1. Introduction
The polar body is an important structure during oogenesis, containing a copy of the genetic information of the oocyte. This structure is located between the cytoplasm and the zona pellucida of the oocyte, and is close to the nucleus [
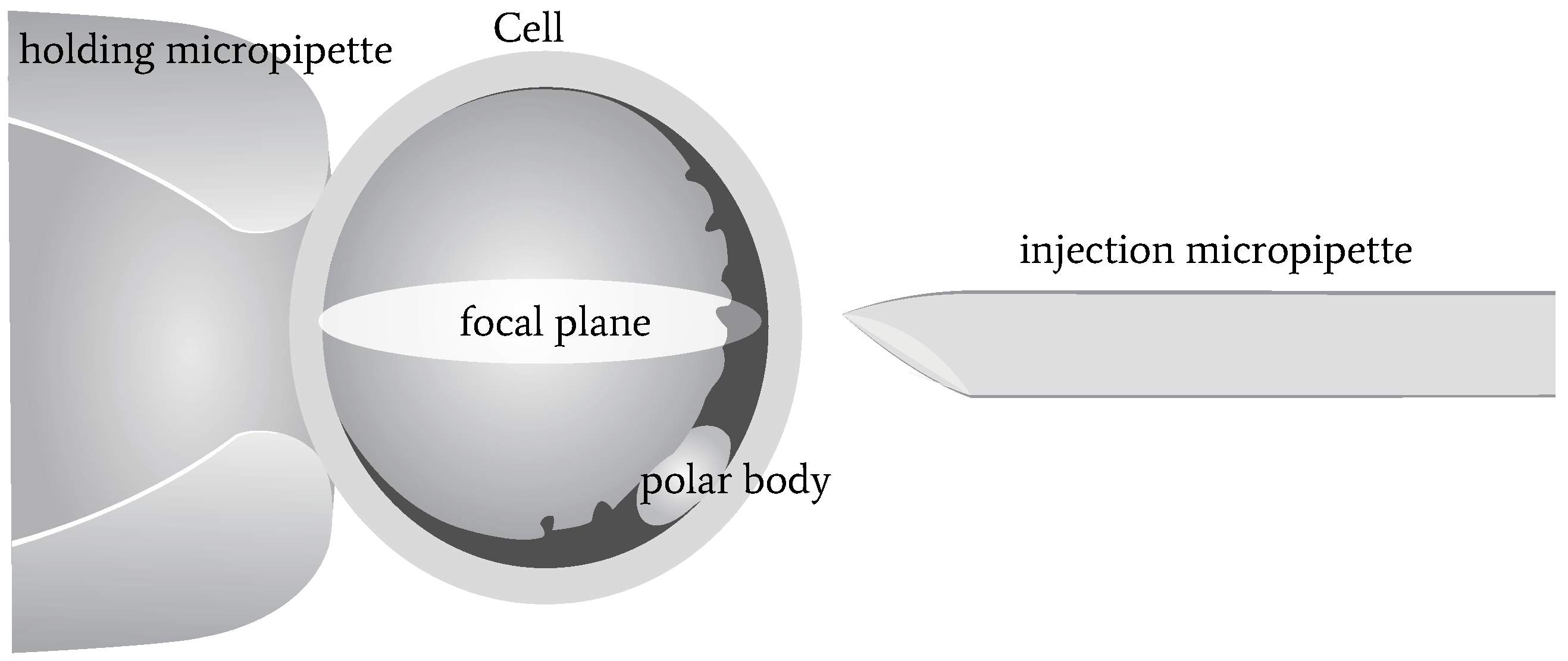
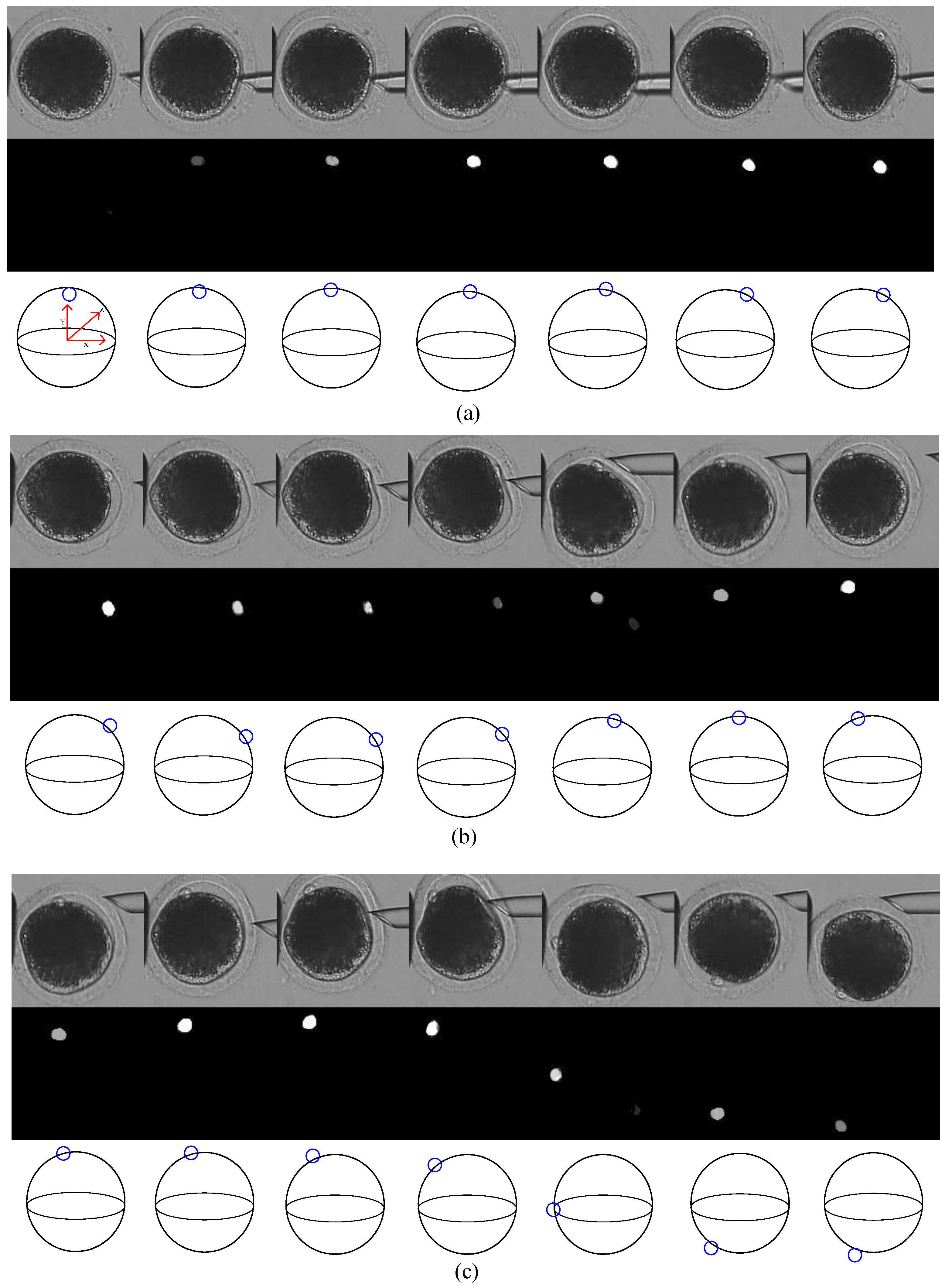
1]. Usually, people use polar-body position to localize the nucleus, since the nucleus is not visible under a bright-field microscope. However, due to cytoplasm occlusion, the polar body can only be observed when it is located near the focal plane. Therefore, the polar body, as well as the oocyte, need to be rotated to the desired position in cell micromanipulations, such as somatic cell nuclear transfer (SCNT), intracytoplasmic sperm injection (ICSC) [
2], and polar-body biopsy [
1].
A three-dimensional illustration of oocyte rotation is shown in
Figure 1. The oocyte is held by a holding micropipette, and rotated by the injection micropipette. Usually, the rotation process can be divided into two stages: In the first stage, the polar body is rotated from being invisible to visible. At the end of this stage, the