Pseudomonas aeruginosa Type III Secretory Toxin ExoU and Its Predicted Homologs
Abstract
:1. Introduction
2. Molecular Biology of the ExoU Cytotoxin
3. ExoU and PLP A2 Activity
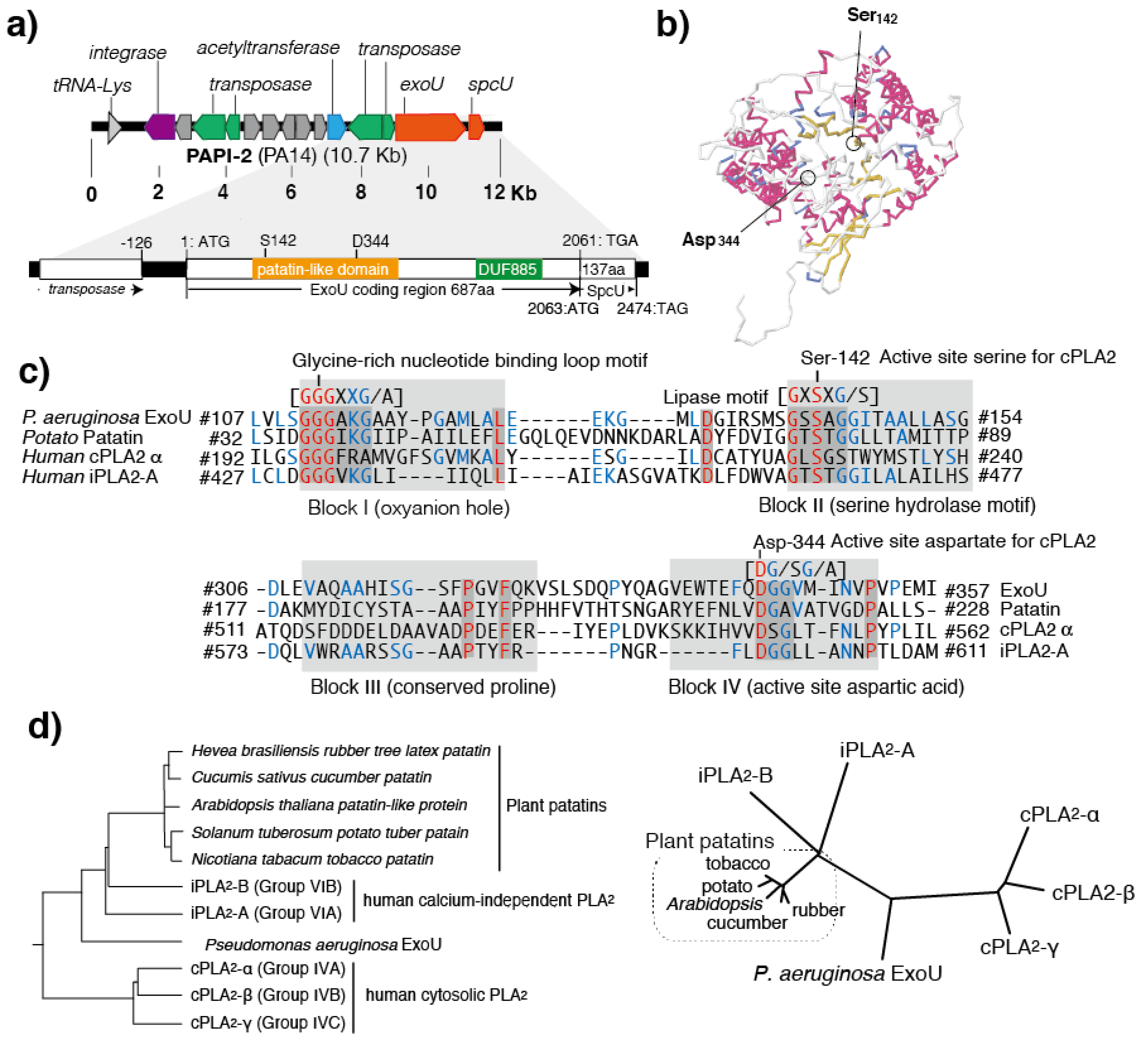
4. Conserved Domains in PLPs
5. Patatins and Phospholipases
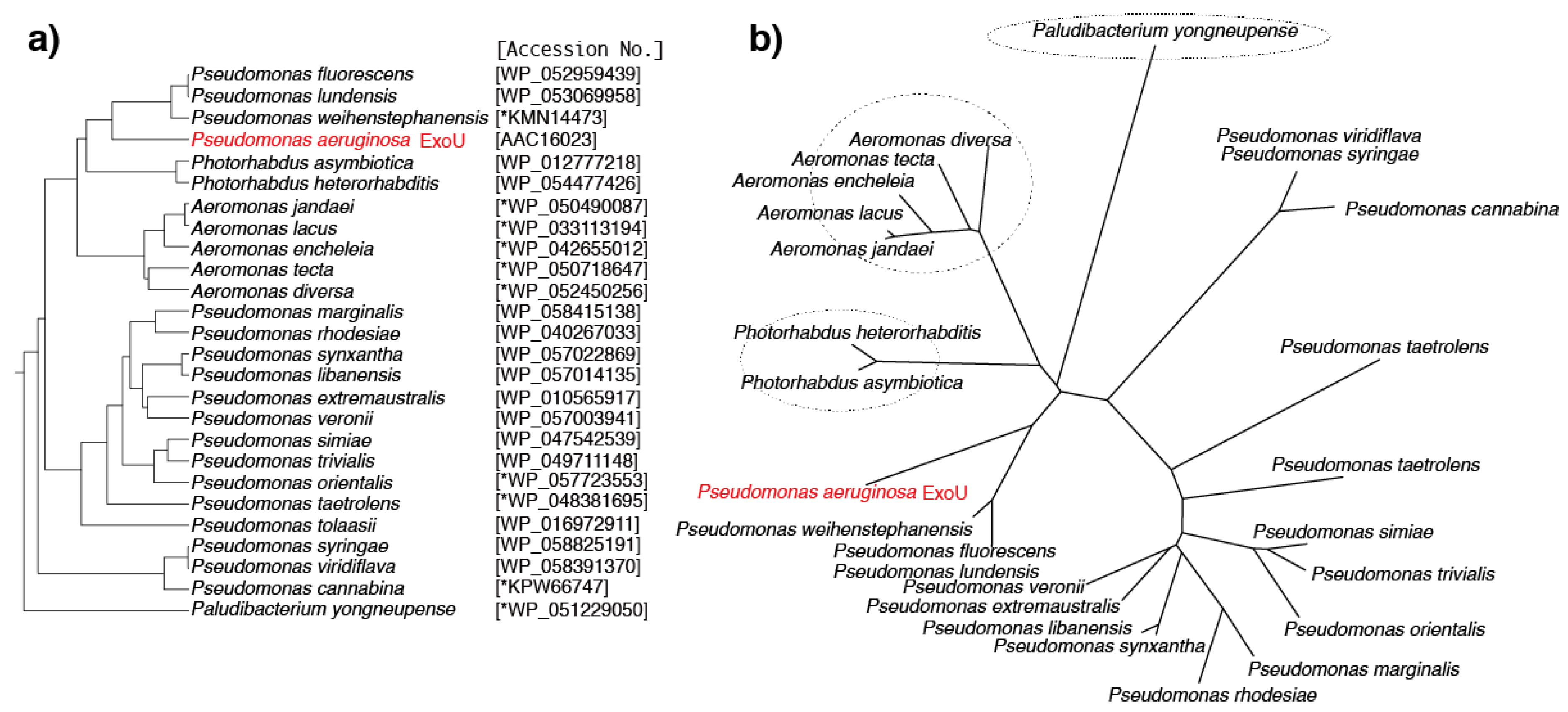
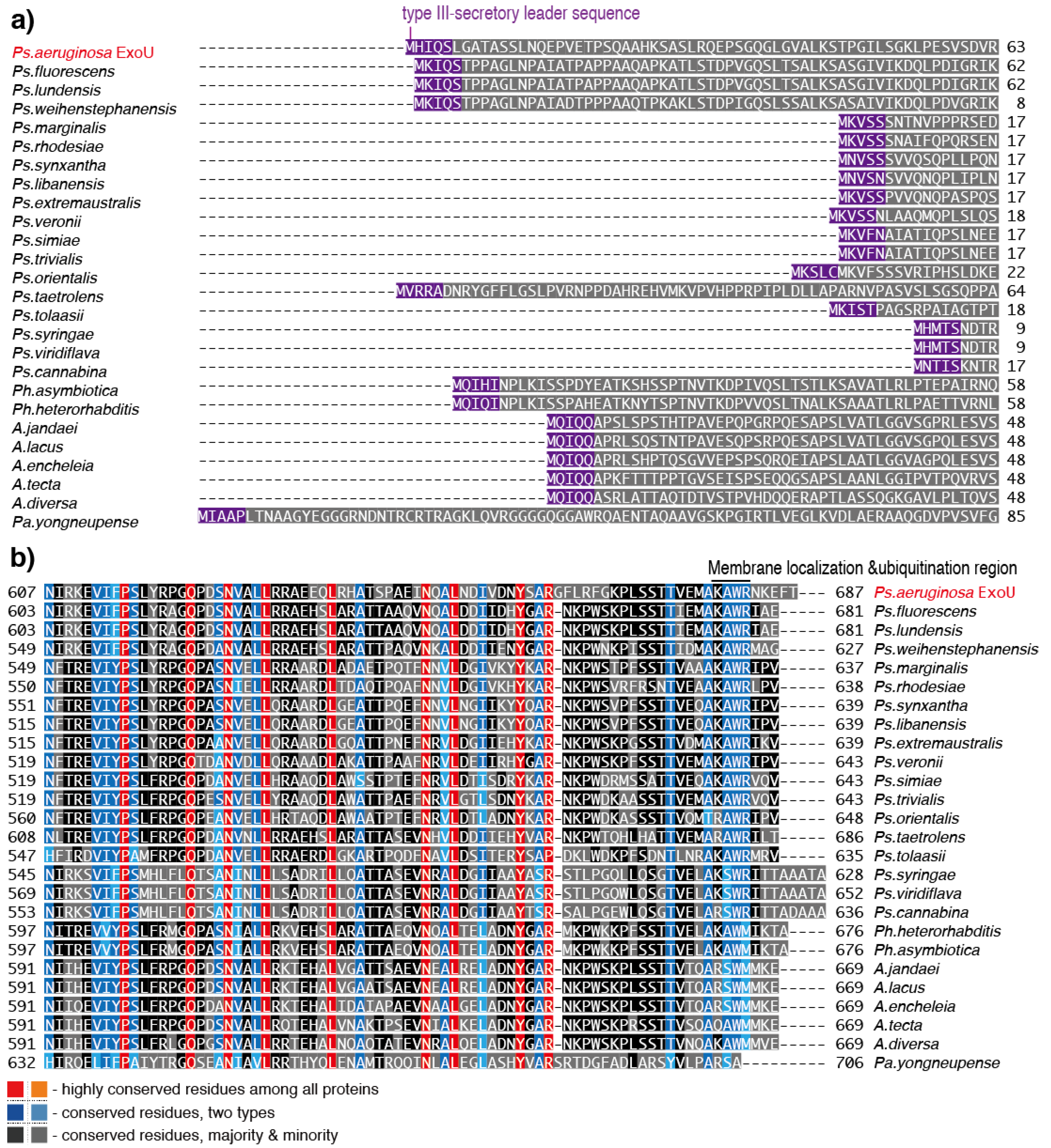
6. Predicted ExoU Homologs in Pseudomonas and Other Gram-Negative Bacteria
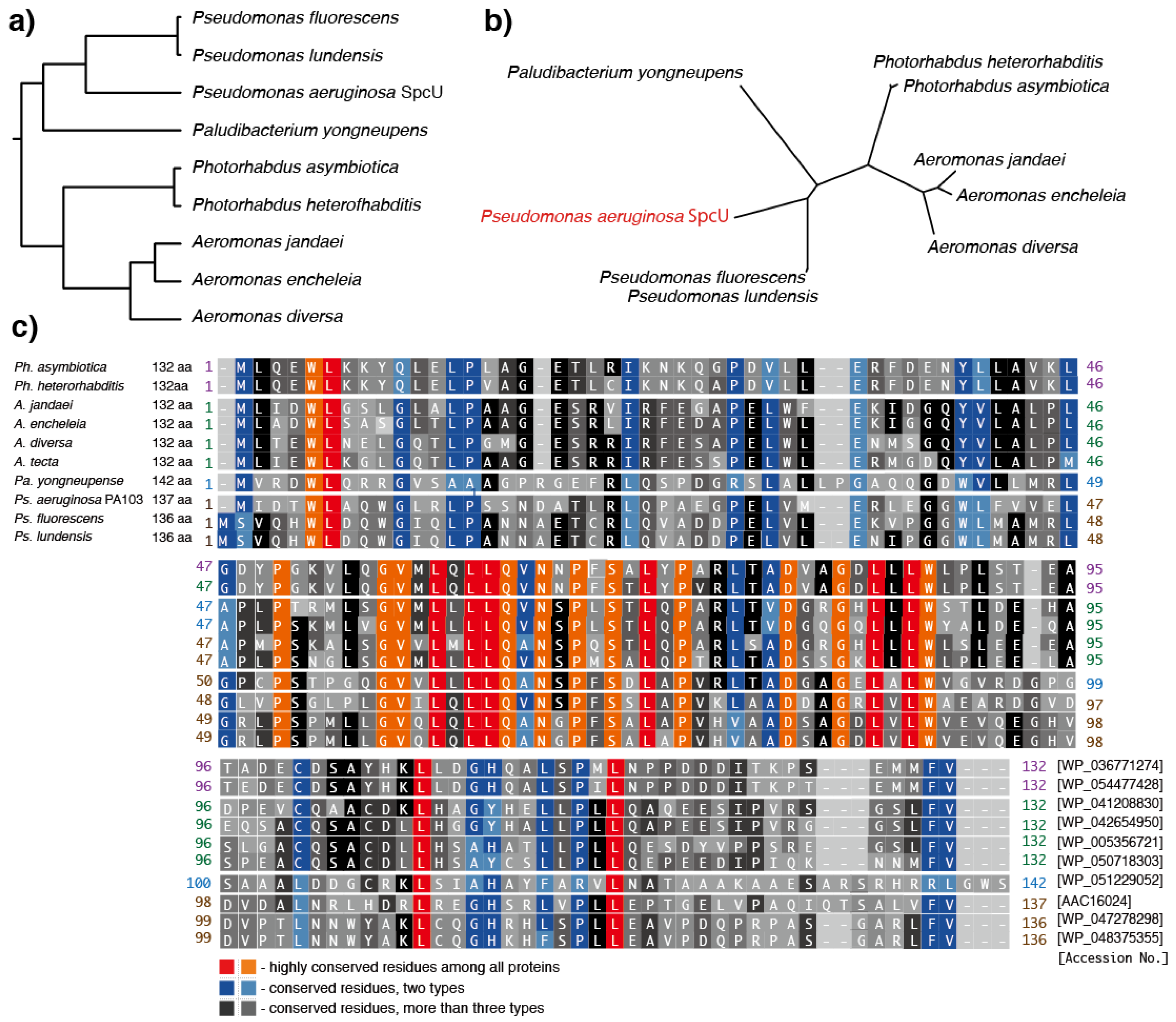
7. Predicted Homologs of the Ps. aeruginosa ExoU Chaperone, SpcU
8. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ramirez-Estrada, S.; Borgatta, B.; Rello, J. Pseudomonas aeruginosa ventilator-associated pneumonia management. Infect. Drug Resist. 2016, 9, 7–18. [Google Scholar] [PubMed]
- Kollef, M.H.; Chastre, J.; Fagon, J.Y.; Francois, B.; Niederman, M.S.; Rello, J.; Torres, A.; Vincent, J.L.; Wunderink, R.G.; Go, K.W.; et al. Global prospective epidemiologic and surveillance study of ventilator-associated pneumonia due to Pseudomonas aeruginosa. Crit Care Med. 2014, 42, 2178–2187. [Google Scholar] [CrossRef] [PubMed]
- Roy-Burman, A.; Savel, R.H.; Racine, S.; Swanson, B.L.; Revadigar, N.S.; Fujimoto, J.; Sawa, T.; Frank, D.W.; Wiener-Kronish, J.P. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 2001, 183, 1767–1774. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.R.; Cobb, E.; Bodi, M.; Mariscal, D.; Valles, J.; Engel, J.N.; Rello, J. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit Care Med. 2002, 30, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Wong-Beringer, A.; Wiener-Kronish, J.; Lynch, S.; Flanagan, J. Comparison of type III secretion system virulence among fluoroquinolone-susceptible and -resistant clinical isolates of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 2008, 14, 330–336. [Google Scholar] [CrossRef] [PubMed]
- El-Solh, A.A.; Hattemer, A.; Hauser, A.R.; Alhajhusain, A.; Vora, H. Clinical outcomes of type III Pseudomonas aeruginosa bacteremia. Crit Care Med. 2012, 40, 1157–1163. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, E.; Bensman, J.; Lou, M.; Agnello, M.; Shriner, K.; Wong-Beringer, A. Risk of developing pneumonia is enhanced by the combined traits of fluoroquinolone resistance and type III secretion virulence in respiratory isolates of Pseudomonas aeruginosa. Crit Care Med. 2014, 42, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Fleiszig, S.M.; Wiener-Kronish, J.P.; Miyazaki, H.; Vallas, V.; Mostov, K.E.; Kanada, D.; Sawa, T.; Yen, T.S.; Frank, D.W. Pseudomonas aeruginosa-mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect. Immun. 1997, 65, 579–586. [Google Scholar] [PubMed]
