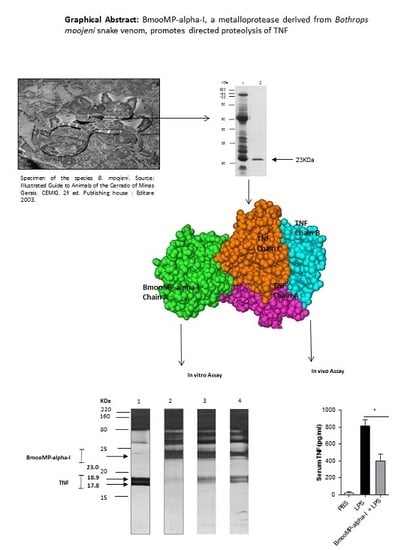
Interaction between TNF and BmooMP-Alpha-I, a Zinc Metalloprotease Derived from Bothrops moojeni Snake Venom, Promotes Direct Proteolysis of This Cytokine: Molecular Modeling and Docking at a Glance
Abstract
:1. Introduction
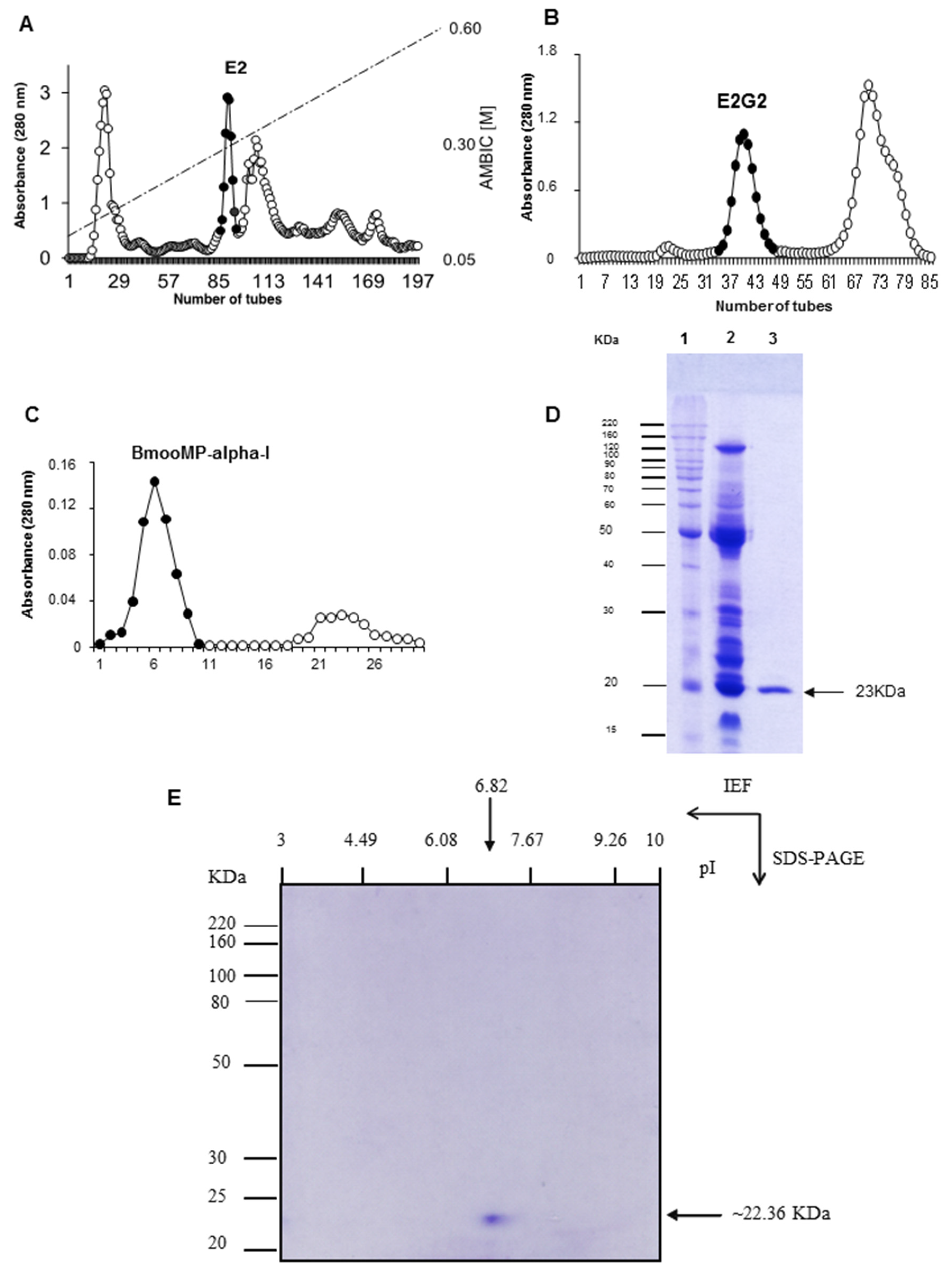
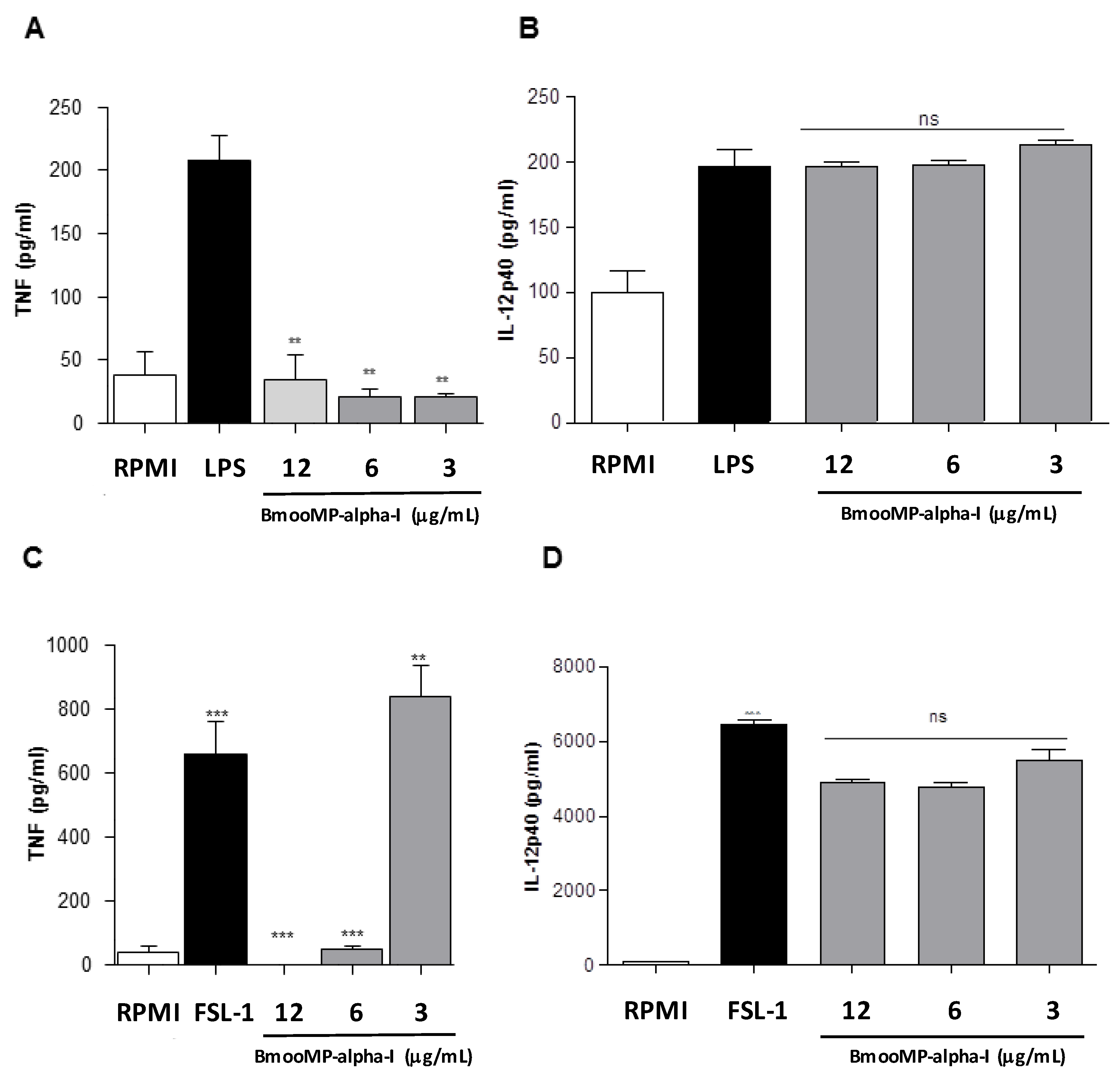
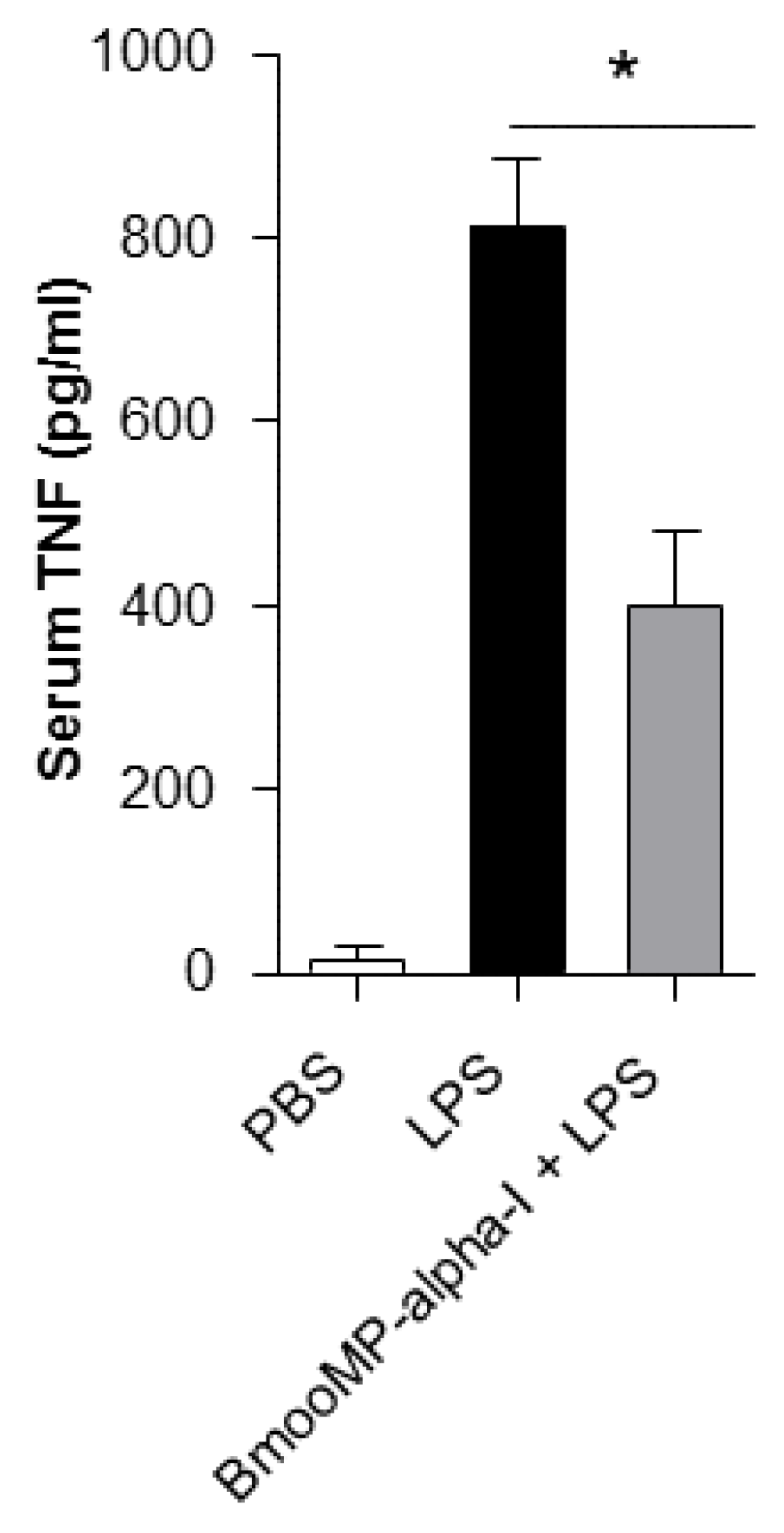
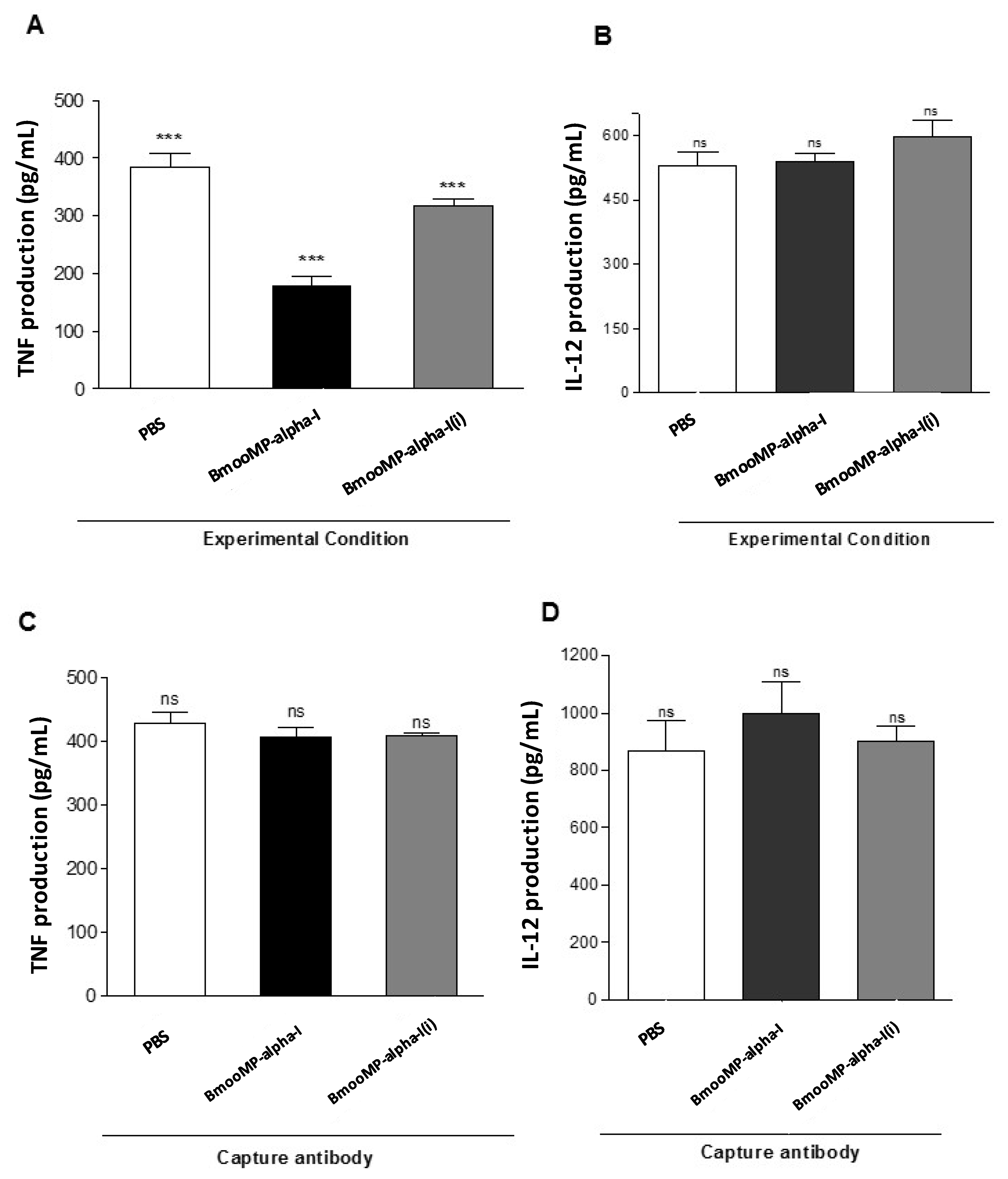
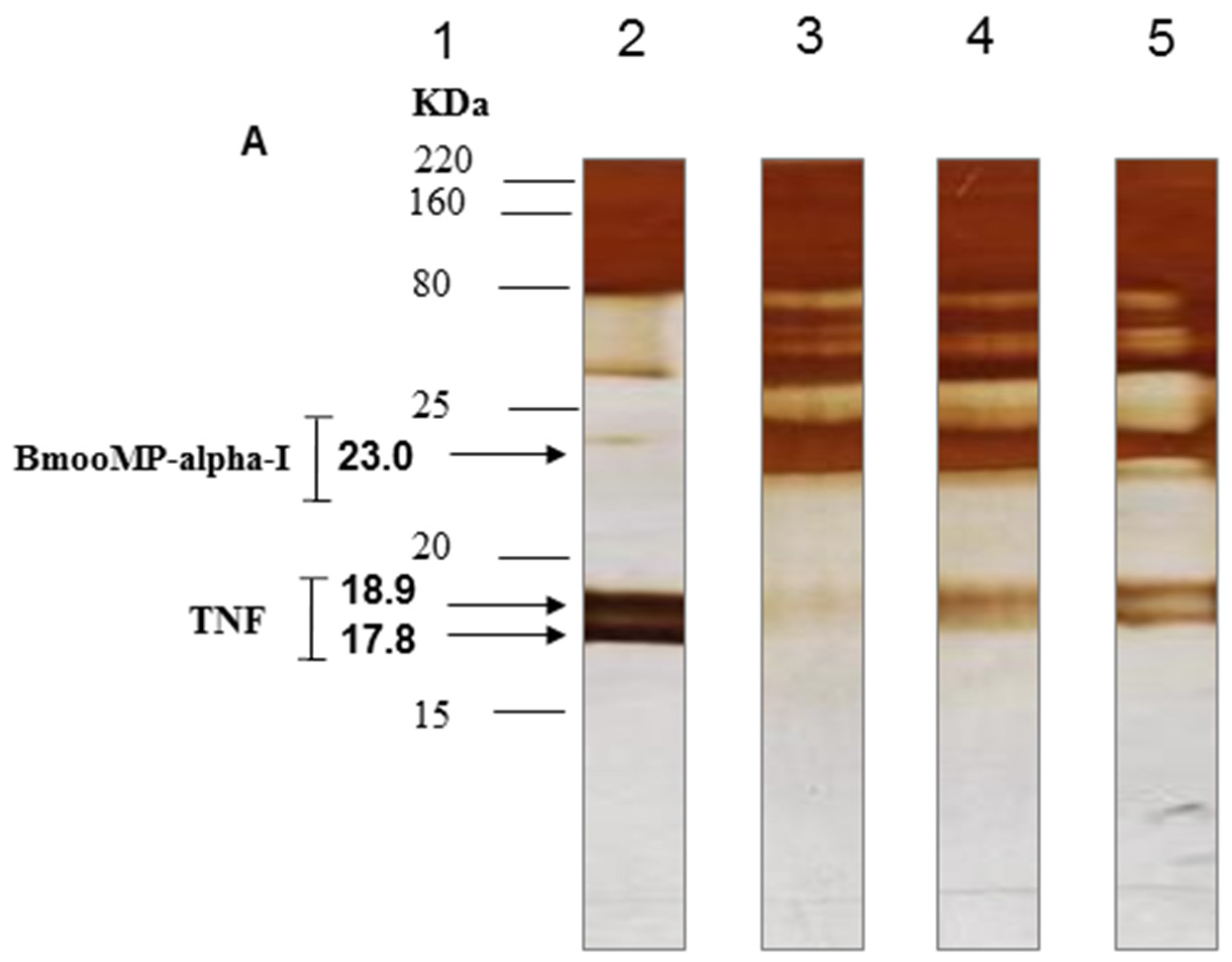
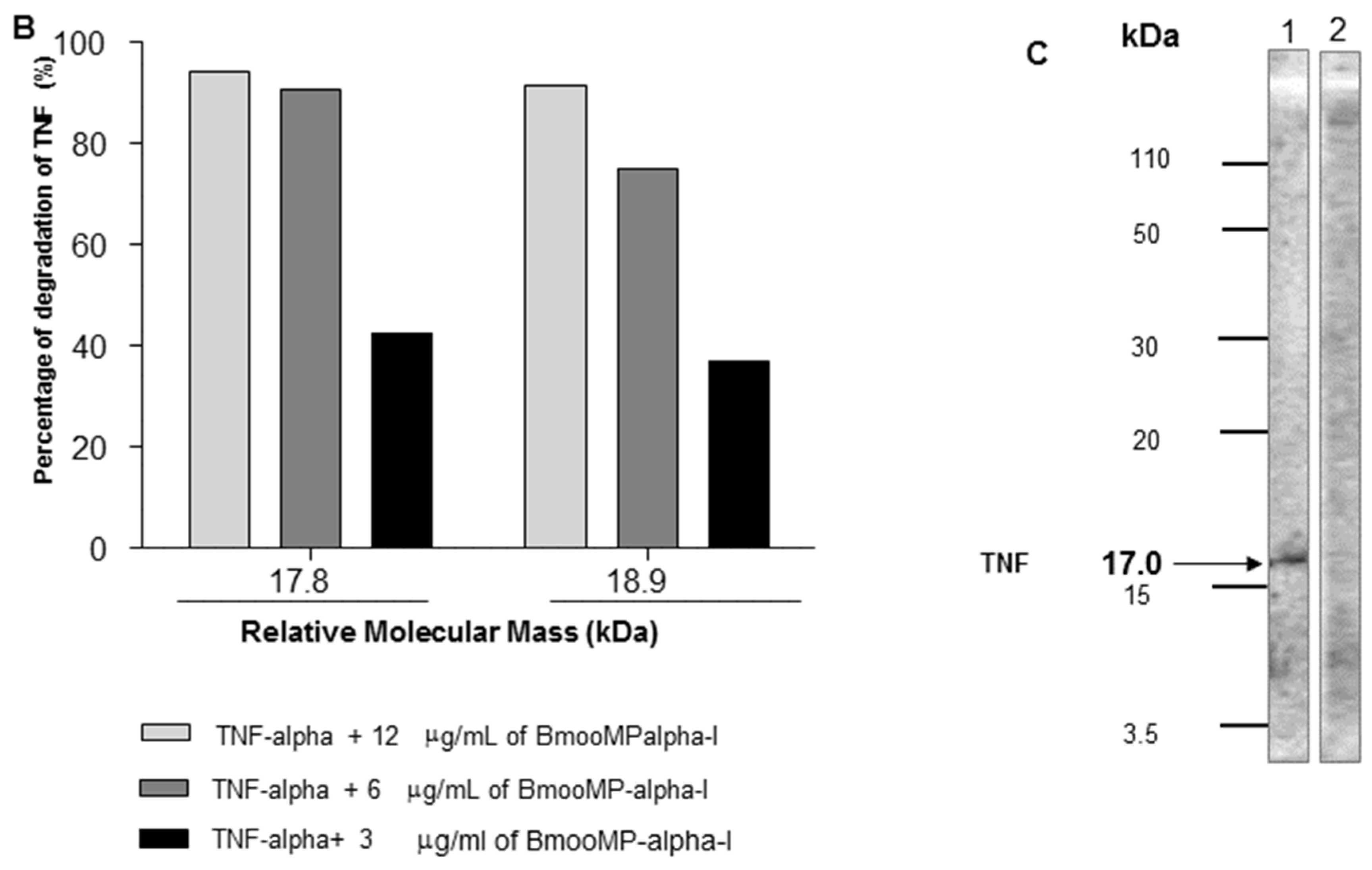
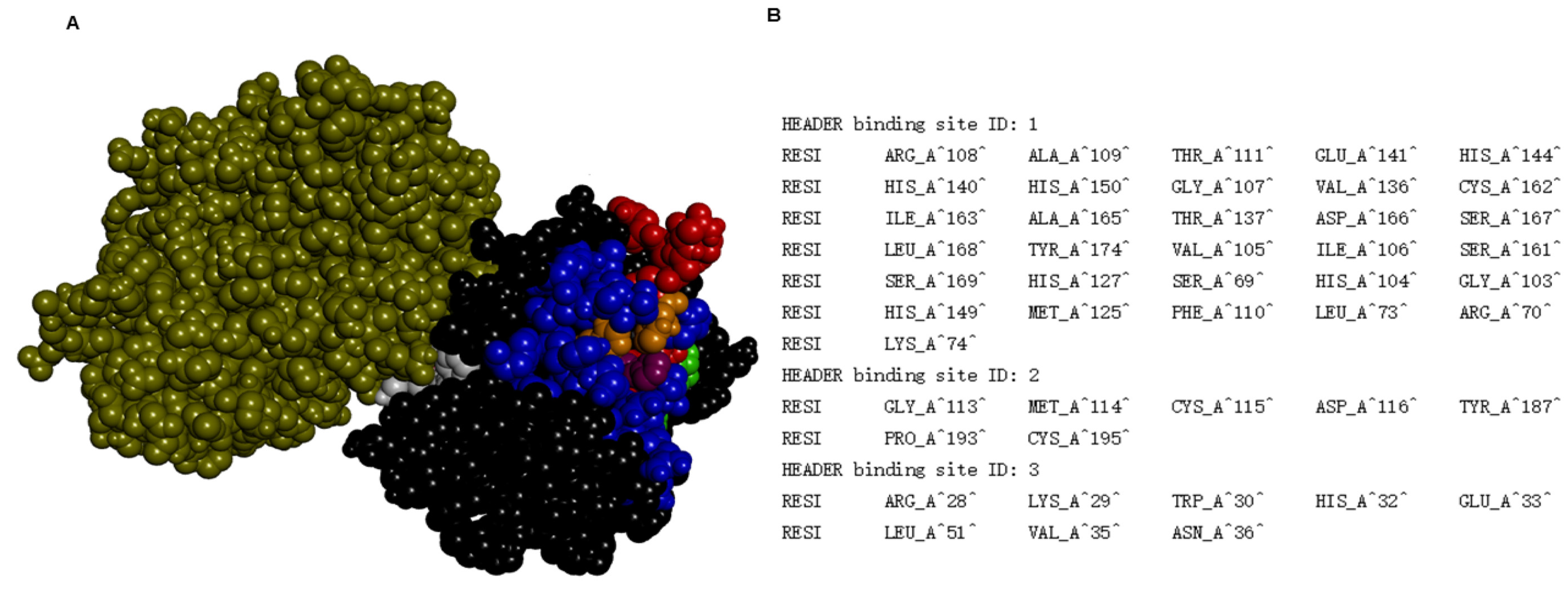
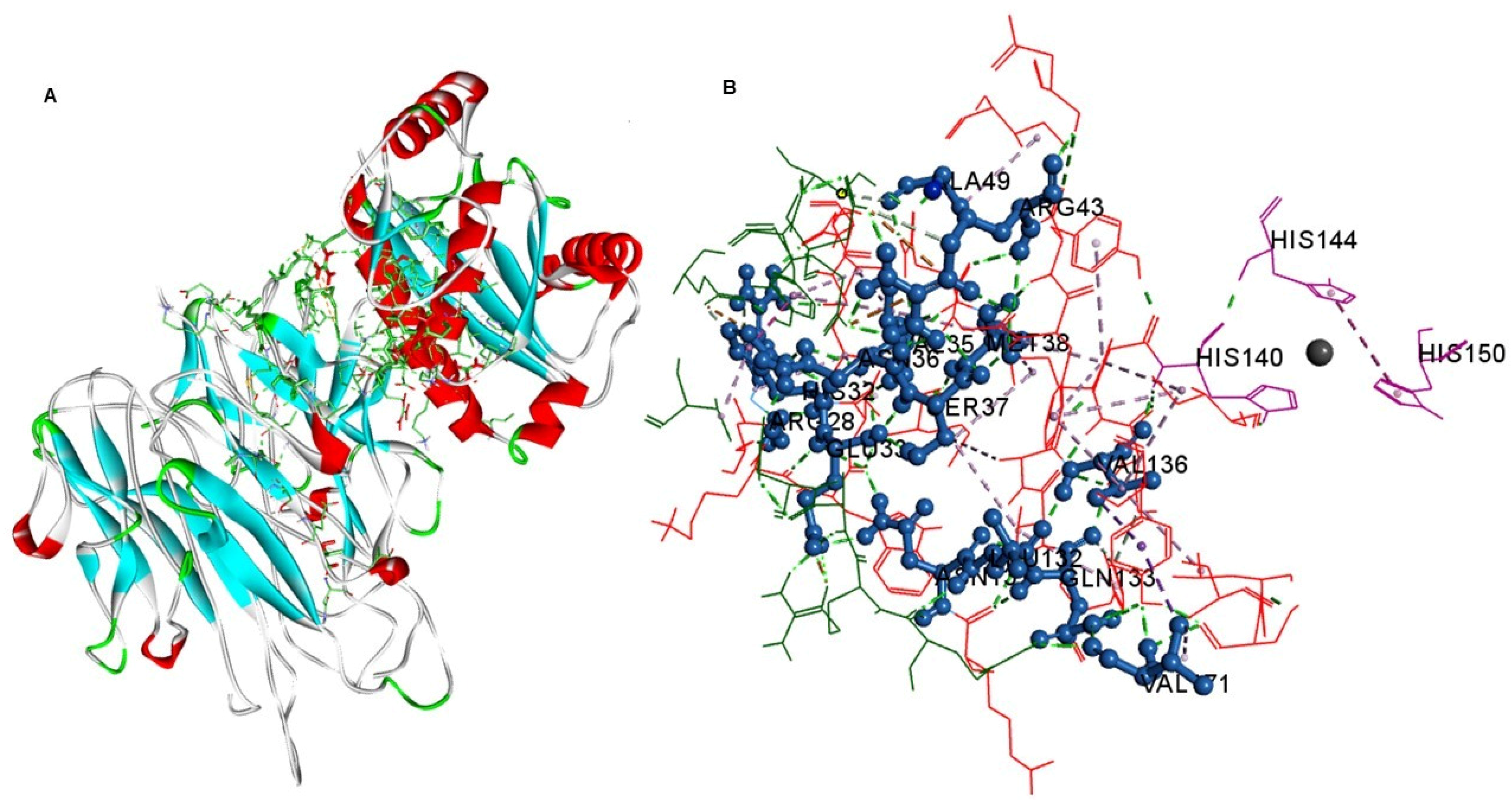
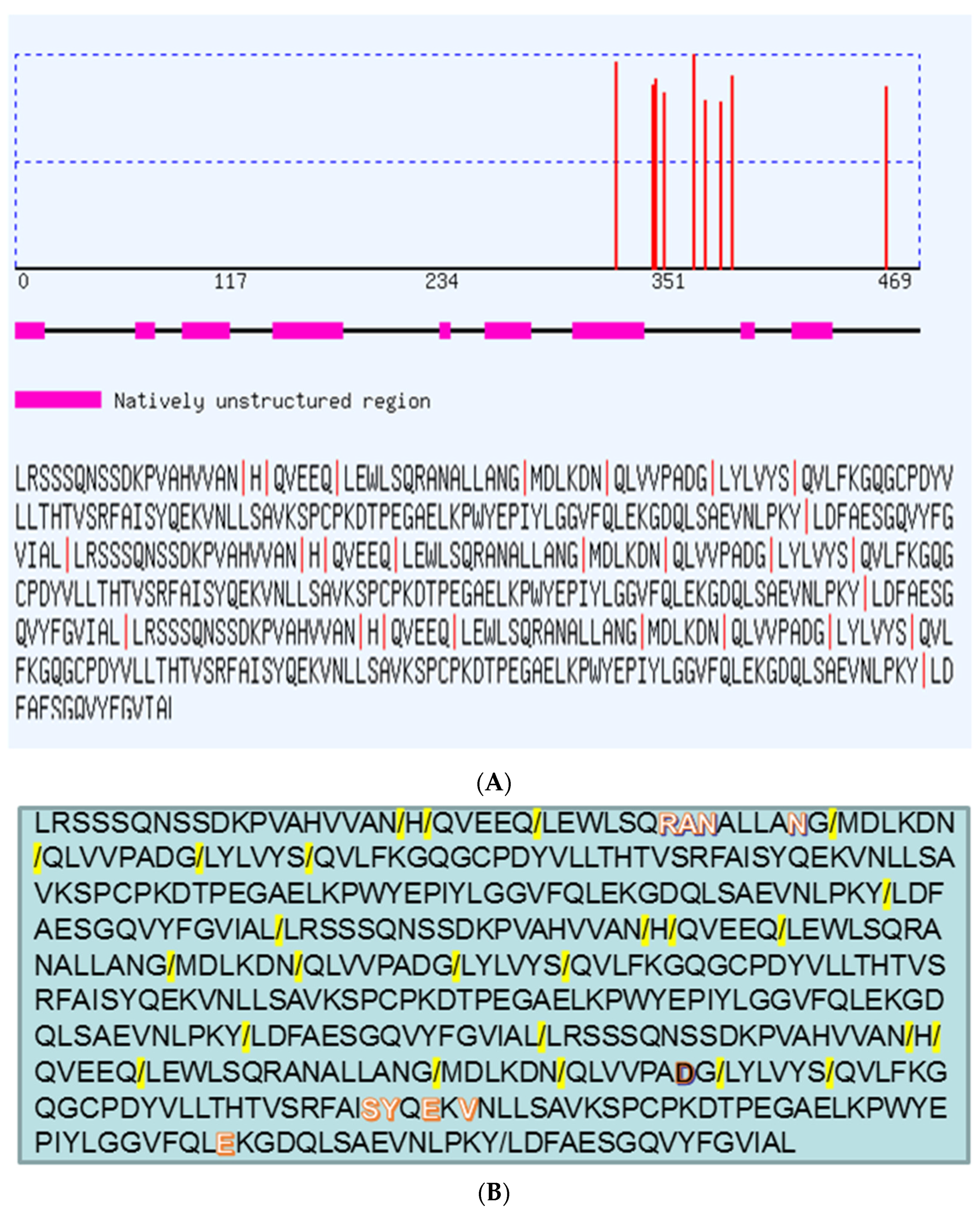
2. Results and Discussion
3. Experimental Section
3.1. Animals
3.2. Crude Venom and Toxin
3.3. Electrophoretic Analysis
3.4. In Vitro Model for Assessment of TNF Production
3.5. In Vivo Model for Assessment of TNF Production
3.6. Cytokine Measurements
3.7. Inhibitory Effect of BmooMP-Alpha-I on TNF Detection
3.8. SDS-PAGE to Assess Proteolytic Effect of BmooMP-Alpha-I on TNF Protein
3.9. Western Blotting for TNF
3.10. Molecular Docking
3.11. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contribution
Conflicts of Interest
Abbreviations
References
- Osta, B.; Benedetti, G.; Miossec, P. Classical and paradoxical effects of TNF-alpha on bone homeostasis. Front. Immunol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Sabio, G.; Davis, R.J. TNF and MAP kinase signalling pathways. Semin. Immunol. 2014, 26, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Wallach, D. The cybernetics of TNF: Old views and newer ones. Semin. Cell. Dev. Biol. 2016, 50, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Ramseyer, V.; Garvin, J.L. Tumor necrosis factor alpha: Regulation of renal function and blood pressure. Am. J. Physiol. Renal. Physiol. 2013, 304, 1231–1242. [Google Scholar] [CrossRef] [PubMed]
