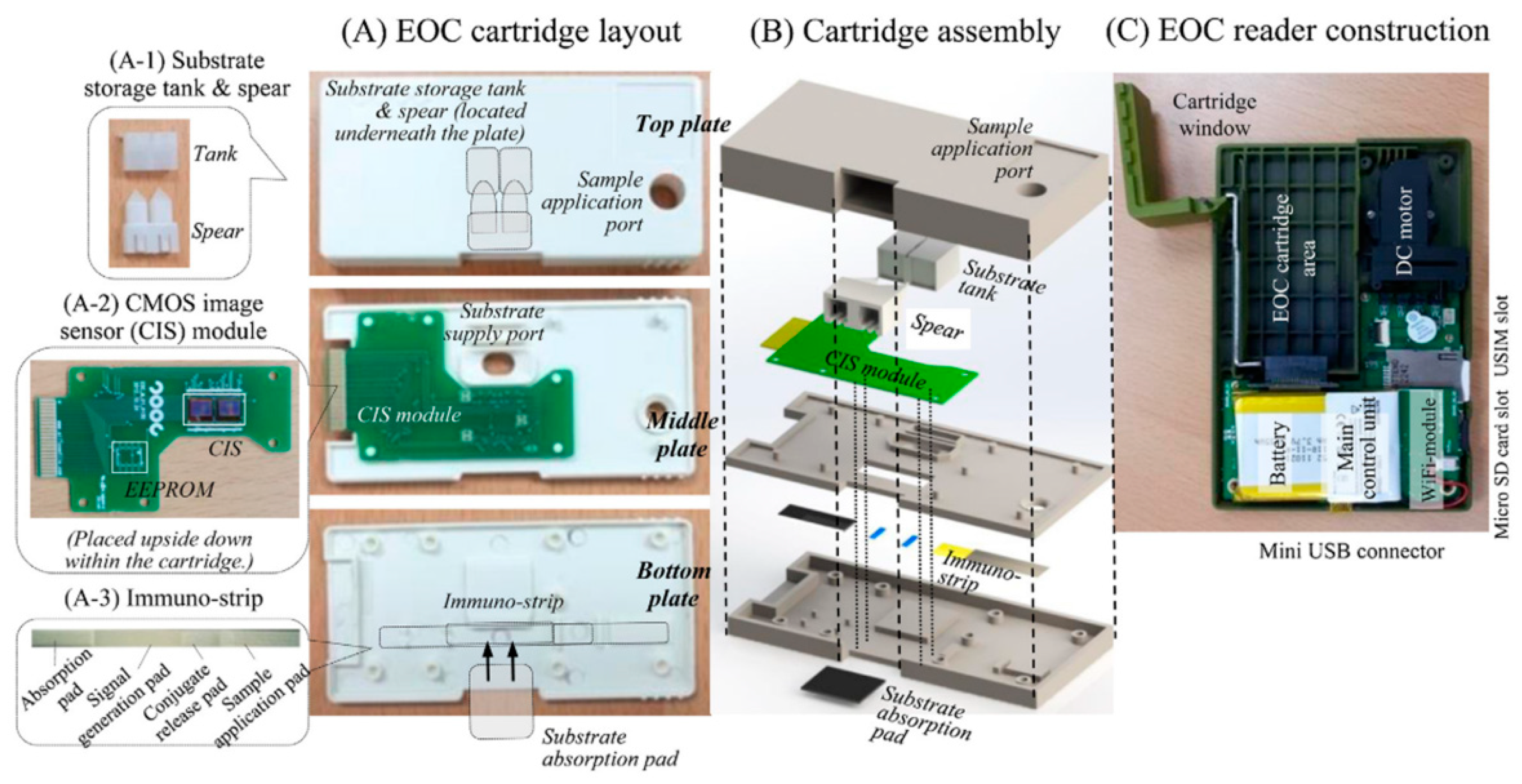
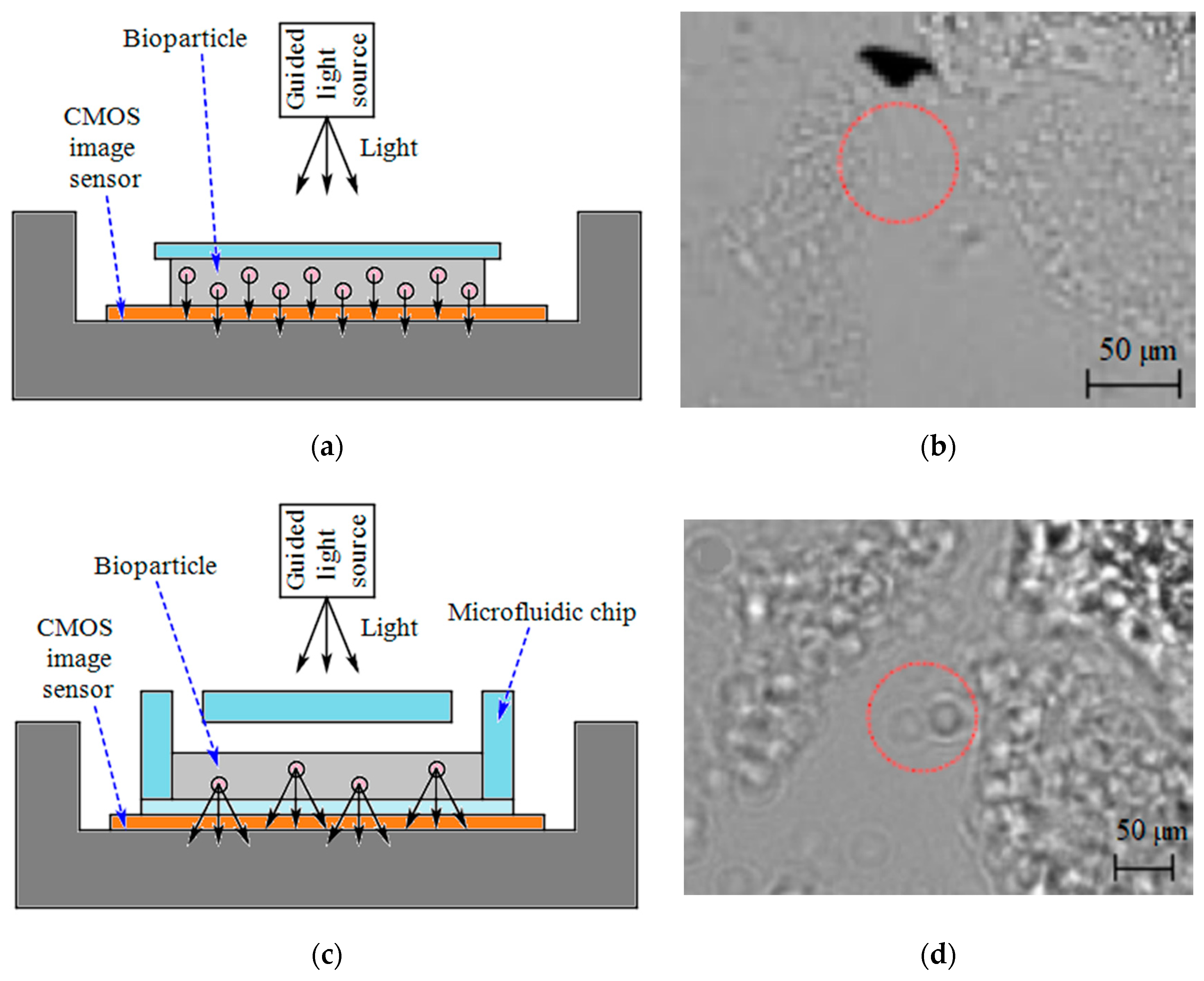

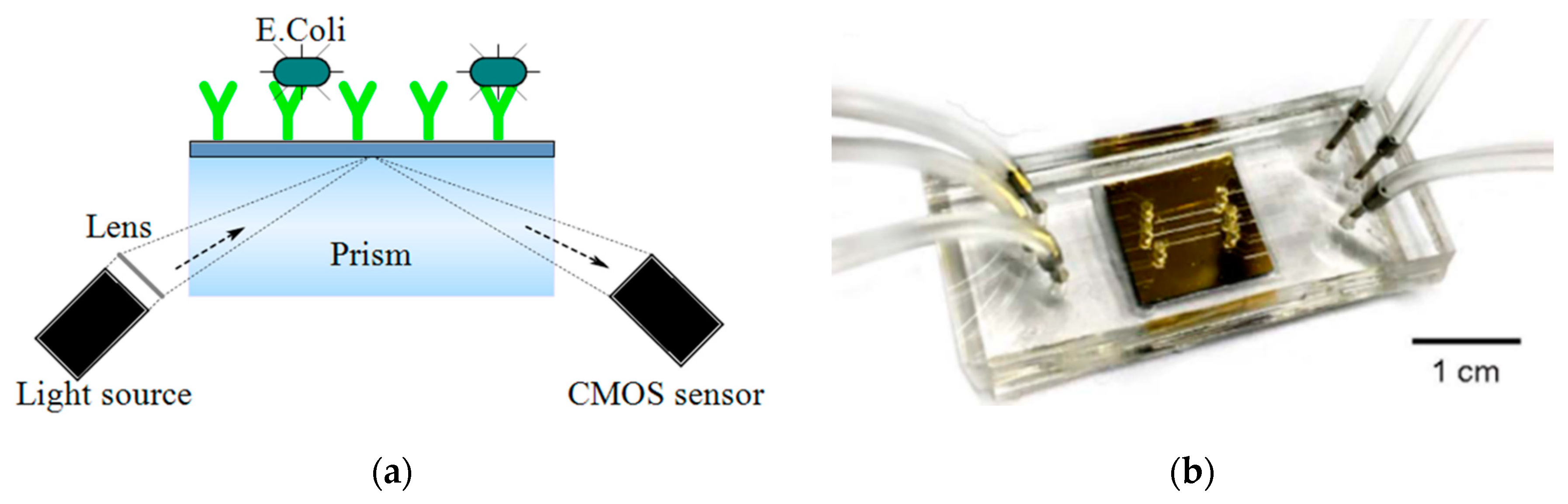


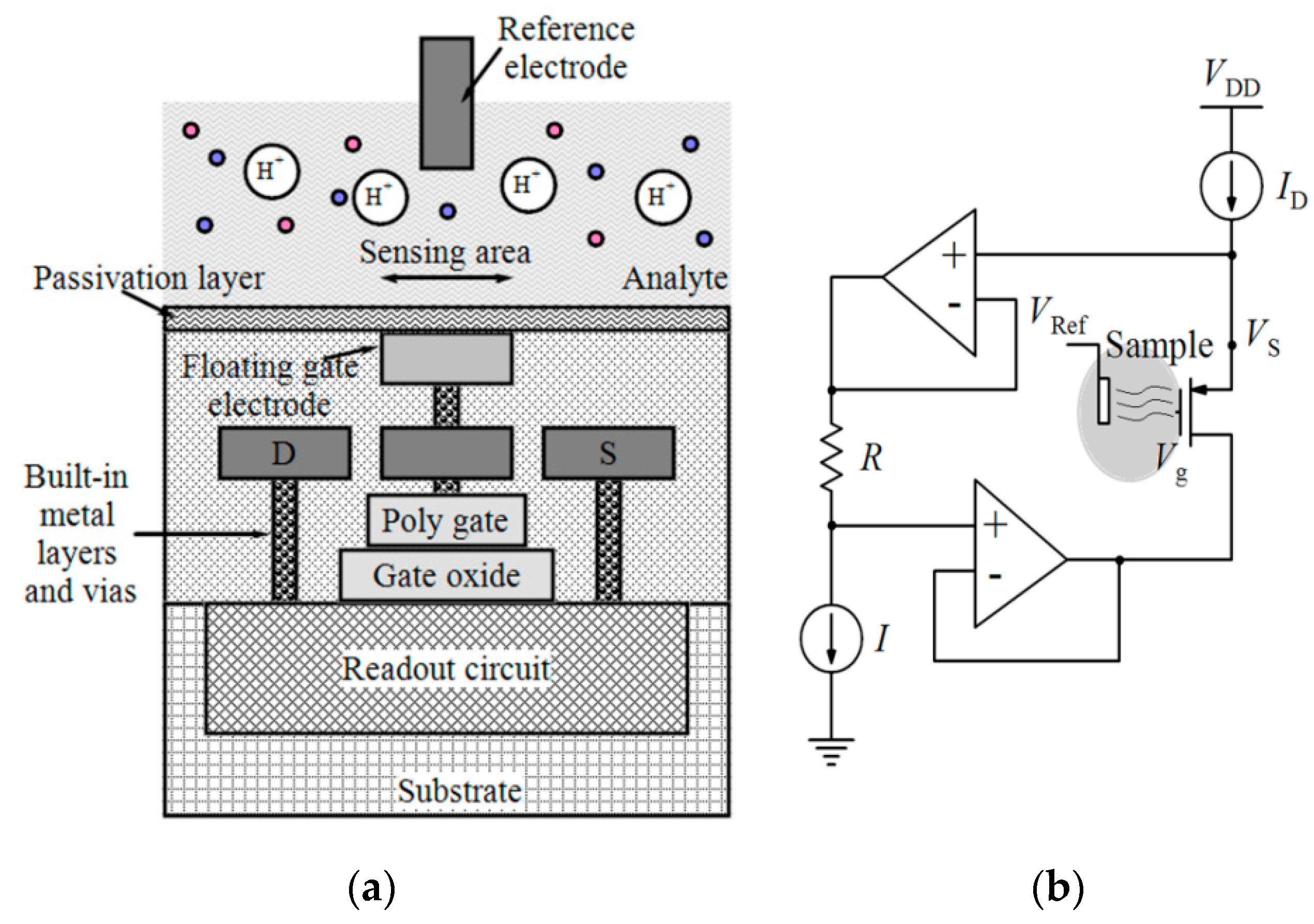

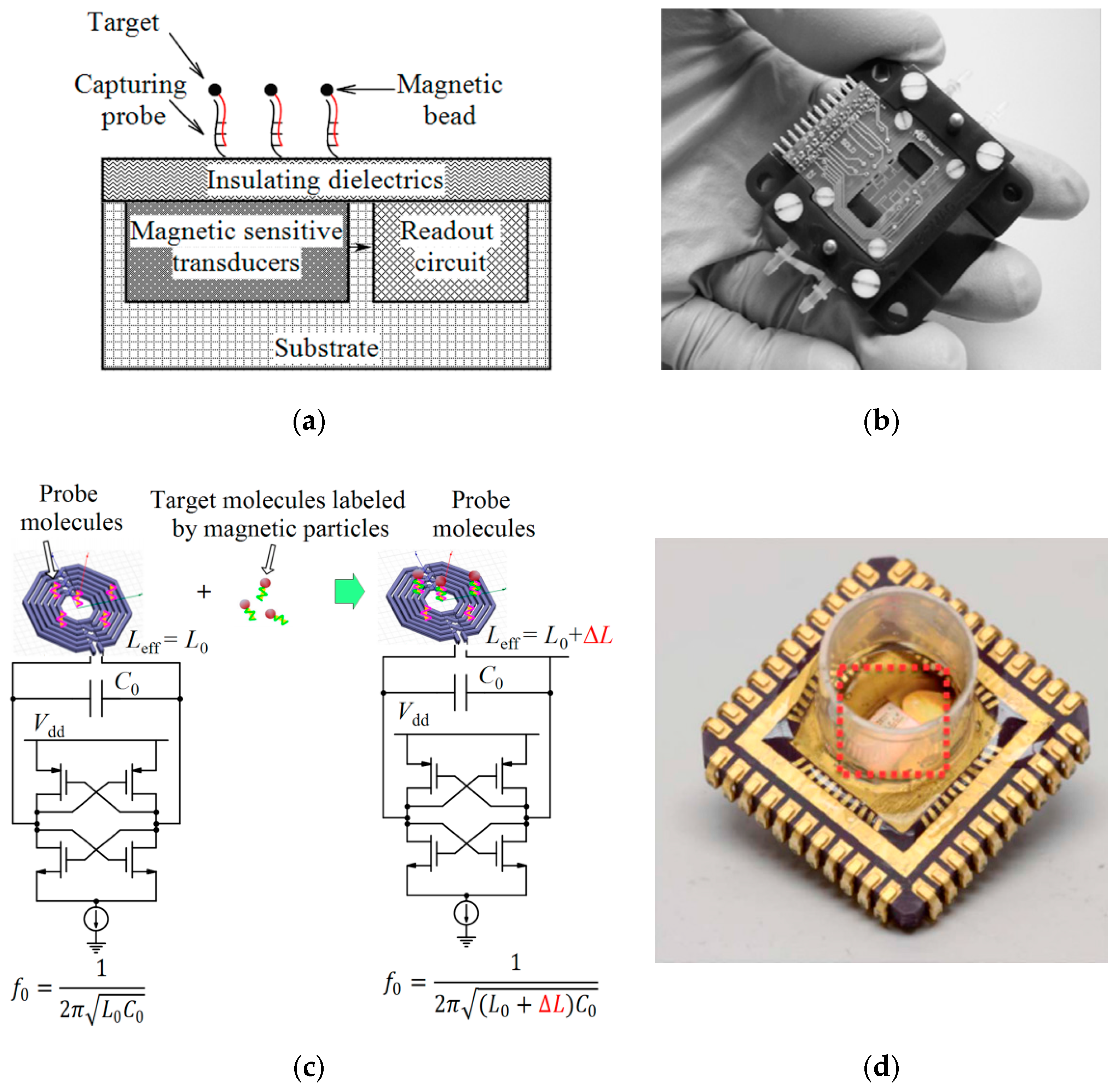
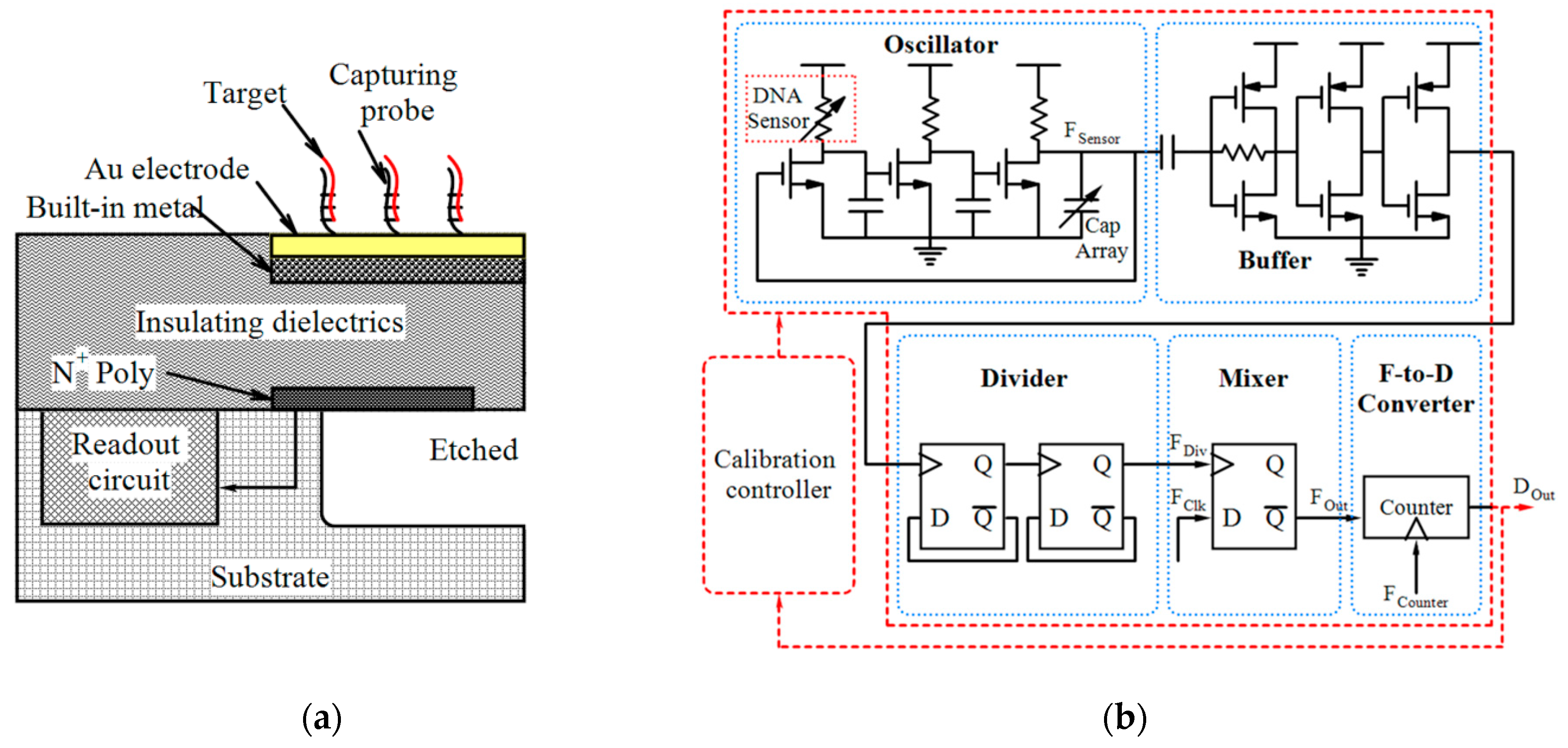
| Device | Target | Specifications | Ref. |
|---|---|---|---|
| Gr-FET based biosensor | SARS-CoV-2 spike protein and SARS-CoV-2 |
| [2] |
| Gr-FET immunosensor | COVID-19 spike protein S1 |
| [3] |
| eCovSens-ultrasensitive PCB-based electrochemical device | nCovid-19 antigen, a spike protein domain 1 of SARS-CoV-2 |
| [8] |
| Dual-Functional Plasmonic Photothermal Biosensor | RdRp-COVID and RdRp-COVID-C |
| [9,10] |
| Cepheid (Xpert Xpress SARS-CoV-2 test) | E gene and N gene |
| [15] |
| On-body skin-integrated sensor system | Key symptoms including cardiac- and respiratory-related signals, coughing and body temperature |
| [16,17] |
| Advantages | Disadvantages |
|---|---|
|
|
| Optical Technique | Application | Tech. | Target | Area | Fluidic Structure | Array/Pixel # | LoD | Other Features | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Laser-induced fluorescence | Detection of B. globigii spores based on the combined use of ELISA and LIF detection | - | B. globigii spores | - | - | 4 × 4 | 0.55 cells/probe | - | [52] |
| Spectrally-multiplexed FRET-on-a-Chip | Detection of marker gene sequences of E. coli and spinal muscular atropy disease | 0.35 µm | Two DNA targets | (Optical filter area) 1.5 × 1.5 mm2 | A PDMS 1 reservoir bonded to a glass cover slip | - | 240 nM and 210 nM | AT 2 = 1 Sec/integration time | [53] |