- Sawa, T.; Ohara, M.; Kurahashi, K.; Twining, S.S.; Frank, D.W.; Doroques, D.B.; Long, T.; Gropper, M.A.; Wiener-Kronish, J.P. In vitro cellular toxicity predicts Pseudomonas aeruginosa virulence in lung infections. Infect Immun. 1998, 66, 3242–3249. [Google Scholar] [PubMed]
- Kurahashi, K.; Kajikawa, O.; Sawa, T.; Ohara, M.; Gropper, M.A.; Frank, D.W.; Martin, T.R.; Wiener-Kronish, J.P. Pathogenesis of septic shock in Pseudomonas aeruginosa pneumonia. J. Clin. Investig. 1999, 104, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Finck-Barbancon, V.; Goranson, J.; Zhu, L.; Sawa, T.; Wiener-Kronish, J.P.; Fleiszig, S.M.; Wu, C.; Mende-Mueller, L.; Frank, D.W. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 1997, 25, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Sawa, T. The molecular mechanism of acute lung injury caused by Pseudomonas aeruginosa: From bacterial pathogenesis to host response. J. Intensiv. Care 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Sawa, T.; Shimizu, M.; Moriyama, K.; Wiener-Kronish, J.P. Association between Pseudomonas aeruginosa type III secretion, antibiotic resistance, and clinical outcome: A review. Crit Care 2014, 18. [Google Scholar] [CrossRef] [PubMed]
- Park, M.H.; Kim, S.Y.; Roh, E.Y.; Lee, H.S. Difference of Type 3 secretion system (T3SS) effector gene genotypes (exoU and exoS) and its implication to antibiotics resistances in isolates of Pseudomonas aeruginosa from chronic otitis media. Auris. Nasus. Larynx 2016. [Google Scholar] [CrossRef] [PubMed]
- de Almeida Silva, K.C.; Calomino, M.A.; Deutsch, G.; de Castilho, S.R.; de Paula, G.R.; Esper, L.M.; Teixeira, L.A. Molecular characterization of multidrug-resistant (MDR) Pseudomonas aeruginosa isolated in a burn center. Burns 2016. [Google Scholar] [CrossRef] [PubMed]
