Culture-Independent Study of the Late-Stage of a Bloom of the Toxic Dinoflagellate Ostreopsis cf. ovata: Preliminary Findings Suggest Genetic Differences at the Sub-Species Level and Allow ITS2 Structure Characterization
Abstract
:1. Introduction
2. Results
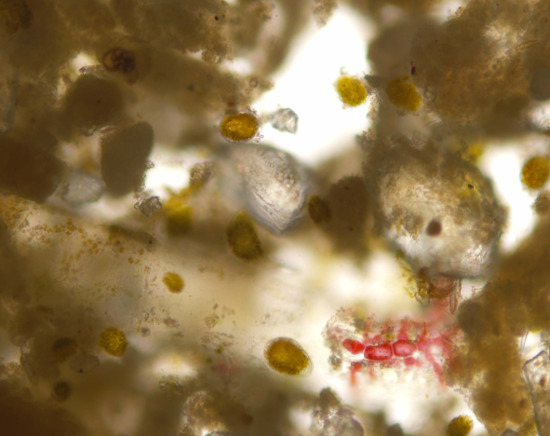
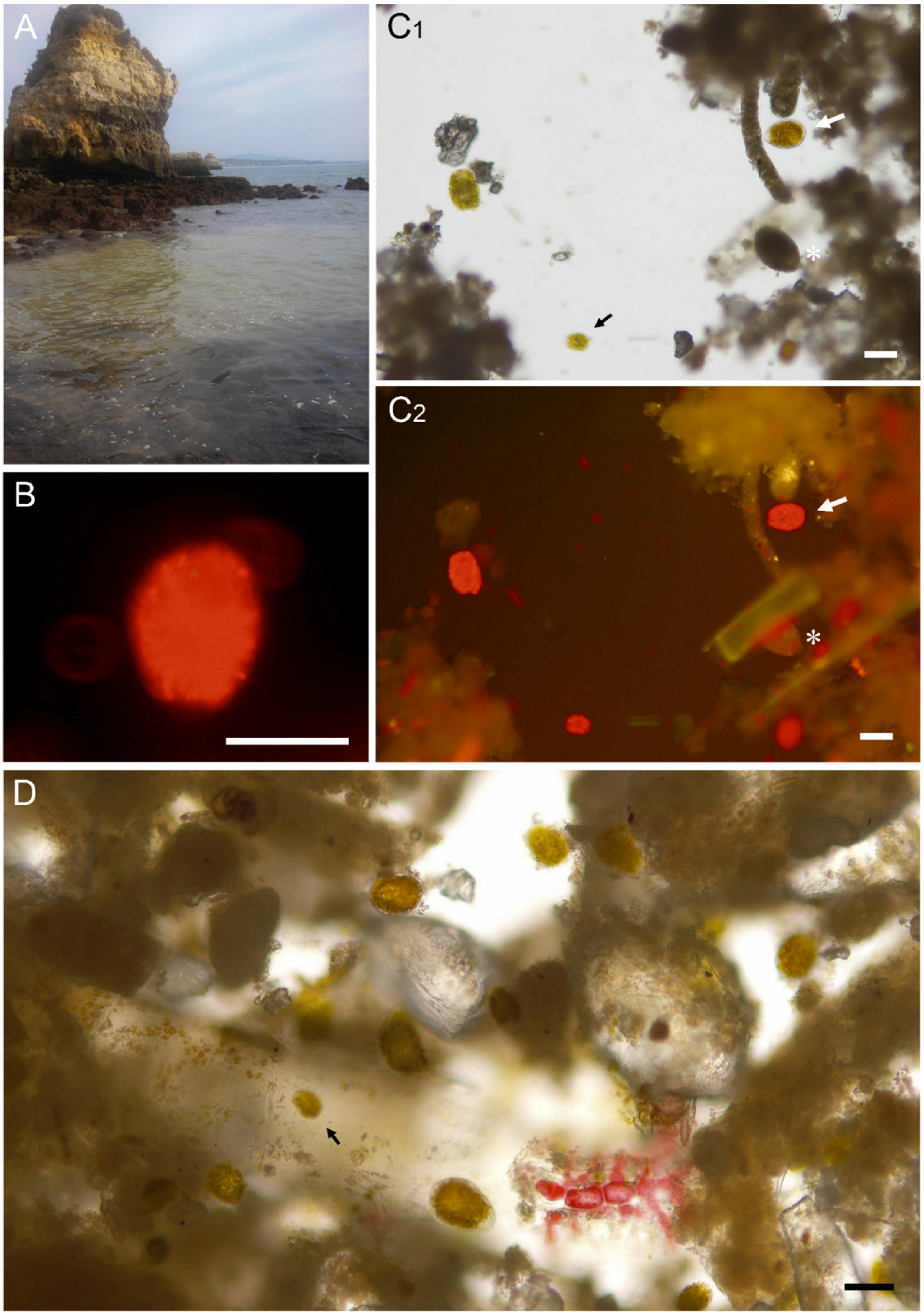
2.1. Morphometric Analysis and Microalgal Community
| Taxa (type of sample) | DV | W | AP | DV/W ratio | DV/AP ratio | Reference |
|---|---|---|---|---|---|---|
| Ostreopsis | 42–105 | 32–75 | - | 1.4 | - | This study |
| O. cf. ovata (environmental) | 26–62 | 13–48 | 14–36 | 1.8 | ~2 | [3] |
| O. cf. ovata (environmental) | 19–75 | 13–60 | 10–31 | - | 2.3 | [4] |
| O. cf. ovata (culture) a | 55–84 | 30–62 | - | 1.2–2 | - | [17] |
| O. cf. ovata (culture) b | 40–60 | 31–49 | - | 1.3 | - | [25] |
| O. cf. ovata (culture) | 27–65 | 19–57 | - | - | <2 | [24] |
| O. cf. ovata (culture) | 27–85 | 17–65 | - | - | - | [26] |
| O. cf. siamensis (environmental) | 36–66 | 24–50 | 14–26 | ~1.4 | ~3 | [3] |
| O. cf. siamensis (environmental) | 50–75 | 38–62 | - | ~1.3 | >4 | [24] |
| O. cf. siamensis (culture) a | 52–75 | 27–57 | - | 1.1–2.1 | - | [17] |
| Taxa | Number of taxa (cells∙L−1) per sampling site a | ||
|---|---|---|---|
| Burgau | Luz | Camilo | |
| Ostreopsis sp. | 2520 (13.1%) | 5480 (19.8%) | 13,040 (18.3%) |
| Other dinoflagellates b | 5880 (30.6%) | 200 (0.7%) | 1520 (2.1%) |
| Diatoms | 5520 (28.7%) | 5840 (21.1%) | 53,040 (74.4%) |
| Cyanobacteria | 2600 (13.5%) | 10,880 (39.3%) | - |
| Other microalgae | 2720 (14.1%) | 5280 (19.1%) | 3680 (5.2%) |
| Total | 19,240 | 27,680 | 71,280 |
2.2. Testing of PCR Primer Sets

2.3. Phylogenetic and Genetic Analyses

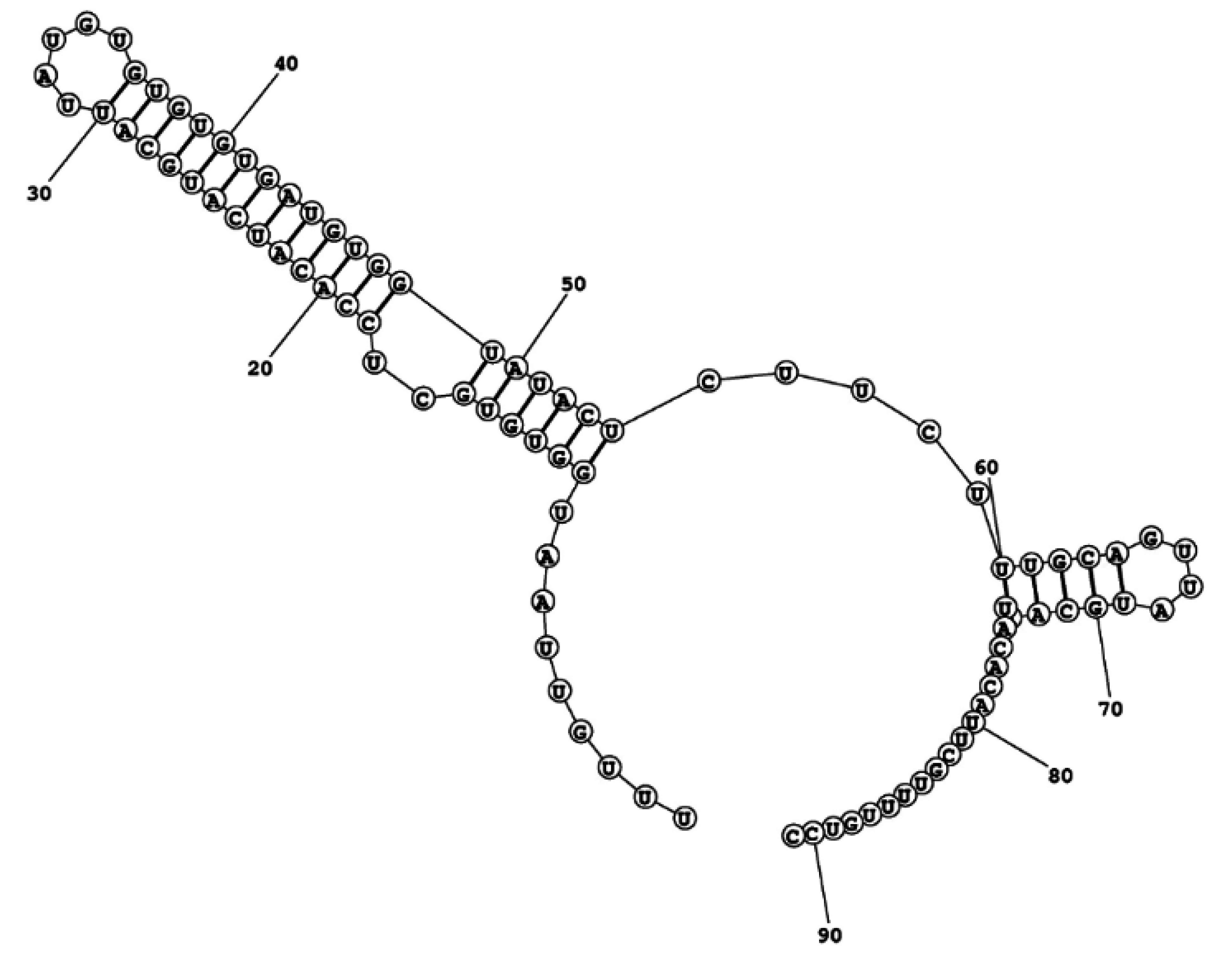
2.4. ITS2 Secondary-Structure Model of Ostreopsis cf. ovata
3. Discussion
3.1. Characterization of the Bloom and Microalgae Composition
3.2. Morphological Characterization, Molecular Identification, and (Phylo)Genetic Analyses
3.3. Evaluation of Oligonucleotides
3.4. Considerations on the ITS2 Characterization
4. Experimental Section
4.1. Sampling Site Locations, and Samples Collection and Processing

4.2. Morphological Analysis and Counting of Microalgae