polar body is located near the focal plane. In the second stage, the polar body is rotated to the desired location, such as 2 or 4 o’ clock. During the whole rotation process, there are three difficulties in polar-body detection, as shown in
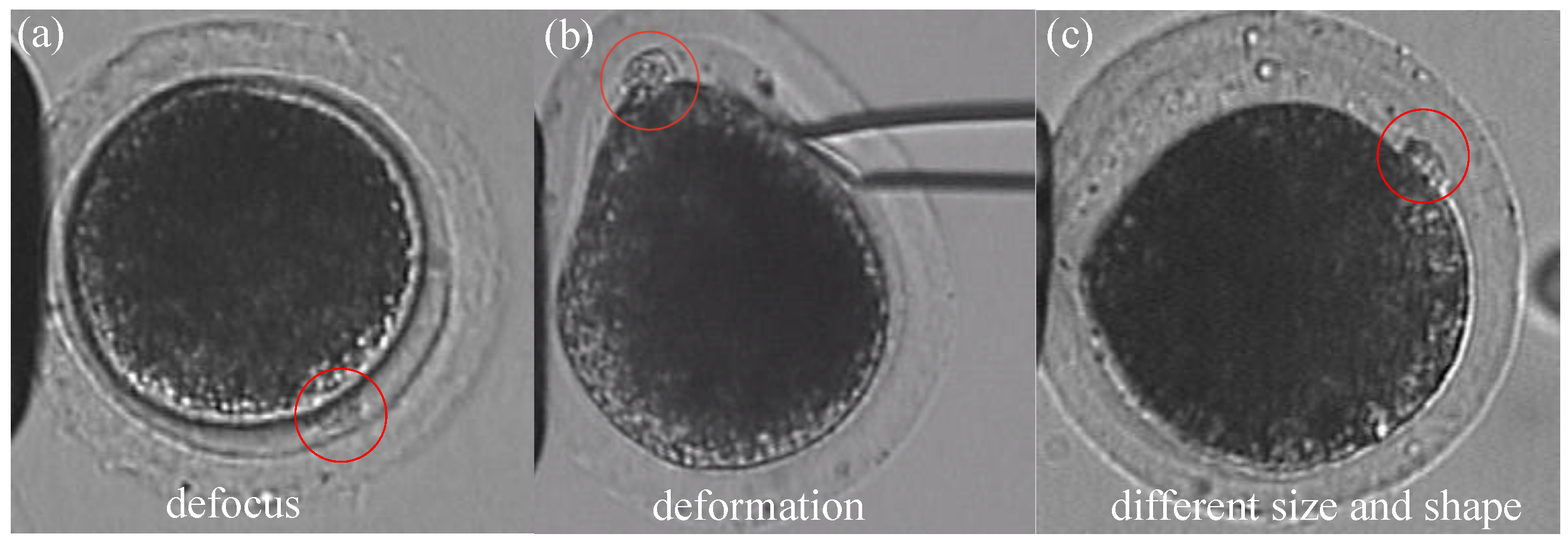
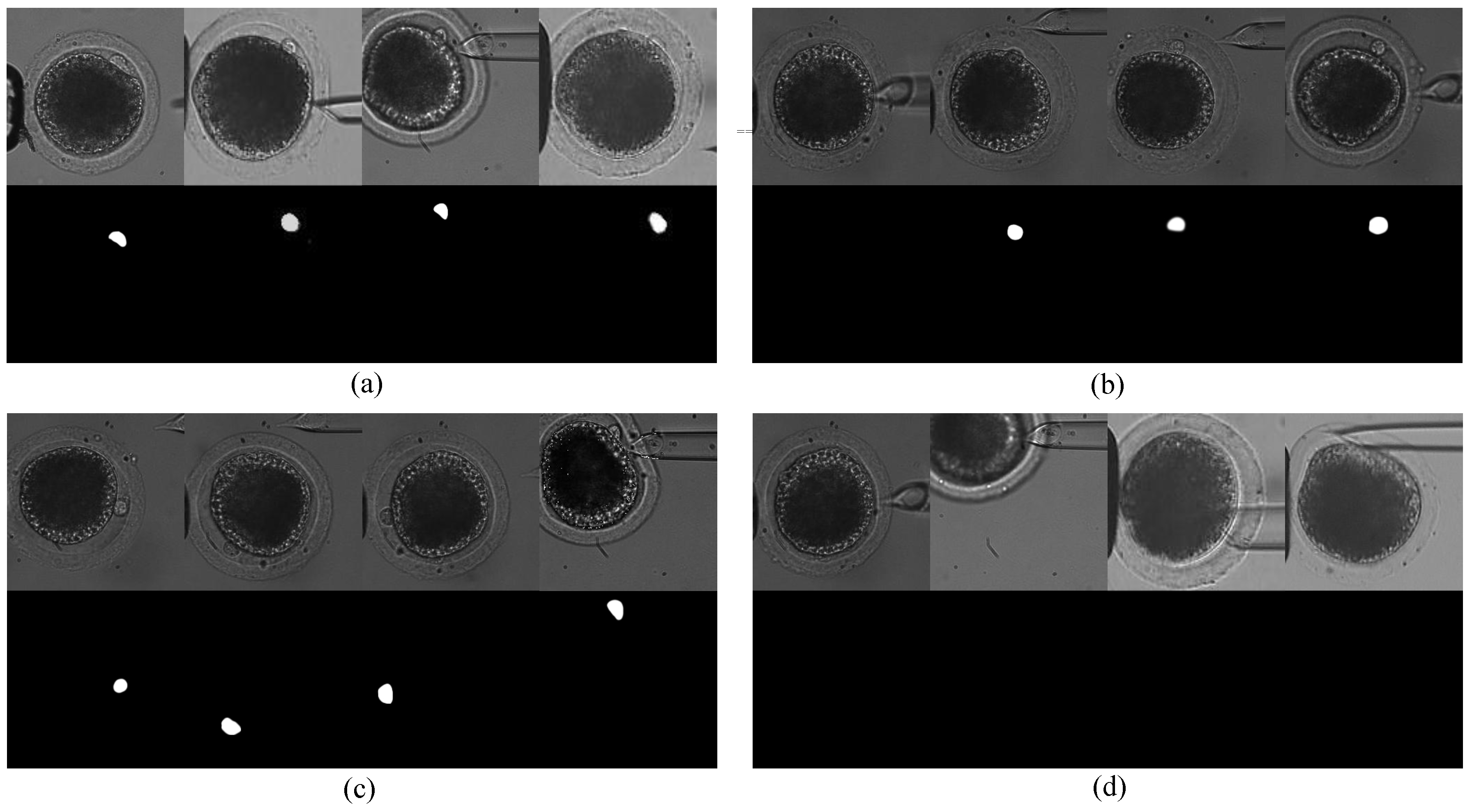
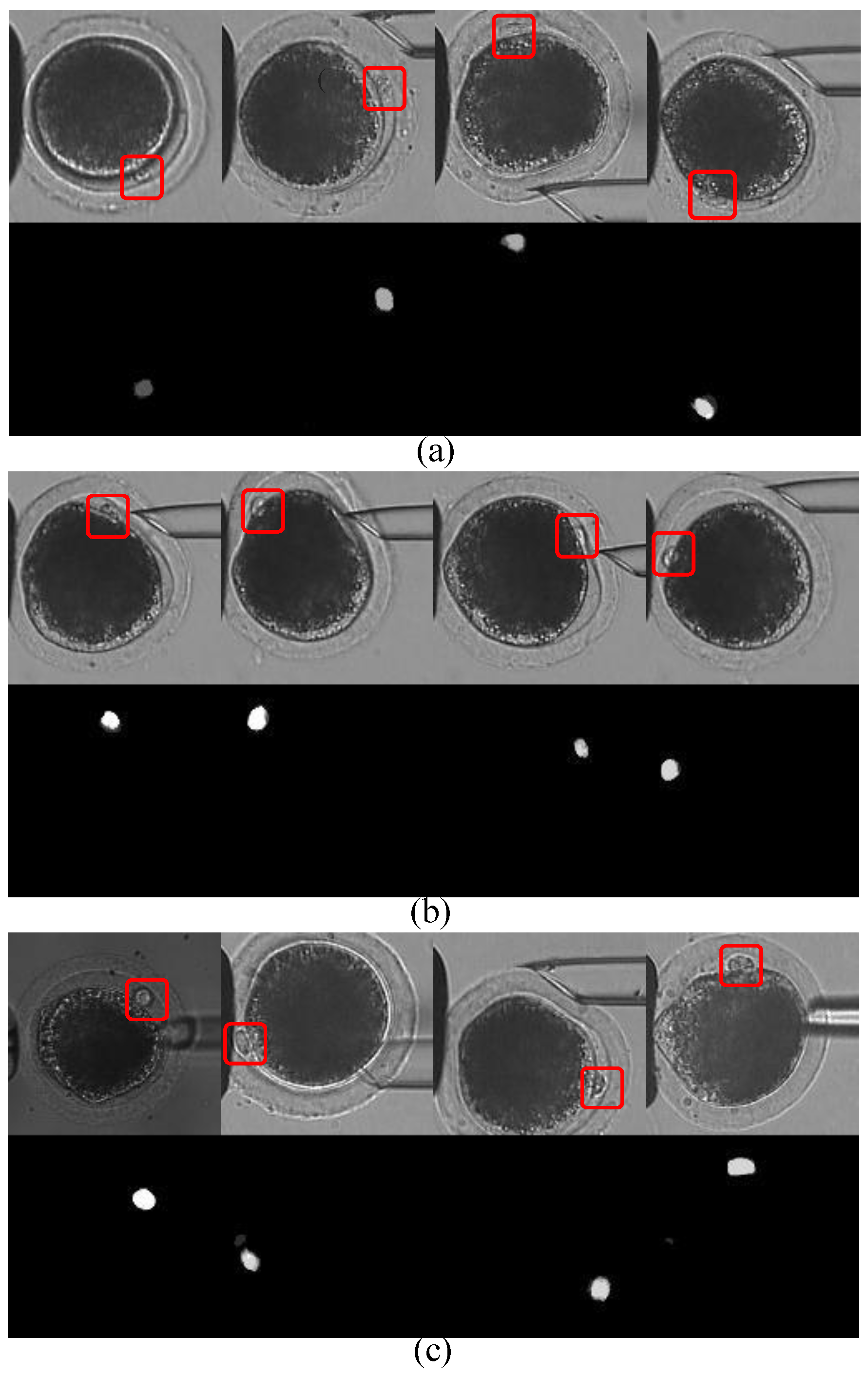
Figure 2: First, the polar body is invisible or defocused most of the time at the first stage. The visual detection of the presence of a polar body or detection in a defocused image is much more difficult than common detection. Second, due to contact with the injection pipette during cell rotation, large-cell deformation is usually generated, easily causing the apparent deformation of the polar body and leading to false detection. Third, polar bodies in different developmental states have different sizes, which requires the detector performing well in a different scale.
In previous methods, polar-body detection was studied as a detection problem at the image level. Leung et al. [
3] utilized image binarization and a circle-fitting algorithm to determine the presence of a polar body. Wang et al. [