- Peña, C.; Cabot, G.; Gómez-Zorrilla, S.; Zamorano, L.; Ocampo-Sosa, A.; Murillas, J.; Almirante, B.; Pomar, V.; Aguilar, M.; Granados, A.; et al. Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin. Infect. Dis. 2015, 60, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Borkar, D.S.; Acharya, N.R.; Leong, C.; Lalitha, P.; Srinivasan, M.; Oldenburg, C.E.; Cevallos, V.; Lietman, T.M.; Evans, D.J.; Fleiszig, S.M. Cytotoxic clinical isolates of Pseudomonas aeruginosa identified during the Steroids for Corneal Ulcers Trial show elevated resistance to fluoroquinolones. BMC Ophthalmol. 2014, 14, 54. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chu, Y.; Wang, P.; Ji, X.; Li, X.; Wang, C.; Peng, Y. Clinical outcomes of multidrug resistant Pseudomonas aeruginosa infection and the relationship with type III secretion system in patients with diabetic foot. Int. J. Low Extrem. Wounds 2014, 13, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Conibear, T.C.; Bandara, R.; Aliwarga, Y.; Stapleton, F.; Willcox, M.D. Type III secretion system-associated toxins, proteases, serotypes, and antibiotic resistance of Pseudomonas aeruginosa isolates associated with keratitis. Curr. Eye Res. 2006, 31, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Housley, N.A.; Winkler, H.H.; Audia, J.P. The Rickettsia prowazekii ExoU homologue possesses phospholipase A1 (PLA1), PLA2, and lyso-PLA2 activities and can function in the absence of any eukaryotic cofactors in vitro. J. Bacteriol. 2011, 193, 4634–4642. [Google Scholar] [CrossRef] [PubMed]
- NCBI. Available online: http://www.ncbi.nlm.nih.gov/ (accessed on 2 October 2016).
- Finck-Barbancon, V.; Yahr, T.L.; Frank, D.W. Identification and characterization of SpcU, a chaperone required for efficient secretion of the ExoU cytotoxin. J. Bacteriol. 1998, 180, 6224–6231. [Google Scholar] [PubMed]
- He, J.; Baldini, R.L.; Deziel, E.; Saucier, M.; Zhang, Q.; Liberati, N.T.; Lee, D.; Urbach, J.; Goodman, H.M.; Rahme, L.G. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc. Natl. Acad. Sci. USA 2004, 101, 2530–2535. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Frank, D.W.; Hillard, C.J.; Feix, J.B.; Pankhaniya, R.R.; Moriyama, K.; Finck-Barbancon, V.; Buchaklian, A.; Lei, M.; Long, R.M.; et al. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 2003, 22, 2959–2969. [Google Scholar] [CrossRef] [PubMed]
- Pankhaniya, R.R.; Tamura, M.; Allmond, L.R.; Moriyama, K.; Ajayi, T.; Wiener-Kronish, J.P.; Sawa, T. Pseudomonas aeruginosa causes acute lung injury via the catalytic activity of the patatin-like phospholipase domain of ExoU. Crit Care Med. 2004, 32, 2293–2299. [Google Scholar] [CrossRef] [PubMed]