| Fluorescence imaging | Mobile nucleic acid amplification testing for C. trachomatis screening using a smartphone CMOS sensor | - | C. trachomatis DNA | - | magnetofluidics | - | - | Sensitivity = 102 to 103 copies of molecular target | [46] |
| Fluorescence sensing in an integrated qPCR system | PoC diagnostics of S. aureus | 0.35 µm CMOS | Target DNA | 4 × 4 mm2 | SU-8 microfluidics | 7 × 8 | - | Power = 33 mW (3.3 V) | [28] |
| Colorimetric-luminance readout for analysis of fluorescence signals | Simultaneous detection of Zika and chikungunya viral RNA via endpoint RT-LAMP and real-time LAMP detection of N. gonorrhoeae using a smartphone CMOS sensor | - | Zika and chikungunya N. gonorrhoeae DNA | - | - | 1000 | 3.5 copies per 10 µL | - | [47] |
| Fluorescence imaging | Rapid and sensitive detection of E. coli O157:H7 using a smartphone CMOS sensor | - | E. coli O157:H7 | - | Corning black flat-bottom 96-well plates | - | 1 CFU/mL | AT = 2 h | [35] |
| Fluorescence imaging | Rapid telemonitoring system for Ebola and Marburg disease surveillance using a smartphone CMOS sensor | - | Ebola and Marburg | - | 3MTM polyester double-sided diagnostic microfluidic medical tape | - | - | AT = 20 min and 15 s Sensitivity ≈ pM ~ ng/mL | [48] |