4] introduced a texture-based method to obtain the position of polar bodies of different animals. Wang et al. [
5] employed image morphology and ellipse fitting to detect the polar body of a mouse oocyte. Chen et al. [
6] introduced a machine-learning method into polar-body detection that applied boundary-curvature information to predict the possible positions of the polar body, and then applied a support vector machine (SVM) algorithm [
7] for classifying the image patches in the possible positions.
There are some limitations to these approaches to detect a polar body during cell rotation. Most of the image-level polar-body detection methods focused on image samples with clear polar bodies, but cannot deal with other situations, such as defocusing or deformation. These methods usually employed shape information to predict the position, and used texture information to verify the prediction, due to the assumption that the polar body was a static object with a fixed shape and texture. However, shape-based detection methods, such as circle- or ellipse-fitting, are not suitable for the rotation process, since polar-body deformation is generated during cell rotation. Meanwhile, texture-based detection methods are not reliable for cell rotation either, since texture varies when a polar body is out of focus. Therefore, polar-body detection is still an unsolved problem on the experiment level, especially when the polar body is not obvious in the view.
Nowadays, deep learning has had great success in computer-vision tasks. Convolutional neural networks (CNN), which is a typical category in deep neural networks, use stacks of convolution layers to extract image features and transform data into very high dimensional and nonlinear spaces. Different CNN structures are shown to be very powerful in image-classification, -detection, and -segmentation tasks [
8,
9,
10,
11,
12,
13]. Deep-learning models have also become increasingly popular in biomedical-image processing, for example, dealing with images with different modalities such as CT, MRI, X-ray, and RGB [
14,
15,
16]. Among these models, U-net [
17] was designed to realize end-to-end biomedical-image segmentation. It can effectively segment touching objects with very few annotated images for training.
