A Rapid Assay to Detect Toxigenic Penicillium spp. Contamination in Wine and Musts
Abstract
:1. Introduction
2. Results
2.1. Set up of Experimental Design
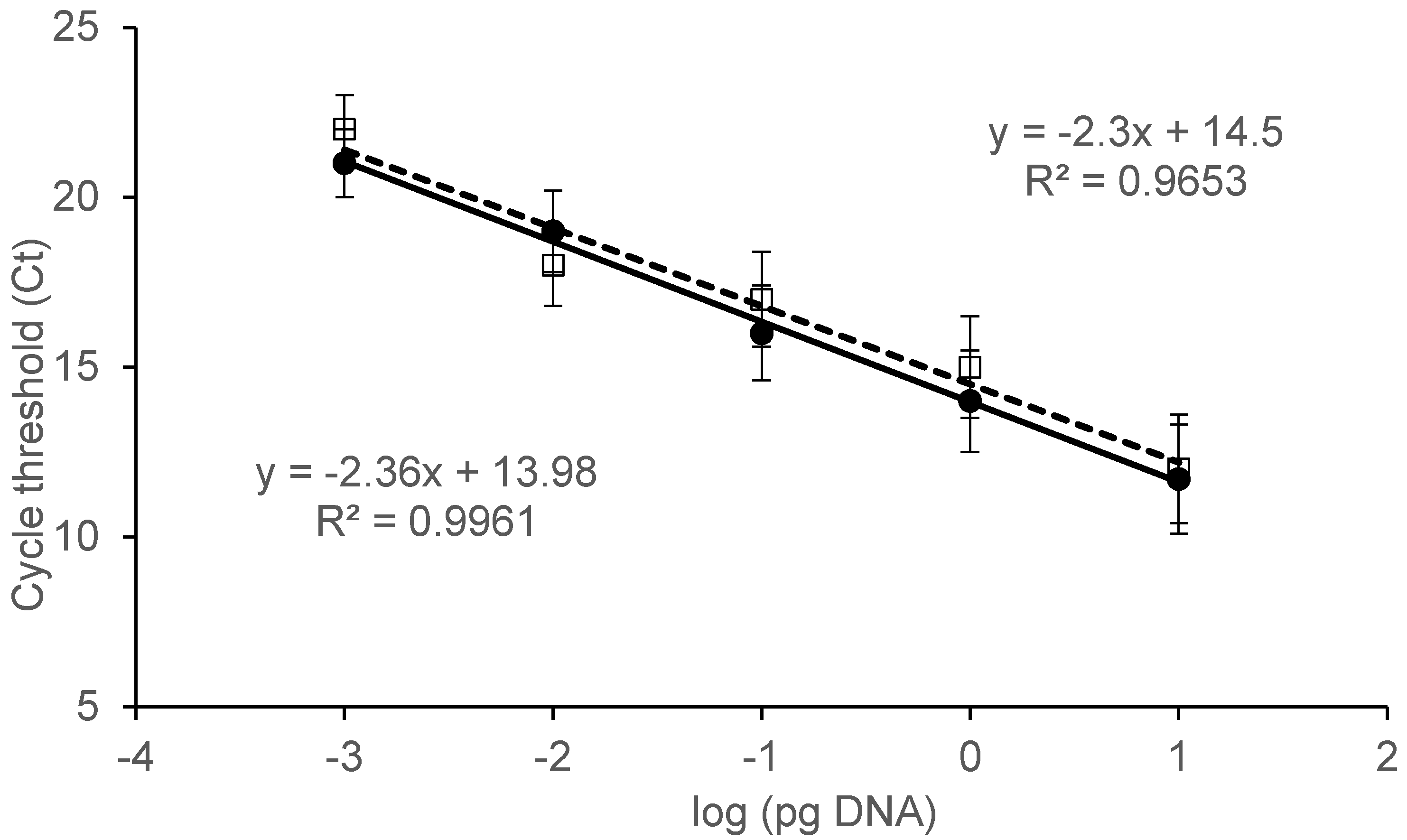
2.2. Specificity and Sensitivity Assay
2.3. Penicillium Detection in Real Samples
2.4. Patulin Quantification in Real Samples
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Sample Collection
5.2. DNA Extraction
5.3. Penicillium Detection System
5.4. Nested One-Tube Real-Time PCR Assays
5.5. Specificity and Sensitivity Assay
5.6. Patulin Evaluation
5.6.1. Chemicals and Reagents
5.6.2. Extraction Procedure
5.6.3. Chromatographic and Mass Spectrometric Conditions
5.7. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- FAOSTAT (The Food and Agriculture Organization Corporate Statistical Database). Statistics on Agriculture, Nutrition, Fisheries, Forestry, Food Aid, Land Use and Population. 2012. Available online: http://faostat.fao.org/ (accessed on 18 January 2016).