| Fluorescence imaging | Simultaneous quantitative detection of H. Pylori using Qdots-labeled Immunochromatiographic test strips | - | H. Pylori | - | Test strip | - | 5 mIU/mL | Specificity = 97% Sensitivity = 95% | [54] |
| Chemiluminescence/fluorescence imaging | Detection of S. pneumoniae 3 by the measurement of IgG antibody concentrations in human blood sera | 0.5 µm | IgG antibody | - | - | 4 × 8 | - | - | [55] |
| Chemiluminescence imaging | Cross-flow immune chromatography based test for the detection of food-borne pathogens | - | S. Typhimurium | 3 × 3 µm2 | EOC cartridge | 1.3 mega pixel | 4.22 × 103 CFU/mL | AT < 6 h | [25] |
| Chemiluminescence imaging | Immunoanalytical analysis for monitoring food contamination | - | Vibrio parahaemolyticus | 3 × 3 µm2 | EOC cartridge | 1.3 mega pixel | 1.4 × 104 CFU/mL | - | [49] |
| Reader for LFIAs | Reader of LFIA for PoC diagnostics of Influenza A nucleoprotein | 0.35 µm | Influenza A nucleoproteins | (Overall pixel array area) 12.28 mm2 | - | 4 × 64 | 0.5–200 ng/mL | FF 4 = 18% TORN 5 = 1.9 mVrms, Total = 21 µW Power/pixel = 0.32 µW | [56] |
| Shadow imaging | E. coli counting for sepsis diagnosis | - | E. coli | - | Microfluidics made of PMMA 6-DSA 7 layers and glass slides | - | - | - | [57] |
| Shadow imaging | ELISA measurements | - | L. monocytogenes and S. typhimurium | - | A commercial 96-well plate | 5M pixel | Density of L. monocytogenes = 5 × 104 cells/mL Density of S. typhimurium = 103 cells/mL | - | [58] |