In this paper, we propose a deep-learning framework to realize cell rotation-oriented polar-body detection. Instead of using shape or texture as feature detector, our method learns the difference between a polar body and the background. In order to separate the two parts, the detection problem is interpreted as image segmentation. Specific configurations were designed for the three problems that arise from cell rotation. In order to overcome the out-of-focus problem, which is a convolving process, we utilized a CNN as the main feature detector, and chose U-net [
17] as the base framework for polar-body segmentation. Then, we improved the U-net by introducing kernels of different sizes for each layer to realize the detection of polar bodies in different sizes. CNN training needs to collect annotated images in different situations. Besides real experimental images, we also designed three kinds of image transformations to enlarge the dataset and simulate more cell-rotation situations, including cell- and polar-body deformation, so that the deformed polar body in cell rotation could be detected. Extensive experiments were performed to validate the effectiveness of the proposed method. The method had 98.7% accuracy on polar-body prediction for 1000 real porcine-oocyte images. The detection results in the whole rotation process with different oocytes show that our method overcomes the defocus and cell-deformation problems very well, meeting the requirement of automatic micromanipulations.
The rest of the paper is organized as follows: First, the polar-body-detection method is proposed in
Section 2. Then, experiment results are illustrated in
Section 3. Finally, the paper is concluded in
Section 4.
2. Deep-Learning-Based Polar-Body Detection
In this section, the proposed method is described in detail. The base segmentation network is illustrated in
Section 2.1, which was designed to overcome the defocused-image problem, and which generalizes to polar bodies with different sizes and shapes. Data augmentation for network training is described in
Section 2.2. Particular image transformation was utilized to simulate more cell-rotation situations and automatically augment the dataset. Finally, the overall flowchart of the polar-body detection method is described in
Section 2.3.
2.1. Segmentation Based on Convolutional Network
Instead of learning the shape or texture as the feature detector, we propose to learn the difference between polar body and background. The problem is interpreted as image segmentation, which segments the polar body from the cell. In optics, microscopic imaging and defocusing are defined as the convolution of the ideal image and point spread function (PSF), which motivates us to introduce a convolutional neural network into polar-body segmentation. We chose U-net, which is a typical CNN for medical-image segmentation, as the base framework, and improved the network for polar bodies in different sizes. By using the improved U-net, we predicted the possibility that each pixel belongs to the polar body region, instead of the original 0–1 pixel-wise classification. This design is more in line with a human-cognitive system, i.e., it would predict higher possibility when the polar body is obvious, and lower possibility when the object is not.
2.1.1. Network Architecture
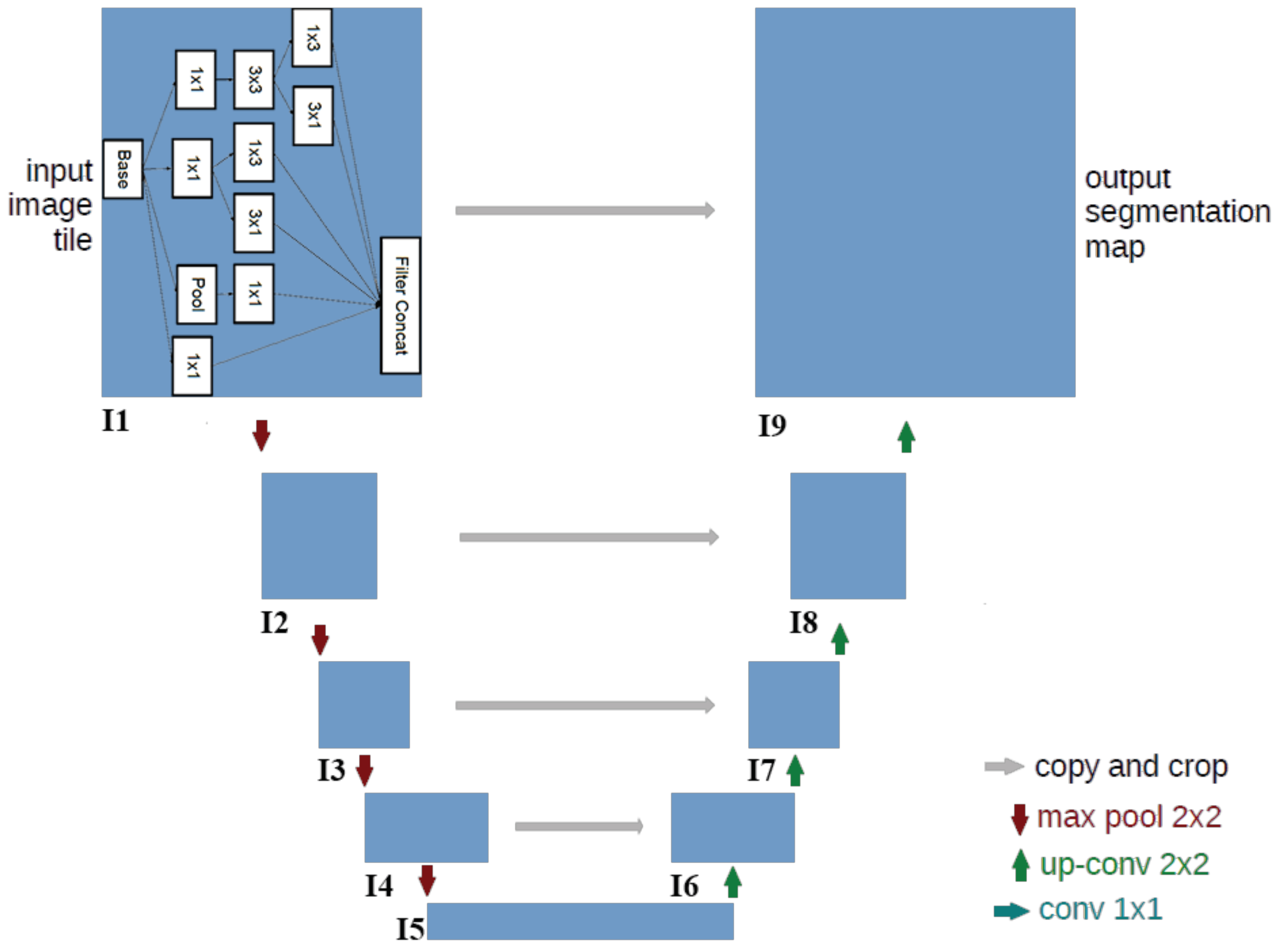
The architecture of the designed CNN for segmentation is illustrated in
Figure 3. The input of the network is the origin image. The output of the network is the segmented map in which the value of each pixel represents the probability that the pixel belongs to the polar-body region. The network includes many convolutional layers that are structured into nine modules represented by the blue rectangles. The modules are critical for detecting features.