- Tamura, M.; Ajayi, T.; Allmond, L.R.; Moriyama, K.; Wiener-Kronish, J.P.; Sawa, T. Lysophospholipase A activity of Pseudomonas aeruginosa type III secretory toxin ExoU. Biochem. Biophys. Res. Commun. 2004, 316, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Coburn, J.; Collier, R.J. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14–3-3 protein family. Proc. Natl. Acad. Sci. USA 1993, 90, 2320–2324. [Google Scholar] [CrossRef] [PubMed]
- Yahr, T.L.; Vallis, A.J.; Hancock, M.K.; Barbieri, J.T.; Frank, D.W. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. USA 1998, 95, 13899–13904. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Feix, J.B.; Frank, D.W. Identification of superoxide dismutase as a cofactor for the Pseudomonas type III toxin, ExoU. Biochemistry 2006, 45, 10368–10375. [Google Scholar] [CrossRef] [PubMed]
- Stirling, F.R.; Cuzick, A.; Kelly, S.M.; Oxley, D.; Evans, T.J. Eukaryotic localization, activation and ubiquitinylation of a bacterial type III secreted toxin. Cell. Microbiol. 2006, 8, 1294–12309. [Google Scholar] [CrossRef] [PubMed]
- Schmalzer, K.M.; Benson, M.A.; Frank, D.W. Activation of ExoU phospholipase activity requires specific C-terminal regions. J. Bacteriol. 2010, 192, 1801–1812. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Schmalzer, K.M.; Sato, H.; Casey, M.; Terhune, S.S.; Haas, A.L.; Feix, J.B.; Frank, D.W. Ubiquitin and ubiquitin-modified proteins activate the Pseudomonas aeruginosa T3SS cytotoxin, ExoU. Mol. Microbiol. 2011, 82, 1454–1467. [Google Scholar] [CrossRef] [PubMed]
- Dessen, A.; Tang, J.; Schmidt, H.; Stahl, M.; Clark, J.D.; Seehra, J.; Somers, W.S. Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell 1999, 97, 349–360. [Google Scholar] [CrossRef]
- Halavaty, A.S.; Borek, D.; Tyson, G.H.; Veesenmeyer, J.L.; Shuvalova, L.; Minasov, G.; Otwinowski, Z.; Hauser, A.R.; Anderson, W.F. Structure of the type III secretion effector protein ExoU in complex with its chaperone SpcU. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- GenomeNet, Kyoto University Bioinformatics Center, Japan. Available online: http://www.genome.jp/tools/clustalw (accessed on 2 October 2016).
- Racusen, D. Lipid acyl hydrolase of patatin. Can. J. Bot. 1984, 62, 1640–1644. [Google Scholar] [CrossRef]