| Holographic on-chip microscope using fiber-optic arrays | Imaging human malaria parasites in thin blood smears | - | P. falciparum | - | Sample tray | - | - | Resolution < 1 µm FoV ≈ 24 mm2 | [43] |
| Holographic on-chip microscope | Computational sensing of HSV | - | HSV | - | A reservoir | 10M pixel | 4 particles/mm2 | FoV ≈ 30 mm2 | [59] |
| Photon counting | PoC testing for the detection of HIV | - | HIV | - | - | 376 × 314 | 10 fg/mL | - | [60] |
| Photon counting | Label-free quantitative immunoassay for Hepatitis B | - | HBV | - | - | 376 × 314 | 10 fg/mL | - | [61] |
| Photon counting | LAMP technique for real-time DNA amplification and detection | - | C. perfringens (DNA) | 5 × 5 mm2 | Polypropylene cylindrical tube | 376 × 314 | 1 fg/µL | AT < 1 h | [27] |
| SPR | Pathogen detection | - | E. coli and S. aureus | 5.3 mm2 | microfluidics made of PMMA-DSA-PMMA-DSA layers and a gold chip | 500 × 582 | 105 to 3.2 × 107 CFUs/mL | - | [62] |
| SPR | Ultrasensitive detection of LAM with plasmonic grating biosensors in clinical samples of HIV negative patients with tuberculosis | - | Tuberculosis | - | ProPlate 24 well slide adapter | - | 1 fg/mL | Sensitivity < 10 fM | [36] |
| Nano-plasmonic | One-step simultaneous detection of C. trachomatis and N. gonorrhoeae in urine | - | C. trachomatis and N. gonorrhoeae | - | PDMS microfluidics | - | LoD = 300 CFU/mL (for C. trachomatis) LoD = 1500 CFU/mL (for N. gonorrhoeae) | - | [63] |
