Cylindrospermopsin Biodegradation Abilities of Aeromonas sp. Isolated from Rusałka Lake
Abstract
:1. Introduction
2. Results
2.1. Activity of Isolated Strains towards CYN and Identification of CYN Biodegraders
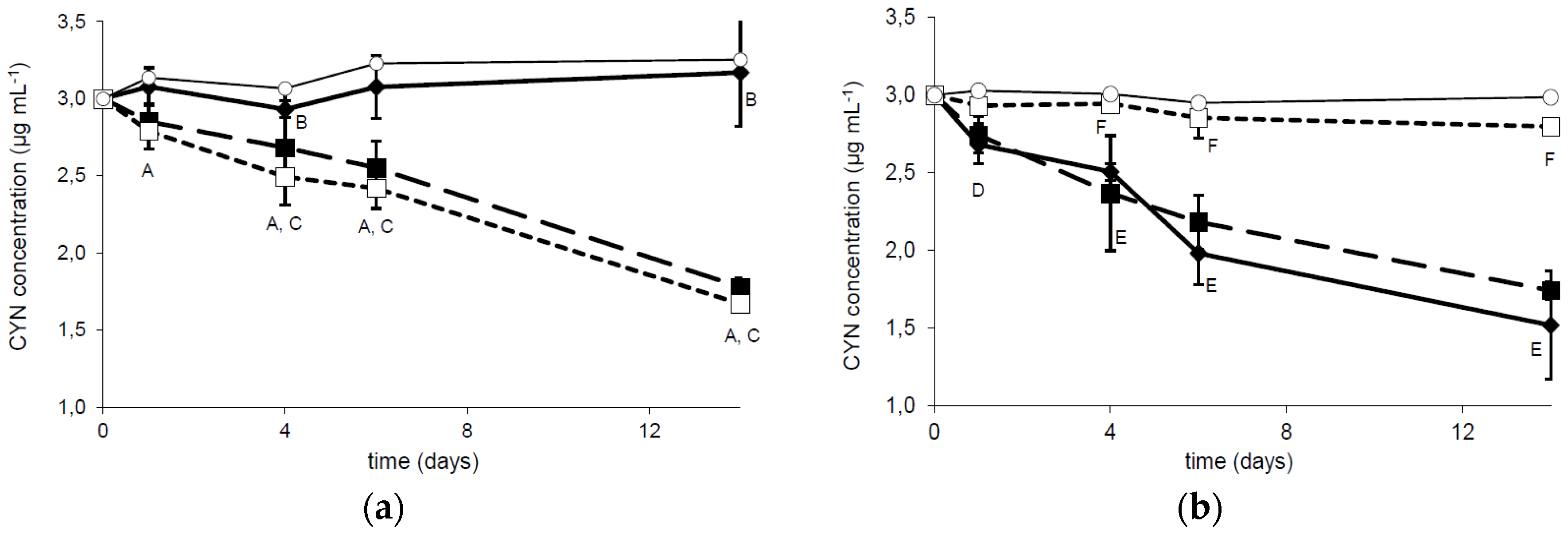
2.2. Biodegradation of CYN at Different Temperatures and pH
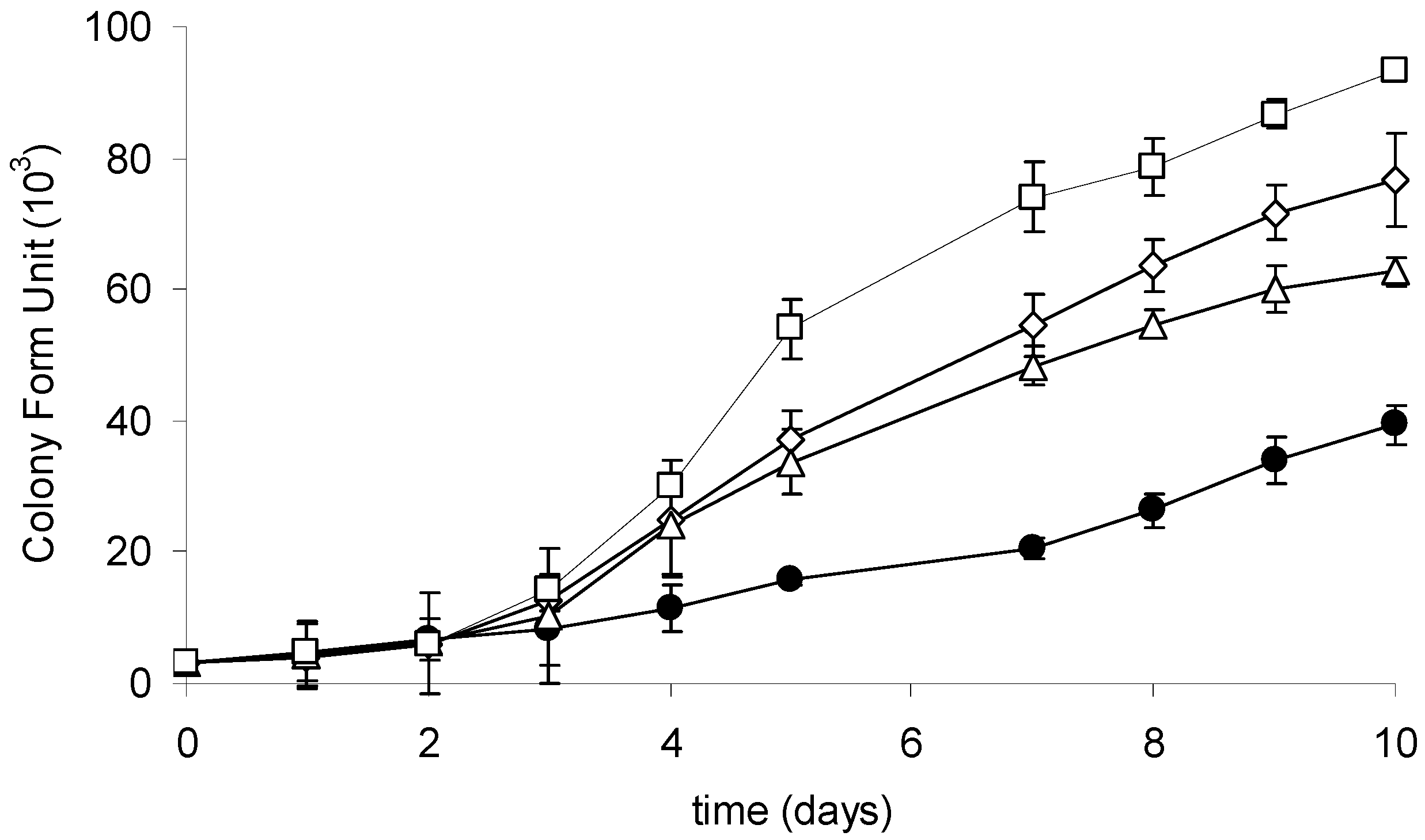
2.3. Growth of the R6 Strain in the Presence of CYN
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Separation of Bacterial Strains
4.2. Phytoplankton Analysis
4.3. Identification of the Bacteria
4.4. CYN Extraction and Preparation of Standard Curve
4.5. Cylindrospermopsin Purification and Analysis by HPLC-Diode Array UV Detection and MS/MS
4.6. Biodegradation Assays
4.7. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| BLAST | Basic Local Alignment Search Tool |
| CYN | cylindrospermopsin |
| MCs | microcystins |
| PCR | polymerase chain reaction |
| rDNA | ribosomal DNA (DNA coding ribosomal RNAs) |
References
- Antunes, J.T.; Leão, P.N.; Vasconcelos, V.M. Cylindrospermopsis raciborskii: Review of the Distribution, Phylogeography, and Ecophysiology of a Global Invasive Species. Front. Microb. 2015. [Google Scholar] [CrossRef] [PubMed]
- Kokociński, M.; Dziga, D.; Spoof, L.; Stefaniak, K.; Jurczak, T.; Mankiewicz-Boczek, J.; Meriluoto, J. First report of the cyanobacterial toxin cylindrospermopsin in the shallow, eutrophic lakes of western Poland. Chemosphere 2009, 74, 669–675. [Google Scholar] [CrossRef] [PubMed]