- Andrews, D.L.; Beames, B.; Summers, M.D.; Park, W.D. Characterization of the lipid acyl hydrolase activity of the major potato (Solanum tuberosum) tuber protein, patatin, by cloning and abundant expression in a baculovirus vector. Biochem. J. 1988, 252, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Kostyal, D.A.; Hickey, V.L.; Noti, J.D.; Sussman, G.L.; Beezhold, D.H. Cloning and characterization of a latex allergen (Hev b7): Homology to patatin, a plant PLA2. Clin. Exp. Immunol. 1998, 112, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Breiteneder, H.; Sowka, S.; Wagner, S.; Krebitz, M.; Hafner, C.; Kinaciyan, T.; Yeang, H.Y.; Scheiner, O. Cloning of the patatin-like latex allergen Hev b7, its expression in the yeast Pichia pastoris and its immunological characterization. Int. Arch. Allergy Immunol. 1999, 118, 309–310. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C. Pathogenesis-related proteins. Plant Mol. Biol. 1985, 4, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Banerji, S.; Flieger, A. Patatin-Like proteins: A new family of lipolytic enzymes present in bacteria? Microbiology 2004, 150, 522–525. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, T.; van Reenen, C.A.; Endo, A.; Tailliez, P.; Pages, S.; Sproer, C.; Malan, A.P.; Dicks, L.M. Photorhabdus heterorhabditis sp. nov., a symbiont of the entomopathogenic nematode Heterorhabditis zealandica. Int. J. Syst. Evol. Microbiol. 2014, 64, 1540–1545. [Google Scholar] [CrossRef] [PubMed]
- Gerrard, J.; Waterfield, N.; Vohra, R.; Ffrench-Constant, R. Human infection with Photorhabdus asymbiotica: An emerging bacterial pathogen. Microbes Infect. 2004, 6, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Tounsi, S.; Blight, M.; Jaoua, S.; de Lima Pimenta, A. From insects to human hosts: Identification of major genomic differences between entomopathogenic strains of Photorhabdus and the emerging human pathogen Photorhabdus asymbiotica. Int. J. Med. Microbiol. 2006, 296, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Costa, S.C.; Girard, P.A.; Brehelin, M.; Zumbihl, R. The emerging human pathogen Photorhabdus asymbiotica is a facultative intracellular bacterium and induces apoptosis of macrophage-like cells. Infect. Immun. 2009, 77, 1022–1030. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, P.; Waterfield, N.R.; Crossman, L.; Corton, C.; Sanchez-Contreras, M.; Vlisidou, I.; Barron, A.; Bignell, A.; Clark, L.; Ormond, D.; et al. Comparative genomics of the emerging human pathogen Photorhabdus asymbiotica with the insect pathogen Photorhabdus luminescens. BMC Genomics 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Mashima, J.; Kodama, Y.; Kosuge, T.; Fujisawa, T.; Katayama, T.; Nagasaki, H.; Okuda, Y.; Kaminuma, E.; Ogasawara, O.; Okubo, K.; et al. DNA data bank of Japan (DDBJ) progress report. Nucleic Acids Res. 2016, 44, D51–D67. [Google Scholar] [CrossRef] [PubMed]
- Janda, J.M.; Abbott, S.L. The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef] [PubMed]
- Beaz-Hidalgo, R.; Latif-Eugenin, F.; Hossain, M.J.; Berg, K.; Niemi, R.M.; Rapala, J.; Lyra, C.; Liles, M.R.; Figueras, M.J. Aeromonas aquatica sp. nov., Aeromonas finlandiensis sp. nov. and Aeromonas lacus sp. nov. isolated from Finnish waters associated with cyanobacterial blooms. Syst. Appl. Microbiol. 2015, 38, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Esteve, C.; Gutierrez, M.C.; Ventosa, A. Aeromonas encheleia sp. nov., isolated from European eels. Int. J. Syst. Bacteriol. 1995, 45, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Murcia, A.J. Phylogenetic positions of Aeromonas encheleia, Aeromonas popoffii, Aeromonas DNA hybridization group 11 and Aeromonas group 501. Int. J. Syst. Bacteriol. 1999, 49, 1403–1408. [Google Scholar] [CrossRef] [PubMed]
- Novakova, D.; Svec, P.; Sedlacek, I. Characterization of Aeromonas encheleia strains isolated from aquatic environments in the Czech Republic. Lett. Appl. Microbiol. 2009, 48, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Demarta, A.; Kupfer, M.; Riegel, P.; Harf-Monteil, C.; Tonolla, M.; Peduzzi, R.; Monera, A.; Jose Saavedra, M.; Martinez-Murcia, A. Aeromonas tecta sp. nov., isolated from clinical and environmental sources. Syst. Appl. Microbiol. 2008, 31, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.W.; Kim, B.Y.; Kim, W.G.; Yoo, K.H.; Yoo, S.H.; Son, J.A.; Weon, H.Y. Paludibacterium yongneupense gen. nov., sp. nov., isolated from a wetland, Yongneup, in Korea. Int. J. Syst. Evol. Microbiol. 2008, 58, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Ajayi, T.; Allmond, L.R.; Sawa, T.; Wiener-Kronish, J.P. Single-Nucleotide-Polymorphism mapping of the Pseudomonas aeruginosa type III secretion toxins for development of a diagnostic multiplex PCR system. J. Clin. Microbiol. 2003, 41, 3526–3531. [Google Scholar] [CrossRef] [PubMed]