| SP-IRIS | Enhanced light microscopy visualization of virus particles from Zika virus to filamentous ebolaviruses | - | Zika, Vesicular stomatitis, Vaccinia, Ebola | - | - | - | 200,000 particles/mm2 | - | [64] |
| Mach− Zehnder Interferometer | Detection of tuberculosis in urine samples using a nanophotonic PoC Platform | - | Tuberculosis | 7.16 × 6.76 mm2 | Polymer microfluidic cartridge | - | 475 pg/mL (27.14 pM) | - | [65] |
| Optical microscopy using angular spatial frequency processing | An image cytometer based on angular spatial frequency processing for the detection of waterborne microorganisms | - | E. Coli, L. pneumophila and Phytoplank | 5.70 × 4.28 mm2 | A fluidic system comprising a barometric pump and a hollow fiber membrane filter | 5M pixel | 0.2 cells/mL | - | [42] |
| Technique | Application | Tech. | Target | Area | Fluidic Structure | Array/pixel # | Power | Other Features | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| ISFET | P. falciparum malaria diagnosis and artemisinin-resistance detection | 0.35 µm | P. falciparum DNA | 0.56 mm2 | Laser-cut acrylic microfluidic chamber | 64 × 64 | - | LoD (LAMP) = 1 Copies/reaction, LoD (pH-LAMP) = 10 Copies/reaction | [81] |
| ISFET | Simultaneous detection of independent DNA sequencing of three bacterial genomes | 0.35 µm | Target DNA (of Vibrio fischeri, E. Coli, R. palustris) | 17.5 × 17.5 mm2 | polycarbonate flow cell | 1.5 M, 7.2 M and 13 M | - | - | [82] |