- Xixi, M.A.; Shengqian, X.U. TNF inhibitor therapy for rheumatoid arthritis. Biomed. Rep. 2013, 1, 177–184. [Google Scholar]
- Takeda, S.; Takeya, H.; Iwanaga, S. Snake venom metalloproteinases: Structure, function and relevance to the mammalian ADAM/ADAMTS family proteins. Biochim. Biophys. Acta 2012, 1824, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Paes Leme, A.F.; Sherman, N.E.; Smalley, D.M.; Sizukusa, L.O.; Oliveira, A.K.; Menezes, M.C.; Fox, J.W.; Serrano, S.M. Hemorrhagic activity of HF3, a snake venom metalloproteinase: Insights from the proteomic analysis of mouse skin and blood plasma. J. Proteome Res. 2012, 11, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Markland, F.S.; Swenson, S. Snake venom metalloproteinases. Toxicon 2013, 62, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Calderon, L.A.; Sobrinho, J.C.; Zaqueo, K.D.; de Moura, A.A.; Grabner, A.N.; Mazzi, M.V.; Nomizo, S.M.A.; Fernandes, C.F.C.; Zuliani, J.P.; Carvalho, B.A.; et al. Antitumoral activity of snake venom proteins: New trends in cancer therapy. BioMed Res. Int. 2014, 2014, 203639. [Google Scholar] [CrossRef] [PubMed]
- Barrett, A.J.; Rawlings, N.D.; Salvesen, G.; Woessner, J.F. Introduction. In Handbook of Proteolytic Enzymes; Rawlings, N.D., Salvesen, G., Eds.; Academic Press: Oxford, UK, 2013; pp. 1–4. [Google Scholar]
- Herrera, C.; Escalante, T.; Voisin, M.B.; Rucavado, A.; Morazá, D.; Macêdo, J.K.; Calvete, J.J.; Sanz, L.; Nourshargh, S.; Gutiérrez, J.M.; et al. Tissue localization and extracellular matrix degradation by PI, PII and PIII snake venom metalloproteinases: Clues on the mechanisms of venom-induced hemorrhage. PLoS Negl. Trop. Dis. 2015, 9, e0003731. [Google Scholar] [CrossRef] [PubMed]