Since the polar body is a local and small structure, which is quite different from the large and salient object of the image, only using traditional convolutional layers is not enough to segment the polar body. In order to realize the detection of polar bodies in diverse sizes, we utilized diverse kernels for each module. Thus, we introduced the inception module in the Inception-v3 [
18] network into U-net, as the module contains multiple small kernels that can scale to different sizes. The details of the module are shown in the I1 box in
Figure 3. The input data, which are represented in the base rectangle (array with shape represented by
), are fed into four branches: three
convolution layers and one pooling layer. The
convolution is used to reduce the
dimension while maintaining the layer’s spatial information (
). Then, different convolutional kernels, sized
,
,
,
are used to extract image features of objects in different scales. Finally, the data processed by the kernels are concatenated together in the
dimension and serve as the output layer of this module. Modules I2–I9 have the same structure.
The overall network has a contraction–expansion architecture in which the first four modules encodes the content information, while the last four modules decodes the location information. From I1 to I5, the spatial size (
) of the network layer reduces and the
increases, which means more filters are used to detect content information. The red arrows in
Figure 3 represent max-pooling, which can reduce the spatial size of the layer and extract the semantic feature. From I5 to I9, regularity is the opposite, and the spatial size is recovered to the original input-image size. Green arrows represent an upconvolution operation that operates on the opposite direction of the convolution. By using this operation, the network restores the image features and enlarges layer size. Segmentation requires precise location information that is preserved in lower-level image features. Therefore, to combine the low- and high-level image features, skip connections (gray arrows) are utilized to concatenate the symmetrical layers of both the convolution layer and the upconvolution layer of the same level.
2.1.2. Loss Function for Network Training
Deep-learning methods require a loss function that represents the optimization direction. In this paper, we designed two kinds of loss functions to train the neural network: segmentation loss and a classification loss.
The first is a segmentation loss called dice coefficient, which is a global loss for the whole image and used to measure the degree of overlapping between ground truth and our predicted result. In Equation (
1),
represents the predicted segmentation mask of the network, and
represents the corresponding reference mask, i.e., the ground truth. The overlapping of the predicted mask and the ideal mask is measured by the ratio between the intersection and union of two masks. The value of this coefficient is between 0 to 1, the larger, the better. However, for network optimization, we need to minimize loss, so the ratio is subtracted by 1.
The second is a classification loss, which is a local loss to measure the correctness of each pixel, and maintain the fineness of the segmented map. The predicted value for each pixel can be considered as the probability that the pixel belongs to the polar-body region, i.e., 1 represents the pixel is predicted as foreground, and 0 represents the pixel that is predicted as background. To realize the classification task, we used categorical cross-entropy loss. As shown in Equation (
2),
N represents the number of pixels in the image,
represents the value for each input pixel, and
represent the expected ground-truth value for this pixel.
h represent the operation of the network.
Both losses measure the difference between the predict mask and the ideal target mask.
focuses on the outline of the mask, while
measures the correctness of all the pixels of the image, no matter if it is in the outline or not. The pixel-level measure helps us to realize more accurate segmentation. To balance the two losses, we also designed different weights (1 and
) for each loss, and combined them to train the whole network. Through an experiment, we chose the coefficients for
and
to be 1 and 0.5, respectively, as shown in Equation (
3). Based on
, we can use a back-propagation algorithm to train the network.