- Kokocinski, M.; Meriluoto, J.; Spoof, L.; Gagala, I.; Jurczak, T.; Rejmonczyk, E.; Mankiewicz-Boczek, J. Distribution and potential producers of cylindrospermopsin in western Poland. Eur. J. Phycol. 2011, 46, 130–131. [Google Scholar]
- Bogially, S.; Bruno, M.; Curini, R.; Di Corcia, A.; Fanali, C.; Laganà, A. Monitoring algal toxins in lake water by liquid chromatography tandem mass spectrometry. Environ. Sci. Technol. 2006, 40, 2917–2923. [Google Scholar] [CrossRef]
- Rücker, J.; Stüken, A.; Nixdorf, B.; Fastner, J.; Chorus, I.; Wiedner, C. Concentrations of particulate and dissolved cylindrospermopsin in 21 Aphanizomenon.-dominated temperate lakes. Toxicon 2007, 50, 800–809. [Google Scholar] [CrossRef] [PubMed]
- Rzymski, P.; Poniedziałek, B. In search of environmental role of cylindrospermopsin: A Review on Global Distribution and Ecology of its Producers. Wat. Res. 2014, 66, 320–337. [Google Scholar] [CrossRef] [PubMed]
- Falconer, I.R. Cyanobacterial Toxins in Drinking Water Supplies: Cylindrospermopsins and Microcystins; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Kinnear, S. Cylindrospermopsin: A Decade of Progress on Bioaccumulation Research. Mar. Drugs 2010, 8, 542–564. [Google Scholar] [CrossRef] [PubMed]