- Bernardes, C.P.; Santos-Filho, N.A.; Costa, T.R.; Gomes, M.S.R.; Torres, F.S.; Costa, J.O.; Borges, M.H.; Richardsond, M.; Santos, D.M.; Pimenta, A.M.C.; et al. Isolation and structural characterization of a new Fibrin(ogen)olytic metalloproteinase from Bothrops moojeni snake venom. Toxicon 2008, 51, 574–584. [Google Scholar] [CrossRef] [PubMed]
- Akao, P.K.; Tonoli, C.C.C.; Navarro, M.S.; Cintra, A.C.O.; Neto, J.R.; Arni, R.K.; Murakami, M.T. Structural studies of BmooMP-alpha-I, a non-hemorrhagic metalloprotease from Bothrops moojeni venom. Toxicon 2010, 55, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Pithayanukul, P.; Leanpolchareanchai, J.; Saparpakorn, P. Molecular docking studies and anti-snake venom metalloproteinase activity of Thai mango seed kernel extract. Molecules 2009, 14, 3198–3213. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, F.; Rodrigues, V.M.; Borges, M.H.; Soares, A.M.; Hamaguchi, A.; Giglio, J.R.; Homsi-Brandeburgo, M.I. Purification and partial characterization of a new proteolytic enzyme from the venom of Bothrops moojeni (Caiçaca). Biochem. Mol. Biol. Int. 1999, 47, 1069–1077. [Google Scholar] [PubMed]
- Serrano, S.M.; Matos, M.F.; Mandelbaum, F.R.; Sampaio, C.A. Basic proteinases from Bothrops moojeni (Caissaca) venom-I. Isolation and activity of two serine proteinases, MSP 1 and MSP 2, on synthetic substrates and on platelet aggregation. Toxicon 1993, 31, 471–481. [Google Scholar] [CrossRef]
- Torres, F.S.; Rates, B.; Gomes, M.T.R.; Salas, C.E.; Pimenta, A.M.C.; Oliveira, F.; Santoro, M.M.; de Lima, M.E. Bmoo FIBMP-I: A new fibrinogenolytic metalloproteinase from Bothrops moojeni snake venom. ISRN Toxicol. 2012, 2012, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Allie, N.; Alexopoulou, L.; Quesniaux, V.J.F.; Fick, L.; Kranidioti, K.; Kollias, G.; Ryffel, B.; Muazzam, J. Protective role of membrane tumour necrosis factor in the host’s resistance to mycobacterial infection. Immunology 2008, 125, 522–534. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, A.; Rajoria, S.; George, L.A.; Mittelman, A.; Suriano, R. Synthetic Toll Like Receptor-4 (TLR-4) agonist peptides as a novel class of adjuvants. PLoS ONE 2012, 7, e30839. [Google Scholar] [CrossRef] [PubMed]
- Croft, M.; Duan, W.; Choi, H.; Eun, S.-Y.; Madireddi, S.; Mehta, A. TNF superfamily in inflammatory disease: Translating basic insights. Trend Immunol. 2012, 33, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Markland, F.S. Snake venoms and the hemostatic system. Toxicon 1998, 36, 1749–1800. [Google Scholar] [CrossRef]
- Cardoso, R.; Homsi-Brandeburgo, M.I.; Rodrigues, V.M.; Santos, W.B.; Souza, G.L.; Prudencio, C.R.; Siquieroli, A.C.; Goulart, L.R. Peptide mimicking antigenic and immunogenic epitope of neuwiedase from Bothrops neuwiedi snake venom. Toxicon 2009, 53, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Harrison, R.A.; Wusterb, W.; Theakston, R.D.G. The conserved structure of snake venom toxins confers extensive immunological cross-reactivity to toxin-specific antibody Theakstona. Toxicon 2003, 41, 441–449. [Google Scholar] [CrossRef]
- Wang, R.; Cai, J.; Huang, Y.; Xu, D.; Sang, H.; Yan, G. Novel recombinant fibrinogenase of Agkistrodon acutus venom protects against LPS-induced DIC. Thromb. Res. 2009, 123, 919–924. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Qiu, P.; Jiang, W.; Cai, X.; Ou, Y.; Su, X.; Cai, J.; Chen, J.; Yin, W.; Yan, G. Recombinant fibrinogenase from Agkistrodon acutus venom protects against sepsis via direct degradation of fibrin and TNF-alpha. Biochem. Pharmacol. 2008, 76, 620–630. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Wang, R.; Jiang, W.; Lin, X.; Qiu, P.; Yan, G. Novel recombinant snake venom metalloprotease from Agkistrodon acutus protects against taurocholate-induced severe acute pancreatitis in rats. Biochimie 2010, 92, 1354–1361. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.Y.; Zou, X. An iterative knowledge-based scoring function for protein-protein recognition. Proteins 2008, 72, 557–579. [Google Scholar] [CrossRef] [PubMed]
- Trellet, M.; Melquiond, A.S.J.; Bonvin, A.M. A unified conformational selection and induced fit approach to protein-peptide docking. PLoS ONE 2013, 8, e58769. [Google Scholar] [CrossRef] [PubMed]
- Krüger, D.M.; Garzón, J.I.; Chacón, P.; Gohlke, H. Drugscore PPI knowledge-based potentials used as scoring and objective function in protein-protein docking. PLoS ONE 2014, 9, e89466. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.; Pierce, B.; Mintseris, J.; Janin, J.; Weng, Z. Protein-protein docking benchmark version 3.0. Proteins 2008, 73, 705–709. [Google Scholar] [CrossRef] [PubMed]
- Apte, S.S.; Parks, W.C. Metalloproteinases: A parade of functions in matrix biology and and outlook for the future. Matrix Biol. 2015, 44–46, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.P.; Hansch, C. Matrix metalloproteinases (MMPs): Chemical-biological functions and (Q) SARs. Bioorg. Med. Chem. 2007, 15, 2223–2268. [Google Scholar] [CrossRef] [PubMed]
- Chellapandi, P. Structural, functional and therapeutic aspects of snake venom metalloproteinases. Mini-Rev. Org. Chem. 2014, 11, 28–44. [Google Scholar] [CrossRef]
- Anand, P.; Nagarajan, D.; Mukherjee, S.; Chandra, N. ABS-Scan: In silico alanine scanning mutagenesis for binding site residues in protein-ligand complex. F1000Research 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Morris, A.L.; MacArthur, M.W.; Hutchinson, E.G.; Thornton, J.M. Stereochemical quality of protein structure coordinates. Proteins Struct. Funct. Bioinform. 1992, 12, 345–364. [Google Scholar] [CrossRef] [PubMed]