4.3. DNA Extraction, PCR Amplification, Cloning and Sequencing
4.4. Phylogenetic and Sequence Analysis
4.5. ITS2 Secondary Structure Modelling
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rhodes, L. World-wide occurrence of the toxic dinoflagellate genus Ostreopsis Schmidt. Toxicon 2011, 57, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Mangialajo, L.; Ganzin, N.; Accoroni, S.; Asnaghi, V.; Blanfuné, A.; Cabrini, M.; Cattaneo-Vietti, R.; Chavanon, F.; Chiantore, M.; Cohu, S.; et al. Trends in Ostreopsis proliferation along the Northern Mediterranean coasts. Toxicon 2011, 57, 408–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aligizaki, K.; Nikolaidis, G. The presence of the potentially toxic genera Ostreopsis and Coolia (Dinophyceae) in the north Aegean sea, Greece. Harmful Algae 2006, 5, 717–730. [Google Scholar] [CrossRef]
- Accoroni, S.; Romagnoli, T.; Pichierri, S.; Colombo, F.; Totti, C. Morphometric analysis of Ostreopsis cf. ovata cells in relation to environmental conditions and bloom phases. Harmful Algae 2012, 19, 15–22. [Google Scholar]
- Hariganeya, N.; Tanimoto, Y.; Yamaguchi, H.; Nishimura, T.; Tawong, W.; Sakanari, H.; Yoshimatsu, T.; Sato, S.; Preston, C.M.; Adachi, M. Quantitative PCR method for enumeration of cells of cryptic species of the toxic marine dinoflagellate Ostreopsis spp. in coastal waters of Japan. PLoS ONE 2013, 8, e57627. [Google Scholar] [CrossRef] [PubMed]
- Faust, M.; Morton, S.; Quod, J. Further SEM study of marine dinoflagellates: The genus Ostreopsis (Dinophyceae). J. Phycol. 1996, 32, 1053–1065. [Google Scholar] [CrossRef]
- Steidinger, K.A.; Tangen, K. Dinoflagellates. In Identifying Marine Phytoplankton; Tomas, C.R., Ed.; Academic Press: San Diego, CA, USA, 1997; pp. 387–584. [Google Scholar]
- Escalera, L.; Benvenuto, G.; Scalco, E.; Zingone, A.; Montresor, M. Ultrastructural features of the benthic dinoflagellate Ostreopsis cf. ovata (Dinophyceae). Protist 2014, 165, 260–274. [Google Scholar] [CrossRef] [PubMed]
- Shears, N.; Ross, P. Blooms of benthic dinoflagellates of the genus Ostreopsis: An increasing and ecologically important phenomenon on temperate reefs in New Zealand and worldwide. Harmful Algae 2009, 8, 916–925. [Google Scholar] [CrossRef]