| ISFET and optical | Genome diagnostics of E. Coli | 0.18 μm | Target DNA | 2.5 × 5 mm2 | A 3D-printed plastic reservoir | 64 × 64 | 105.6 mW | Sensitivity = 26.2 mV/pH FPNR 1 = 4% to 0.3% Speed = 1200 fps 2 | [83] |
| ISFET | E. coli Screening | 65 nm | E. Coli | 5 × 5 mm2 | PDMS cylindrical reservoir | 512 × 128 | Pixel array = 80.6 mW Analog blocks = 108.4 mW Digital blocks = 6.5 mW | Sensitivity = 123.8 mV/pH Resolution = 0.01 pH Density range = 14 to 140 CFU/mL Screening time = 4 h | [37] |
| AptaFET | Detection of P. falciparum glutamate dehydrogenase in serum samples | 0.7 µm | P. falciparum glutamate dehydrogenase | - | - | - | - | LoD = 48.6 pM Dynamic range = 100 fM to 10 nM | [84] |
| Potassium-sensitive FET sensor | Detection of E. Coli | 0.18 µm | E. Coli | 1.5 × 0.6 mm2 | A dark chamber | 6 | - | AT < 30 Min | [33] |
| Potassium-sensitive FET sensor | Rapid bacterial detection | 0.13 µm | S. aureus, E. Coli | 1.6 × 1.6 mm2 | - | 2 | - | LoD = 103 bacteria/mL | [85] |
| Impedimetric | AIV detection | 0.35 µm 2P4M | Viral target DNA | - | - | 4 × 4 | - | LoD ~ 6.14 fg/mL | [86] |
| Impedimetric | Detection of Zika virus | 0.18 µm | Zika Virus oligonucleotide | 3 × 4 mm2 | - | 16 × 20 | 63 mW | - | [87] |
| Capacitive | Bacteria detection in saline buffers | 0.25 µm | S. epidermidis | 220 × 230 µm2 | - | 1 | 29 mW | LoD = 10 fF (107 CFU/mL) Sensitivity = 11 kHz/fF (@ 254 MHz @ 17.5 pF) | [38] |
| Capacitive | Detection of single bacterial cell | 0.25 µm | S. epidermidis | 14 × 16 µm2 | - | 16 × 16 | 29 µW | LoD = 450 aF (~ 7 bact.) Sensitivity = 55 mV/fF (22 mV/bact.) | [80] |