- Ünlü, A. Computational prediction of actin-actin interaction. Mol. Biol. Rep. 2014, 41, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, D.N.; Kondo, M.Y.; Oliveira, L.C.; Honorato, R.V.; Zanphorlin, L.M.; Coronado, M.A.; Araújo, M.S.; Mottae, G.; Veroneze, C.L.; Andrade, S.S.; et al. P-I class metalloproteinase from Bothrops moojeni venom is a post-proline cleaving peptidase with kininogenase activity: Insights into substrate selectivity and kinetic behavior. Biochim. Biophys. Acta 2014, 1844, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Zelanis, A.; Huesgen, P.F.; Oliveira, A.K.; Tashima, A.K.; Serrano, S.M.; Overall, C.M. Snake venom serine proteinases specificity mapping by proteomic identification of cleavage sites. J. Proteom. 2015, 113, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Gunasekaran, K.; Ma, B.Y.; Nussinov, R. Is allostery an intrinsic property of all dynamic proteins? Proteins Struct. Funct. Bioinform. 2004, 57, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W.; Serrano, S.M. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon 2005, 45, 969–985. [Google Scholar] [CrossRef] [PubMed]
- Udi, Y.; Fragai, M.; Grossman, M.; Mitternacht, S.; Arad-Yellin, R.; Calderone, V.; Melikian, M.; Toccafondi, M.; Berezovsky, I.N.; Luchinat, C.; et al. Unraveling hidden regulatory sites in structurally homologous metalloproteases. J. Mol. Biol. 2013, 425, 2330–2346. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S. Structure-function relationship of modular domains of P-III class snake venom metalloproteinases. In Venom Genomics and Proteomics; Gopalakrishnakone, P., Calvete, J.J., Eds.; Springer International Publishing AG: Cham, Switzerland, 2016; pp. 185–209. [Google Scholar]