- European Commission. Regulation n. 479/2008. 2008. Available online: http://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2008:148:0001:0061:IT:PDF (accessed on 18 January 2016).
- Majerus, P.; Hain, J.; Kölb, C. Patulin in grape must and new, still fermenting wine (Federweißer). Mycotoxin Res. 2008, 24, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, K. Occurrence of ochratoxin A in commodities and processed food—A review of EU occurrence data. Food Addit. Contam. 2005, 1, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Sanzani, S.M.; Reverberi, M.; Fanelli, C.; Ippolito, A. Detection of ochratoxin A using molecular beacons and real-time PCR thermal cycler. Toxins 2015, 7, 812–820. [Google Scholar] [CrossRef] [PubMed]
- Díaz, G.A.; Yañez, L.; Latorre, B.A. Low occurrence of patulin-producing strains of Penicillium in grapes and patulin degradation during winemaking in Chile. Am. J. Enol. Vitic. 2011, 62, 542–546. [Google Scholar] [CrossRef]
- Picco, A.M.; Rodolfi, M. Assessments of indoor fungi in selected wineries of Oltrepo Pavese (Northern Italy) and Sottoceneri (Switzerland). Am. J. Enol. Vitic. 2004, 55, 355–362. [Google Scholar]
- Frisvad, J.C.; Smedsgaard, J.; Larsen, T.O.; Samson, R.A. Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud. Mycol. 2004, 49, 201–241. [Google Scholar]
- Sanzani, S.M.; Montemurro, C.; di Rienzo, V.; Solfrizzo, M.; Ippolito, A. Genetic structure and natural variation associated with host of origin in Penicillium expansum strains causing blue mould. Int. J. Food Microbiol. 2013, 165, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, J. The toxicological hazards of patulin. BIBRA Bull. 1993, 32, 3–4. [Google Scholar]
- WHO. Evaluation of certain food additives and contaminants. In 44th Report of the Joint FAO/WHO Expert Committee on Food Additives; Technical Report Series; WHO: Geneva, Switzerland, 1995; Volume 859, pp. 36–38. [Google Scholar]
- European Commission. Commission Regulation (EC) No 1881/2006 setting maximum levels for certain contaminants in foodstuffs. Off. J. Eur. Union 2006, L364, 5–24. [Google Scholar]
- Ministry of Health, Labour and Welfare of Japan (MHLW). Notification No. 369 (26 November 2003). Available online: http://www.mhlw.go.jp/english/ (accessed on 20 January 2016).
- Paterson, R.R.M.; Archer, S.; Kozakiewicz, Z.; Lea, A.; Locke, T.; O’Grady, E. A gene probe for the patulin metabolic pathway with potential for use in patulin and novel disease control. Biocontrol Sci. Technol. 2000, 10, 509–512. [Google Scholar] [CrossRef]
- Paterson, R.R.M. The isoepoxydon dehydrogenase gene of patulin biosynthesis in cultures and secondary metabolites as candidate PCR inhibitors. Mycol. Res. 2004, 108, 1431–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dombrink-Kurtzman, M.A. The sequence of the isoepoxydon dehydrogenase gene of the patulin biosynthetic pathway in Penicillium species. Antonie van Leeuwenhoek 2007, 91, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Sanzani, S.M.; Reverberi, M.; Punelli, M.; Ippolito, A.; Fanelli, C. Study on the role of patulin on pathogenicity and virulence of Penicillium expansum. Int. J. Food Microbiol. 2012, 153, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, A.; Luque, M.I.; Andrade, M.J.; Rodríguez, M.; Asensio, M.A.; Córdoba, J.J. Development of real-time PCR methods to quantify patulin-producing molds in food products. Food Microbiol. 2011, 28, 1190–1199. [Google Scholar]