| Capacitive | Bacteria growth monitoring | 0.18 µm | E. Coli | 100 × 100 µm2 | DWFP microfluidics | 1 | - | LoD = 107 CFU/mL Sensitivity = 255 mV/fF | [26] |
| Conductometric | Growth monitoring and sensing of bacteria | 0.35 µm | E. Coli | 190 × 220 µm2 | ABP cultureware | 8 | 1.85 mW | Concentration = 4 × 102 to 4 × 104 CFU/mL | [88] |
| Amperometric | Direct counting of bacterial and HeLa cells | 0.6 µm | Bacteria-sized microbeads and HeLa cells | (for 1024 × 1024) 7.48 mm2 | - | 1024 × 1024, 4 × 4, 16 × 16 | - | - | [89] |
| Amperometric | Bacteria Counting | 0.6 µm 2P3M | Bacterial-sized microbeads | 7.6 ×7.1 mm2 | - | 1024 × 1024 | 9.5 mW | Detection limit = 106 cells | [24] |
| Poly-Si NW | Biomolecular detection | 0.35 2p4M | HBV DNA and cTnI | 2.5 × 2.5 mm2 | A plastic reservoir | - | - | LoD = 10 fM (With post-etching) | [90] |
| Coulostatic discharge sensing | Rubella and mumps virus detection | 0.18 µm | Rubella and mumps virus capsid protein | 5 × 5 mm2 | A well | 64 × 64 | 95 mW | LoD = 100 nM | [91] ([92]) |
| Technique | Application | Tech. | Target | Area | Fluidic Structure | Array/Pixel # | Power | Other Features | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Magnetic (Hall sensor) | Diagnosis of infectious disease (Dengue) | 0.25 µm CMOS | Antigen of purified mouse IgG and human anti-dengue virus IgG | 2.5 × 2.5 mm2 | Integrated gold-plated 150-μL sample wells | 1024 | - | - | [104] |