- Takeda, S. ADAM and ADAMTS family proteins and snake venom metalloproteinases: A structural overview. Toxins 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Mukaiyama, A.; Itoh, Y.; Nagase, H.; Thogersen, I.B.; Enghild, J.J.; Sasaguri, Y.; Mori, Y. Degradation of Interleukin 1-beta by matrix metalloproteinases. J. Biol. Chem. 1996, 271, 14657–14660. [Google Scholar] [PubMed]
- Sisto, M.; Lisi, S.; Lofrument, D.D.; Frassanito, M.A.; Cucci, L.; D’Amore, S.; Mitolo, V.; D’Amore, M. Induction of TNF-alpha-converting enzyme-ectodomain shedding by pathogenic autoantibodies. Int. Immunol. 2009, 12, 1341–1349. [Google Scholar]
- Mohammed, F.F.; Smookler, D.S.; Khokh, A.R. Metalloproteinases, inflammation, and rheumatoid arthritis. Ann. Rheum. Dis. 2003, 62, 43–47. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Marim, F.M.; Silveira, T.N.; Lima, D.S., Jr.; Zamboni, D.S. A method for generation of bone marrow-derived macrophages from cryopreserved mouse bone marrow cells. PLoS ONE 2010, 5, e15263. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 277, 680–689. [Google Scholar] [CrossRef]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [PubMed]
- RCSB Protein Data Bank. Available online: www.pdb.org (accessed on 10 June 2016).
- ModRefiner. Available online: http://zhanglab.ccmb.med.umich.edu/ModRefiner (accessed on 12 June 2016).
- Cluspro program. Available online: http://cluspro.bu.edu/login.php (accessed on 11 June 2016).
- MetaPocket 2.0. Available online: http://projects.biotec.tu-dresden.de/metapocket/index.php (accessed on 10 June 2016).
- The PDB sum platform. Available online: http://www.ebi.ac.uk/pdbsum (accessed on 9 June 2016).
- Drugscore PPI 2.2. Available online: http://cpclab.uni-duesseldorf.de/dsppi/main.php (accessed on 12 June 2016).