- Pfannkuchen, M.; Godrijan, J.; Pfannkuchen, D.; Ivesa, L.; Kruzic, P.; Ciminiello, P.; Dell’Aversano, C.; Iacovo, E.; Fattorusso, E.; Forino, M.; et al. Toxin-producing Ostreopsis cf. ovata are likely to bloom undetected along coastal areas. Environ. Sci. Technol. 2012, 46, 5574–5582. [Google Scholar]
- Ramos, V.; Vasconcelos, V. Palytoxin and analogs: Biological and ecological effects. Mar. Drugs 2010, 8, 2021–2037. [Google Scholar] [CrossRef] [PubMed]
- Deeds, J.R.; Schwartz, M.D. Human risk associated with palytoxin exposure. Toxicon 2010, 56, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Amzil, Z.; Sibat, M.; Chomerat, N.; Grossel, H.; Marco-Miralles, F.; Lemee, R.; Nezan, E.; Sechet, V. Ovatoxin-a and palytoxin accumulation in seafood in relation to Ostreopsis cf. ovata blooms on the French Mediterranean coast. Mar. Drugs 2012, 10, 477–496. [Google Scholar]
- Amorim, A.; Veloso, V.; Penna, A. First detection of Ostreopsis cf. siamensis in Portuguese coastal waters. Harmful Algae News 2010, 42, 6–7. [Google Scholar]
- Laza-Martinez, A.; Orive, E.; Miguel, I. Morphological and genetic characterization of benthic dinoflagellates of the genera Coolia, Ostreopsis and Prorocentrum from the south-eastern Bay of Biscay. Eur. J. Phycol. 2011, 46, 45–65. [Google Scholar] [CrossRef]
- David, H.; Ganzedo, U.; Laza-Martínez, A.; Orive, E. Relationships between the presence of Ostreopsis (Dinophyceae) in the Atlantic coast of the Iberian Peninsula and sea-surface temperature. Cryptogam. Algol. 2012, 33, 199–207. [Google Scholar] [CrossRef]
- David, H.; Laza-Martínez, A.; Miguel, I.; Orive, E. Ostreopsis cf. siamensis and Ostreopsis cf. ovata from the Atlantic Iberian eninsula: Morphological and phylogenetic characterization. Harmful Algae 2013, 30, 44–55. [Google Scholar]
- Silva, A.; Brotas, V.; Orive, E.; Neto, A.I. First records of Ostreopsis heptagona, O. cf. siamensis and O. cf. ovata—In the Azores archipelago, Portugal. Harmful Algae News 2010, 42, 1–2. [Google Scholar]
- Penna, A.; Fraga, S.; Battocchi, C.; Casabianca, S.; Giacobbe, M.; Riobó, P.; Vernesi, C. A phylogeographical study of the toxic benthic dinoflagellate genus Ostreopsis Schmidt. J. Biogeogr. 2010, 37, 830–841. [Google Scholar] [CrossRef] [Green Version]