- Chiswell, R.K.; Shaw, G.R.; Eaglesham, G.K.; Smith, M.J.; Norris, R.L.; Seawright, A.A.; Moore, M.R. Stability of cylindrospermopsin, the toxin from the cyanobacterium Cylindrospermopsis. raciborskii: Effect of pH, Temperature, and Sunlight on Decomposition. Environ. Toxicol. 1999, 14, 155–165. [Google Scholar] [CrossRef]
- Humpage, A.R.; Falconer, I.R. Oral toxicity of the cyanobacterial toxin cylindrospermopsin in male Swiss albino mice: Determination of no Observed Adverse Effect Level for Deriving a Drinking Water Guideline Value. Environ. Toxicol. 2003, 18, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, K.; Masui, H.; Uemura, H.; Mori, Y.; Harada, K.I. Analysis of microcystins in sediments using MMPB method. Toxicon 2001, 39, 687–692. [Google Scholar] [CrossRef]
- Klitzke, S.; Apelt, S.; Weiler, C.; Fastner, J.; Chorus, I. Retention and degradation of the cyanobacterial toxin cylindrospermopsin in sediments - the role of sediment preconditioning and DOM composition. Toxicon 2010, 55, 999–1007. [Google Scholar] [CrossRef] [PubMed]
- Klitzke, S.; Beusch, C.; Fastner, J. Sorption of the cyanobacterial toxins cylindrospermopsin and anatoxin-a to sediments. Water Res. 2011, 45, 1338–1346. [Google Scholar] [CrossRef] [PubMed]
- Senogles, P.; Smith, M.; Shaw, G. Physical, Chemical and Biological Methods for the Degradation of the Cyanobacterial Toxin, Cylindrospermopsin. In Proceedings of the Water Quality Technology Conference, Seattle, WA, USA, 10–14 November 2002.