| Magnetic (Frequency-shift based sensing) | Detection of an Interferon-γ protein (relevant for tuberculosis diagnostics) and a 31 bp DNA oligomer | 0.13 µm CMOS | Interferon-γ protein and a 31 bp DNA oligomer | 2.95 × 2.56 µm2 | A polypropylene well | 8 | 165 mW | LoD (Interferon-γ) = 1 pM, LoD (DNA) = 100 pM | [105] ([106]) |
| Magnetic (Magnetoresistive) | Detection of the pathogen E. coli O157:H7 in food and clinical samples | - | E. coli O157:H7 | 16 × 21 mm2 | 4 parallel SU-8 microchannels | 16 magnetoresistive meanders in groups of 4 | - | Specificity = 105 CFU/mL | [39] |
| Peizo-resistive (cantilever-based sensor) | HBV Detection | 0.35 µm Bio-MEMS CMOS | 19 base HBV DNA | 30.4 mm2 | - | 18 | Receive: 12.9 mW Transmit: 18.6 mW Sleep mode: 225 µW | LoD < 1 pM | [40] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Forouhi, S.; Ghafar-Zadeh, E. Applications of CMOS Devices for the Diagnosis and Control of Infectious Diseases. Micromachines 2020, 11, 1003. https://doi.org/10.3390/mi11111003
Forouhi S, Ghafar-Zadeh E. Applications of CMOS Devices for the Diagnosis and Control of Infectious Diseases. Micromachines. 2020; 11(11):1003. https://doi.org/10.3390/mi11111003
Chicago/Turabian StyleForouhi, Saghi, and Ebrahim Ghafar-Zadeh. 2020. "Applications of CMOS Devices for the Diagnosis and Control of Infectious Diseases" Micromachines 11, no. 11: 1003. https://doi.org/10.3390/mi11111003