- Tannous, J.; Atoui, A.; El Khoury, A.; Kantar, S.; Chdid, N.; Oswald, I.P.; Puel, O.; Lteif, R. Development of a real-time PCR assay for Penicillium expansum quantification and patulin estimation in apples. Food Microbiol. 2015, 50, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Bergerová, E.; Brežná, B.; Kuchta, T. A novel method with improved sensitivity for the detection of peanuts based upon single-tube nested real-time polymerase chain reaction. Eur. Food Res. Technol. 2011, 232, 1087–1091. [Google Scholar] [CrossRef]
- Costa, J.; Mafra, I.; Kuchta, T.; Oliveira, M.B.P.P. Single-tube nested real-time PCR as a new highly sensitive approach to trace hazelnut. J. Agric. Food Chem. 2012, 60, 8103–8110. [Google Scholar] [CrossRef] [PubMed]
- Costa, J.; Oliveira, M.B.P.P.; Mafra, I. Novel approach based on single-tube nested real-time PCR to detect almond allergens in foods. Food Res. Int. 2013, 51, 228–235. [Google Scholar] [CrossRef]
- Serey, R.A.; Torres, R.; Latorre, B.A. Pre- and post-infection activity of new fungicides against Botrytis cinerea and other fungi causing decay of table grapes. Cien. Investig. Agric. 2007, 34, 215–224. [Google Scholar] [CrossRef]
- Lorenzini, M.; Mainente, F.; Zapparoli, G.; Cecconi, D.; Simonato, B. Post-harvest proteomics of grapes infected by Penicillium during withering to produce Amarone wine. Food Chem. 2016, 199, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Tosi, E.; Fedrizzi, B.; Azzolini, M.; Finato, F.; Simonato, B.; Zapparoli, G. Effects of noble rot on must composition and aroma profile of Amarone wine produced by the traditional grape withering protocol. Food Chem. 2012, 130, 370–375. [Google Scholar] [CrossRef]
- Lorenzini, M.; Azzolini, M.; Tosi, E.; Zapparoli, G. Postharvest grape infection of Botrytis cinerea and its interactions with other moulds under withering conditions to produce noble-rotten grapes. J. Appl. Microbiol. 2013, 114, 762–770. [Google Scholar] [CrossRef] [PubMed]
- Lachhab, N.; Sanzani, S.M.; Bahouaoui, M.A.; Boselli, M.; Ippolito, A. Effect of some protein hydrolysates against gray mould of table and wine grapes. Eur. J. Plant Pathol. 2015, 144, 821–830. [Google Scholar] [CrossRef]
- Schena, L.; Li Destri Nicosia, M.G.; Sanzani, S.M.; Faedda, R.; Ippolito, A.; Cacciola, S.O. Development of quantitative PCR detection methods for phytopathogenic fungi and oomycetes. J. Plant Pathol. 2013, 95, 7–24. [Google Scholar]
- Kullman, B.; Tamm, H.; Kullman, K. Fungal Genome Size Database. 2005. Available online: http://www.zbi.ee/fungal-genomesize (accessed on 18 January 2016).
- SCOOP-Report. Assessment of Dietary Intake of Patulin by Population of the EU Member States (Task 3.2.8). 2002. Available online: http://ec.europa.eu/food/fs/scoop/3.2.8_en.pdf (accessed on 22 January 2016).