| Species | NCBI GenBank Accession No. | DNA Sequence Length (bp) | Protein Sequence Length (aa) | Predicted Protein Size (kDa) | Alignment Score against ExoU (%) | |
|---|---|---|---|---|---|---|
| Protein | Genome | |||||
| Pseudomonas aeruginosa (ExoU) | AAC16023 | U97065 | 2064 | 687 | 73.9 | - |
| Pseudomonas weihenstephanensis | *KMN14473 | JYLF01000003 | 2046 | 681 | 72.9 | 60 |
| Pseudomonas fluorescens | WP_052959439 | NZ_LCYS01000046 | 2046 | 681 | 72.7 | 60 |
| Pseudomonas lundensis | WP_053069958 | NZ_JYKY01000004 | 2046 | 681 | 72.7 | 59 |
| Pseudomonas marginalis | WP_058415138 | NZ_LKGY01000112 | 1914 | 637 | 67.9 | 43 |
| Pseudomonas rhodesiae | WP_040267033 | NZ_CCYI01000012 | 1917 | 638 | 68.7 | 42 |
| Pseudomonas synxantha | WP_057022869 | NZ_JYLJ01000004 | 1920 | 639 | 68.9 | 45 |
| Pseudomonas libanensis | WP_057014135 | NZ_JYLH01000017 | 1920 | 639 | 69.2 | 45 |
| Pseudomonas extremaustralis | WP_010565917 | NZ_AHIP01000021 | 1920 | 639 | 68.1 | 44 |
| Pseudomonas veronii | WP_057003941 | NZ_JYLL01000003 | 1932 | 643 | 69.1 | 45 |
| Pseudomonas simiae | WP_047542539 | NZ_JRMC01000004 | 1932 | 643 | 69.2 | 44 |
| Pseudomonas trivialis | WP_049711148 | NZ_CP011507 | 1932 | 643 | 69.0 | 44 |
| Pseudomonas orientalis | *WP_057723553 | NZ_JYLM01000004 | 1947 | 648 | 70.0 | 41 |
| Pseudomonas taetrolens | *WP_048381695 | NZ_JYLA01000004 | 2061 | 686 | 74.2 | 41 |
| Pseudomonas tolaasii | WP_016972911 | NZ_AJXG01000265 | 1911 | 635 | 68.4 | 42 |
| Pseudomonas syringae | WP_058825191 | NZ_LKEM01000062 | 1890 | 629 | 68.1 | 46 |
| Pseudomonas viridiflava | WP_058391370 | NZ_LKEJ01000112 | 1959 | 652 | 70.6 | 44 |
| Pseudomonas cannabina | *KPW66747 | LJPX01000541 | 1887 | 628 | 68.1 | 45 |
| Photorhabdus asymbiotica | WP_012777218 | NC_012962 | 2061 | 676 | 73.5 | 52 |
| Photorhabdus heterorhabditis | WP_054477426 | NZ_LJCS01000013 | 2061 | 676 | 73.7 | 51 |
| Aeromonas jandaei | *WP_050490087 | NZ_JFDL01000003 | 2010 | 669 | 71.1 | 48 |
| Aeromonas lacus | *WP_033113194 | NZ_JRGM01000022 | 2010 | 669 | 71.2 | 48 |
| Aeromonas encheleia | *WP_042655012 | NZ_CDDI01000033 | 2010 | 669 | 71.2 | 48 |