- Smith, M.J.; Shaw, G.R.; Eaglesham, G.K.; Ho, L.; Brookes, J.D. Elucidating the factors influencing the biodegradation of cylindrospermopsin in drinking water sources. Environ. Toxicol. 2008, 23, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Wormer, L.; Cires, A.; Carrasco, D.; Quesada, A. Cylindrospermopsin is not degradated by co-occuring natural bacterial communities during a 40-day study. Harmful Algae 2008, 7, 206–213. [Google Scholar] [CrossRef]
- Mohamed, Z.A.; Alamri, S.A. Biodegradation of cylindrospermopsin toxin by microcystin- degrading bacteria isolated from cyanobacterial blooms. Toxicon 2012, 60, 1390–1395. [Google Scholar] [CrossRef] [PubMed]
- Nybom, S.M.; Salminen, S.J.; Meriluoto, J.A. Specific strains of probiotic bacteria are efficient in removal of several different cyanobacterial toxins from solution. Toxicon 2008, 52, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Gołdyn, R.; Podsiadłowski, S.; Kowalczewska-Madura, K.; Dondajewska, R.; Szeląg-Wasielewska, E.; Budzyńska, A.; Domek, P.; Romanowicz-Brzozowska, W. Functioning of the Lake Rusałka ecosystem in Poznań (western Poland). Oceanol. Hydrobiol. Stud. 2010, 3, 65–80. [Google Scholar] [CrossRef]
- Kokociński, M.; Soininen, J. Environmental factors related to the occurrence of Cylindrospermopsis. raciborskii (Nostocales, Cyanophyta) at the north-eastern limit of its geographical range. Eur. J. Phycol. 2012, 47, 12–21. [Google Scholar] [CrossRef]
- Kokociński, M.; Mankiewicz-Boczek, J.; Jurczak, T.; Spoof, L.; Meriluoto, J.; Rejmonczyk, E.; Hautala, H.; Vehniäinen, M.; Pawełczyk, J.; Soininen, J. Aphanizomenon. gracile (Nostocales), a cylindrospermopsin -producing cyanobacterium in Polish lakes. Environ. Sci. Pollut. Res. Int. 2013, 20, 5243–5264. [Google Scholar] [CrossRef] [PubMed]
- Janda, J.M.; Abbott, S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, Perils, and Pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef] [PubMed]
- Mankiewicz-Boczek, J.; Gagala, I.; Jurczak, T.; Jaskulska, A.; Pawelczyk, J.; Dziadek, J. Bacteria homologus to Aeromonas. capable of microcystin degradation. Open Life Sci. 2015, 10, 119–129. [Google Scholar] [CrossRef]
- Dziga, D.; Wasylewski, M.; Wladyka, B.; Nybom, S.; Meriluoto, J. Microbial degradation of microcystins. Chem. Res. Toxicol. 2013, 26, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More Models, New Heuristics and Parallel Computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, J.; Codd, G.A. Solid Phase Extraction of Cylindrospermopsin in Filtered Water Samples. In Cyanobacterial Monitoring and Cyanotoxin Analysis; Meriluoto, J., Codd, G.A., Eds.; Åbo Akademi University Press: Turku, Finland, 2005; pp. 93–104. [Google Scholar]


© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dziga, D.; Kokocinski, M.; Maksylewicz, A.; Czaja-Prokop, U.; Barylski, J. Cylindrospermopsin Biodegradation Abilities of Aeromonas sp. Isolated from Rusałka Lake. Toxins 2016, 8, 55. https://doi.org/10.3390/toxins8030055
Dziga D, Kokocinski M, Maksylewicz A, Czaja-Prokop U, Barylski J. Cylindrospermopsin Biodegradation Abilities of Aeromonas sp. Isolated from Rusałka Lake. Toxins. 2016; 8(3):55. https://doi.org/10.3390/toxins8030055
Chicago/Turabian StyleDziga, Dariusz, Mikolaj Kokocinski, Anna Maksylewicz, Urszula Czaja-Prokop, and Jakub Barylski. 2016. "Cylindrospermopsin Biodegradation Abilities of Aeromonas sp. Isolated from Rusałka Lake" Toxins 8, no. 3: 55. https://doi.org/10.3390/toxins8030055
APA StyleDziga, D., Kokocinski, M., Maksylewicz, A., Czaja-Prokop, U., & Barylski, J. (2016). Cylindrospermopsin Biodegradation Abilities of Aeromonas sp. Isolated from Rusałka Lake. Toxins, 8(3), 55. https://doi.org/10.3390/toxins8030055