- Fredlund, E.; Gidlund, A.; Sulyok, M.; Börjesson, T.; Krska, R.; Olsen, M.; Lindblad, M. Deoxynivalenol and other selected Fusarium toxins in Swedish oats—Occurrence and correlation to specific Fusarium species. Int. J. Food Microbiol. 2013, 167, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Moake, M.M.; Padilla-Zakour, O.I.; Worobo, R.W. Comprehensive review of patulin control methods in foods. Compr. Rev. Food Sci. Food Saf. 2005, 4, 8–21. [Google Scholar] [CrossRef]
- Sanzani, S.M.; Susca, A.; Mastrorosa, S.; Solfrizzo, M. Patulin risk associated with blue mould of pome fruit marketed in southern Italy. Qual. Assur. Saf. Crops Food 2016. [Google Scholar] [CrossRef]
- Andersen, B.; Smedsgaard, J.; Frisvad, J.C. Penicillium expansum: Consistent production of patulin, chaetoglobosins, and other secondary metabolites in culture and their natural occurrence in fruit products. J. Agric. Food Chem. 2004, 52, 2421–2428. [Google Scholar] [CrossRef] [PubMed]
- Di Rienzo, V.; Miazzi, M.M.; Fanelli, V.; Savino, V.; Pollastro, S.; Colucci, F.; Miccolupo, A.; Blanco, A.; Pasqualone, A.; Montemurro, C. An enhanced analytical procedure to discover table grape DNA adulteration in industrial musts. Food Control 2016, 60, 124–130. [Google Scholar] [CrossRef]
- Sanzani, S.M.; Schena, L.; De Cicco, V.; Ippolito, A. Early detection of Botrytis cinerea latent infections as a tool to improve postharvest quality of table grapes. Postharvest Biol. Technol. 2012, 68, 64–71. [Google Scholar] [CrossRef]
- Mehndiratta, M.; Palanichamy, J.K.; Ramalingam, P.; Pal, A.; Das, P.; Sinha, S.; Chattopadhyay, P. Fluorescence acquisition during hybridization phase in quantitative real-time PCR improves specificity and signal-to-noise ratio. BioTechniques 2008, 45, 625–634. [Google Scholar] [CrossRef] [PubMed]

| Isolate Code | Organism | Presence of Amplification Product |
|---|---|---|
| Pex6 | Penicillium expansum | + |
| Pex29 | Penicillium chrysogenum | + |
| Pex30 | Penicillium crustosum | + |
| A64 | Alternaria alternata | - |
| FV52 | Botrytis cinerea | - |
| FV509 | Monilia laxa | - |
| FV139 | Phellinus ignarius | - |
| FV366 | Sclerotinia sclerotiorum | - |
| FV155 | Cladosporium spp. | - |
| FV406 | Aspergillus spp. | - |
| FV150 | Rhizopus stolonifer | - |
| FV126 | Fusarium spp. | - |
| VV1 | Vitis vinifera | - |
| Sample Code | Type | Storage | DNA Concentration (pg/μL) |
|---|---|---|---|
| C10 | White must | Cistern | - |
| C11a | White wine | Cistern | 1.007 |
| C11b | White must | Cistern | - |
| C12a | Red must | Cistern | 0.010 |
| C12b | White must | Cistern | - |
| C13 | White must | Cistern | - |
| C14 | White must | Cistern | - |
| C15 | White must | Cistern | - |
| C19 | White must | Cistern | - |
| C21 | White must | Cistern | - |
| C22 | White must | Cistern | - |
| C23 | White must | Cistern | - |
| C35 | White wine | Cistern | - |
| C43 | White must | Cistern | - |
| C47 | White must | Cistern | - |
| C48 | White must | Cistern | - |
| C52 | White must | Cistern | - |
| C53 | Red must | Cistern | - |
| C55 | Red must | Cistern | 0.016 |
| C56 | Red must | Cistern | - |
| C57 | Red must | Cistern | 0.003 |
| C59 | Red must | Cistern | 0.006 |
| SS5 | White must | Silos | 0.011 |
| SS8 | Red must | Silos | 0.029 |
| SS10 | Red wine | Silos | 1.0074 |
| SS13 | Red wine | Silos | - |
| SS14 | Red must | Silos | 0.010 |
| SS15 | White must | Silos | 0.002 |
| SS17 | Red wine | Silos | 0.002 |
| SS19 | Red wine | Silos | - |
| SS21 | White wine | Silos | 0.002 |
| SS25 | Red must | Silos | 1.96 |
| SS26 | Red must | Silos | 2.634 |
| SS27 | Red must | Silos | - |
| SS28 | Red must | Silos | - |
| SS29 | Red must | Silos | 0.014 |