2.2. Data Collection and Augmentation for CNN Training
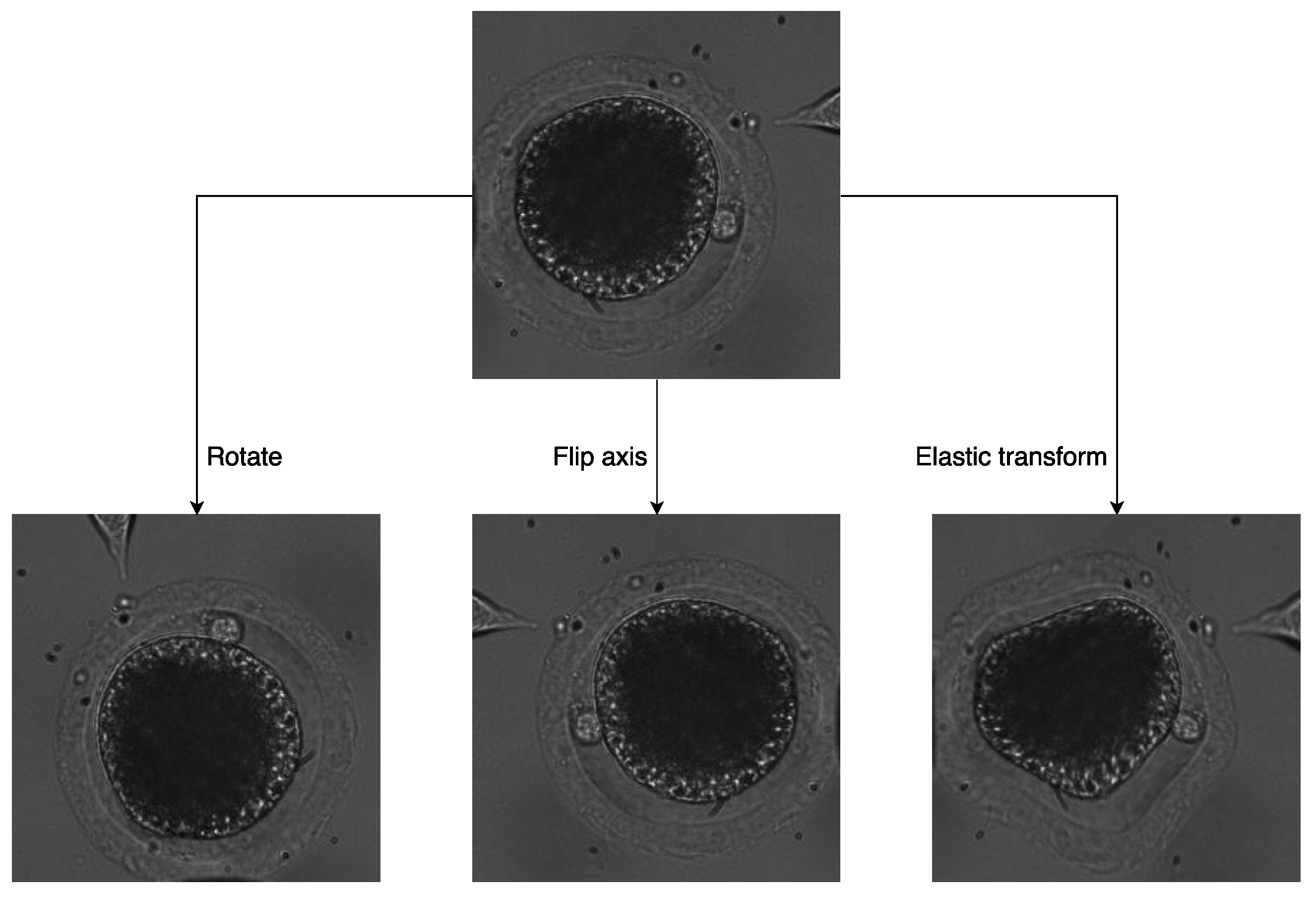
During cell rotation, polar bodies can be observed at different positions in various situations. Especially when the injection micropipette comes into contact with the oocyte, cell deformation generates and finally leads to polar-body deformation. This problem is hard to overcome through network design. Usually, studies improve algorithm performance by data augmentation for network training. In this paper, we designed three particular kinds of image-transformation methods to simulate more situations in the rotation process.
First, two kinds of image-transformation methods were utilized to simulate oocyte rotation without deformation. For oocytes rotating in the focal plane, we used random-image rotation with angles ranging from −90 to 90 to transform the image. For oocytes rotating vertically to the focal plane, image-axis flipping was used to transform the image.
In addition, elastic transform [
19] was applied to simulate more deformation situations. The transform was created by generating a displacement matrix of pixels, i.e., computing a new target location with respect to the original location for every pixel. The process of the elastic transform is listed below:
Generate random displacement fields for each pixel:
Convolve displacement fields with a Gaussian of standard deviation
and mean value
:
Generate deformed image according to the new displacements on original image:
Examples of the three transformations of the same image are shown in
Figure 4.
2.3. Polar-Body Detection Process
The polar-body detection method is divided into two parts: CNN training, and polar-body position prediction. In training stage, the network operates on the collected and augmented dataset, and uses the loss values as feedback to update its parameters, until the loss value reaches a lower value, which means the network has been trained well.