- Ciminiello, P.; Dell’Aversano, C.; Iacovo, E.D.; Fattorusso, E.; Forino, M.; Tartaglione, L.; Yasumoto, T.; Battocchi, C.; Giacobbe, M.G.; Amorim, A.; et al. Investigation of toxin profile of Mediterranean and Atlantic strains of Ostreopsis cf. siamensis (Dinophyceae) by liquid chromatography–high resolution mass spectrometry. Harmful Algae 2013, 23, 19–27. [Google Scholar]
- Totti, C.; Accoroni, S.; Cerino, F.; Cucchiari, E.; Romagnoli, T. Ostreopsis ovata bloom along the Conero Riviera (northern Adriatic Sea): Relationships with environmental conditions and substrata. Harmful Algae 2010, 9, 233–239. [Google Scholar] [CrossRef]
- Cúrdia, J.; Monteiro, P.; Afonso, C.M.L.; Santos, M.N.; Cunha, M.R.; Gonçalves, J.M.S. Spatial and depth-associated distribution patterns of shallow gorgonians in the Algarve coast (Portugal, NE Atlantic). Helgol. Mar. Res. 2012, 67, 521–534. [Google Scholar] [CrossRef]
- David, H.; Moita, M.T.; Laza-Martínez, A.; Silva, A.; Mateus, M.; Pablo, H.; Orive, E. First bloom of Ostreopsis cf. ovata in the continental Portuguese coast. Harmful Algae News 2012, 45, 12–13. [Google Scholar]
- Penna, A.; Vila, M.; Fraga, S.; Giacobbe, M.; Andreoni, F.; Riobó, P.; Vernesi, C. Characterization of Ostreopsis and Coolia (Dinophyceae) isolates in the western Mediterranean Sea based on morphology, toxicity and internal transcribed spacer 5.8S rDNA sequences. J. Phycol. 2005, 41, 212–225. [Google Scholar] [CrossRef]
- Accoroni, S.; Romagnoli, T.; Pichierri, S.; Totti, C. New insights on the life cycle stages of the toxic benthic dinoflagellate Ostreopsis cf. ovata. Harmful Algae 2014, 34, 7–16. [Google Scholar] [CrossRef]
- Bravo, I.; Vila, M.; Casabianca, S.; Rodriguez, F.; Rial, P.; Riobó, P.; Penna, A. Life cycle stages of the benthic palytoxin-producing dinoflagellate Ostreopsis cf. ovata (Dinophyceae). Harmful Algae 2012, 18, 24–34. [Google Scholar] [CrossRef] [Green Version]
- Penna, A.; Bertozzini, E.; Battocchi, C.; Galluzzi, L.; Giacobbe, M.G.; Vila, M.; Garces, E.; Lugliè, A.; Magnani, M. Monitoring of HAB species in the Mediterranean Sea through molecular methods. J. Plankton Res. 2007, 29, 19–38. [Google Scholar] [CrossRef]