- Platform Prosper. Available online: https://prosper.erc.monash.edu.au/home.html (accessed on 10 June 2016).

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, M.C.; Lopes Silva, T.; Silva, M.V.; Mota, C.M.; Santiago, F.M.; Fonseca, K.C.; Oliveira, F.; Mineo, T.W.P.; Mineo, J.R. Interaction between TNF and BmooMP-Alpha-I, a Zinc Metalloprotease Derived from Bothrops moojeni Snake Venom, Promotes Direct Proteolysis of This Cytokine: Molecular Modeling and Docking at a Glance. Toxins 2016, 8, 223. https://doi.org/10.3390/toxins8070223
Silva MC, Lopes Silva T, Silva MV, Mota CM, Santiago FM, Fonseca KC, Oliveira F, Mineo TWP, Mineo JR. Interaction between TNF and BmooMP-Alpha-I, a Zinc Metalloprotease Derived from Bothrops moojeni Snake Venom, Promotes Direct Proteolysis of This Cytokine: Molecular Modeling and Docking at a Glance. Toxins. 2016; 8(7):223. https://doi.org/10.3390/toxins8070223
Chicago/Turabian StyleSilva, Maraisa Cristina, Tamires Lopes Silva, Murilo Vieira Silva, Caroline Martins Mota, Fernanda Maria Santiago, Kelly Cortes Fonseca, Fábio Oliveira, Tiago Wilson Patriarca Mineo, and José Roberto Mineo. 2016. "Interaction between TNF and BmooMP-Alpha-I, a Zinc Metalloprotease Derived from Bothrops moojeni Snake Venom, Promotes Direct Proteolysis of This Cytokine: Molecular Modeling and Docking at a Glance" Toxins 8, no. 7: 223. https://doi.org/10.3390/toxins8070223
APA StyleSilva, M. C., Lopes Silva, T., Silva, M. V., Mota, C. M., Santiago, F. M., Fonseca, K. C., Oliveira, F., Mineo, T. W. P., & Mineo, J. R. (2016). Interaction between TNF and BmooMP-Alpha-I, a Zinc Metalloprotease Derived from Bothrops moojeni Snake Venom, Promotes Direct Proteolysis of This Cytokine: Molecular Modeling and Docking at a Glance. Toxins, 8(7), 223. https://doi.org/10.3390/toxins8070223