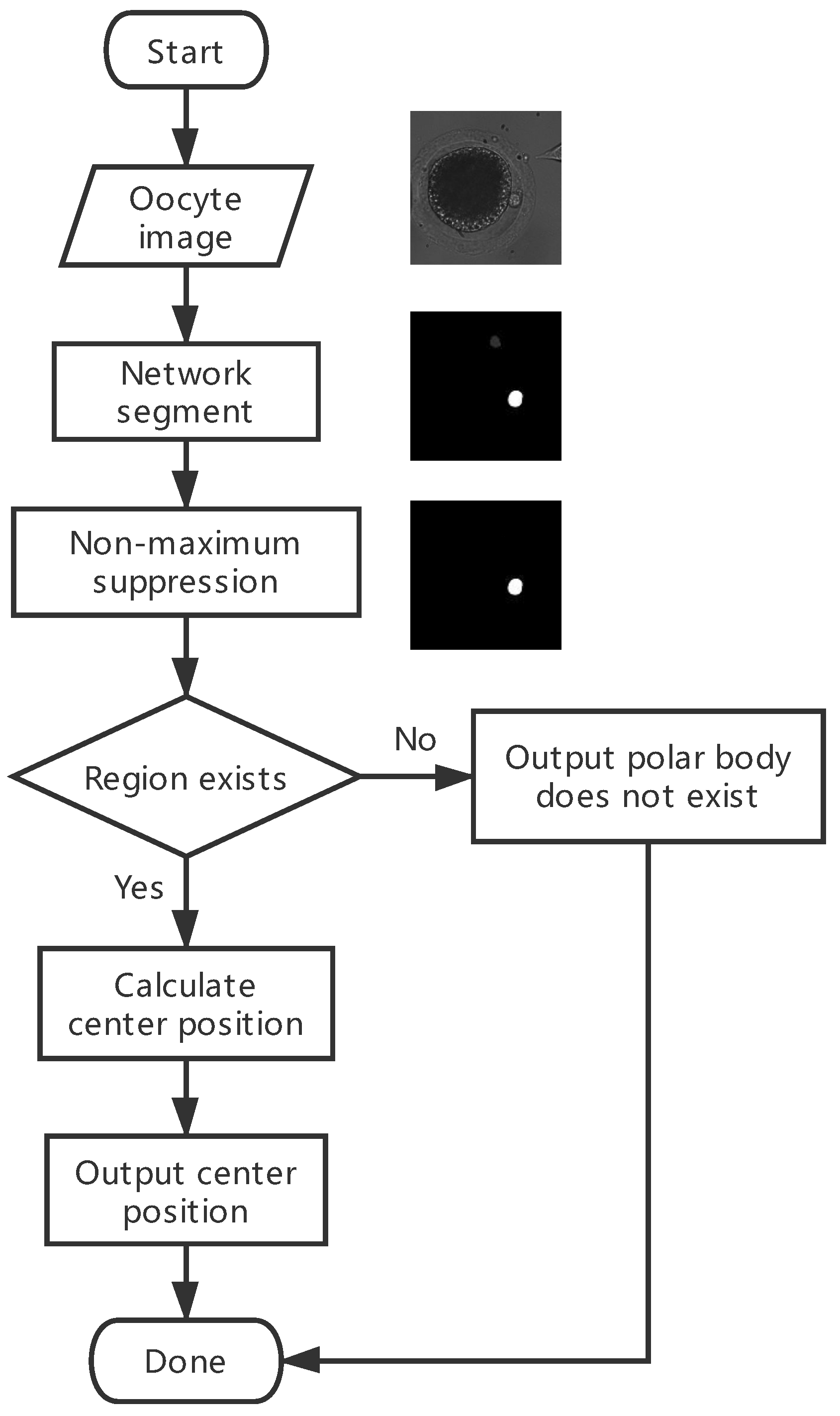
Figure 5 shows the flowchart of the prediction stage. Detailed steps are listed as follows:
Feed the oocyte image (referred to as the first picture in the right) into the trained network; then, a segmented map in black and white is acquired (the second picture).
Perform nonmaximum suppression to obtain the most area of the polar body possible. Most of the time, the segmented map has multiple connected regions that represent all possible areas of the polar body. Nonmaximum suppression is used to suppress the areas of lower possibilities. In non-maximum suppression, two constraints are utilized. First is the maximum value in the region. We set the threshold to 0.5 and ignored the regions with a max value lower than 0.5, as the pixel value of the segmented result represents the possibility. Second is the threshold of pixel numbers for the area. After that, areas of the satisfied regions are calculated.
Judge if there is a satisfied region. If yes, calculate the mean value of all pixel locations in this region as the center of the polar body; otherwise, put out that there is no polar body in the image.
4. Conclusions
In this paper, we proposed a framework to realize polar-body detection for automatic cell manipulation. Different from previous image-level detection methods, we aim to overcome problems in a real cell-rotation process, including out of focus, deformation, and different sizes of polar bodies. In our method, we first converted detection into image segmentation with a deep convolutional network, and then improved the network by introducing small and different-sized convolutional kernels, so that the method could be applied to detect defocused polar bodies and polar bodies in different sizes. Moreover, we designed three kinds of image-transformation methods to augment the training dataset for the network and simulate more cell-rotation situations, so that the deformed polar body in cell rotation would be detected.
Extensive experiments were performed on 1000 images of 74 porcine oocytes. The accuracies on defocused images, deformation images, and images of oocytes in different development states are 98.4%, 97.1%, and 98.6%, respectively. The overall accuracy of 1000 images was 98.7%, higher than all pervious methods. Our method also performed well during cell rotation, which meets the requirement of automatic biological cell-manipulation tasks. Our method costs 0.10 s for each image on the GPU server, while the industrial computer that is now used for cell manipulations is slower than the GPU server. Therefore, we still need to improve the speed of the algorithm and add GPU into the industrial computer to apply the method in real-time cell manipulations in the future.