| Aeromonas tecta | *WP_050718647 | NZ_CDCA01000025 | 2010 | 669 | 71.5 | 46 |
| Aeromonas diversa | *WP_052450256 | NZ_CDCE01000029 | 2010 | 669 | 71.7 | 47 |
| Paludibacterium yongneupense | *WP_051229050 | NZ_AUGZ01000008 | 2121 | 706 | 74.3 | 45 |
| Species | NCBI GenBank Accession No. | DNA Sequence Length (bp) | Protein Sequence Length (aa) | Predicted Protein Size (kDa) | Alignment Score against SpcU (%) | |
|---|---|---|---|---|---|---|
| Protein | Genome | |||||
| Pseudomonas aeruginosa (SpcU) | AAC16024 | U97065 | 414 | 137 | 14.9 | - |
| Pseudomonas fluorescens | WP_047278298 | NZ_LCYS01000046 | 411 | 136 | 14.9 | 48.5294 |
| Pseudomonas lundensis | WP_048375355 | NZ_JYKY01000004 | 411 | 136 | 14.9 | 47.7941 |
| Photorhabdus asymbiotica | WP_036771274 | NZ_JONO01000032 | 399 | 132 | 14.8 | 31.8182 |
| Photorhabdus heterorhabditis | WP_054477428 | NZ_LJCS01000013 | 399 | 132 | 14.9 | 31.0606 |
| Aeromonas jandaei | WP_041208830 | NZ_JFDL01000003 | 399 | 132 | 14.4 | 28.7879 |
| Aeromonas encheleia | WP_042654950 | NZ_CDDI01000033 | 399 | 132 | 14.2 | 31.0606 |
| Aeromonas tecta | WP_050718303 | NZ_CDCA01000025 | 399 | 132 | 14.6 | 28.0303 |
| Aeromonas diversa | WP_005356721 | NZ_CDCE01000029 | 399 | 132 | 14.4 | 28.7879 |
| Paludibacterium yongneupense | WP_050718303 | NZ_AUGZ01000008 | 429 | 142 | 15.0 | 33.5766 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sawa, T.; Hamaoka, S.; Kinoshita, M.; Kainuma, A.; Naito, Y.; Akiyama, K.; Kato, H. Pseudomonas aeruginosa Type III Secretory Toxin ExoU and Its Predicted Homologs. Toxins 2016, 8, 307. https://doi.org/10.3390/toxins8110307
Sawa T, Hamaoka S, Kinoshita M, Kainuma A, Naito Y, Akiyama K, Kato H. Pseudomonas aeruginosa Type III Secretory Toxin ExoU and Its Predicted Homologs. Toxins. 2016; 8(11):307. https://doi.org/10.3390/toxins8110307
Chicago/Turabian StyleSawa, Teiji, Saeko Hamaoka, Mao Kinoshita, Atsushi Kainuma, Yoshifumi Naito, Koichi Akiyama, and Hideya Kato. 2016. "Pseudomonas aeruginosa Type III Secretory Toxin ExoU and Its Predicted Homologs" Toxins 8, no. 11: 307. https://doi.org/10.3390/toxins8110307
APA StyleSawa, T., Hamaoka, S., Kinoshita, M., Kainuma, A., Naito, Y., Akiyama, K., & Kato, H. (2016). Pseudomonas aeruginosa Type III Secretory Toxin ExoU and Its Predicted Homologs. Toxins, 8(11), 307. https://doi.org/10.3390/toxins8110307