| SS33 | White must | Silos | 0.003 |
| SS34 | White must | Silos | 0.010 |
| SS36 | White wine | Silos | - |
| SS39 | White must | Silos | 0.056 |
| SS42 | Red must | Silos | - |
| SS44 | Red wine | Silos | - |
| SS45 | White wine | Silos | 0.002 |
| SS47 | Red wine | Silos | - |
| SS48 | Red wine | Silos | - |
| SS51 | Red must | Silos | 0.056 |
| SS52 | Red must | Silos | - |
| SS73 | White wine | Silos | - |
| SS75 | Red wine | Silos | - |
| SS77 | Red wine | Silos | - |
| T2 | Red wine | Tank | 0.034 |
| T7 | Red must | Tank | 0.183 |
| T11 | Red must | Tank | - |
| T13 | Red must | Tank | 0.034 |
| T15 | Red must | Tank | - |
| T17 | White wine | Tank | 0.006 |
| T20 | Red wine | Tank | - |
| T21 | Red wine | Tank | 0.011 |
| T23 | Red wine | Tank | 0.010 |
| T24 | White wine | Tank | - |
| T25 | Red wine | Tank | - |
| T26 | Red wine | Tank | - |
| T27 | Red wine | Tank | - |
| T28 | Red wine | Tank | 0.065 |
| T31 | Red wine | Tank | 0.070 |
| T32 | White wine | Tank | 0.042 |
| T33 | Red wine | Tank | 0.010 |
| T35 | Red wine | Tank | 0.309 |
| T36 | Red wine | Tank | - |
| T38 | Red wine | Tank | 0.029 |
| T40 | White wine | Tank | 0.001 |
| T41 | White wine | Tank | 0.023 |
| T44 | Red wine | Tank | - |
| T45 | Red wine | Tank | - |
| T48 | White wine | Tank | 0.014 |
| T49 | Red wine | Tank | - |
| T51 | Red wine | Tank | - |
| T52 | Red must | Tank | - |
| T55 | Red must | Tank | 0.016 |
| T58 | Red wine | Tank | 0.010 |
| T70 | Red wine | Tank | 0.070 |
| Sample Code | Type | Penicillium DNA Concentration (pg/μL) | Patulin Concentration (μg/L) |
|---|---|---|---|
| SS8 | Red must | 0.029 | 0 |
| SS10 | Red wine | 1.0074 | 173 |
| SS14 | Red must | 0.010 | 277 |
| SS17 | Red wine | 0.002 | 397 |
| SS21 | White wine | 0.002 | 27 |
| SS29 | Red must | 0.014 | 154 |
| SS33 | White must | 0.003 | 778 |
| SS34 | White must | 0.010 | 0 |
| SS39 | White must | 0.056 | 65 |
| T13 | Red must | 0.034 | 60 |
| T17 | White wine | 0.006 | 1911 |
| T21 | Red wine | 0.011 | 712 |
| T35 | Red wine | 0.309 | 82 |
| T40 | White wine | 0.001 | 681 |
| T41 | White wine | 0.023 | 0 |
| T48 | White wine | 0.014 | 0 |
| T58 | Red wine | 0.010 | 669 |
| ID | Q1 Mass (Da) | Q3 Mass (Da) | Dwell (msec) | DP | EP | CE | CXP |
|---|---|---|---|---|---|---|---|
| Patulin 1 | 152.9 | 109.0 | 5.00 | −45.00 | −10.00 | −13.00 | −7.00 |
| Patulin 2 | 152.9 | 80.9 | 5.00 | −45.00 | −10.00 | −15.00 | −7.00 |
| Dinoseb 1 | 239.0 | 133.9 | 5.00 | −120.00 | −8.00 | −58.68 | −10.00 |
| Dinoseb 2 | 239.0 | 163.1 | 5.00 | −120.00 | −8.00 | −41.78 | −10.00 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanzani, S.M.; Miazzi, M.M.; Di Rienzo, V.; Fanelli, V.; Gambacorta, G.; Taurino, M.R.; Montemurro, C. A Rapid Assay to Detect Toxigenic Penicillium spp. Contamination in Wine and Musts. Toxins 2016, 8, 235. https://doi.org/10.3390/toxins8080235
Sanzani SM, Miazzi MM, Di Rienzo V, Fanelli V, Gambacorta G, Taurino MR, Montemurro C. A Rapid Assay to Detect Toxigenic Penicillium spp. Contamination in Wine and Musts. Toxins. 2016; 8(8):235. https://doi.org/10.3390/toxins8080235
Chicago/Turabian StyleSanzani, Simona Marianna, Monica Marilena Miazzi, Valentina Di Rienzo, Valentina Fanelli, Giuseppe Gambacorta, Maria Rosaria Taurino, and Cinzia Montemurro. 2016. "A Rapid Assay to Detect Toxigenic Penicillium spp. Contamination in Wine and Musts" Toxins 8, no. 8: 235. https://doi.org/10.3390/toxins8080235
APA StyleSanzani, S. M., Miazzi, M. M., Di Rienzo, V., Fanelli, V., Gambacorta, G., Taurino, M. R., & Montemurro, C. (2016). A Rapid Assay to Detect Toxigenic Penicillium spp. Contamination in Wine and Musts. Toxins, 8(8), 235. https://doi.org/10.3390/toxins8080235