- Saldarriaga, J.F.; Taylor, M.F.J.R.; Cavalier-Smith, T.; Menden-Deuer, S.; Keeling, P.J. Molecular data and the evolutionary history of dinoflagellates. Eur. J. Protistol. 2004, 40, 85–111. [Google Scholar] [CrossRef]
- Litaker, R.W.; Vandersea, M.W.; Kibler, S.R.; Reece, K.S.; Stokes, N.A.; Lutzoni, F.M.; Yonish, B.A.; West, M.A.; Black, M.N.D.; Tester, P.A. Recognizing dinoflagellate species using ITS rDNA sequences. J. Phycol. 2007, 43, 344–355. [Google Scholar] [CrossRef]
- Stern, R.F.; Andersen, R.A.; Jameson, I.; Kupper, F.C.; Coffroth, M.A.; Vaulot, D.; Le Gall, F.; Véron, B.; Brand, J.J.; Skelton, H.; et al. Evaluating the ribosomal internal transcribed spacer (ITS) as a candidate dinoflagellate barcode marker. PLoS ONE 2012, 7, e42780. [Google Scholar] [CrossRef] [PubMed]
- Pin, L.; Teen, L.; Ahmad, A.; Usup, G. Genetic diversity of Ostreopsis ovata (Dinophyceae) from Malaysia. Mar. Biotechnol. 2001, 3, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Nishimura, T.; Uehara, K.; Sakanari, H.; Tawong, W.; Hariganeya, N.; Smith, K.; Rhodes, L.; Yasumoto, T.; Taira, Y.; et al. Phylogeography of Ostreopsis along West Pacific coast, with special reference to a novel clade from Japan. PLoS ONE 2011, 6, e27983. [Google Scholar] [CrossRef] [PubMed]
- News about the Algarve’s Bloom on the Portuguese Public TV Channel’s Website (RTP). Available online: http://www.webcitation.org/6YUtUTvot (accessed on 13 May 2015). (In Portuguese)
- Honsell, G.; Bonifacio, A.; De Bortoli, M.; Penna, A.; Battocchi, C.; Ciminiello, P.; Dell’Aversano, C.; Fattorusso, E.; Sosa, S.; Yasumoto, T.; et al. New insights on cytological and metabolic features of Ostreopsis cf. ovata Fukuyo (Dinophyceae): A multidisciplinary approach. PLoS ONE 2013, 8, e57291. [Google Scholar]
- Scholin, C.; Herzog, M.; Sogin, M.; Anderson, D. Identification of group- and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). II. Sequence analysis of a fragment of the LSU rRNA gene. J. Phycol. 1994, 30, 999–1011. [Google Scholar] [CrossRef]
- Keller, A.; Schleicher, T.; Schultz, J.; Muller, T.; Dandekar, T.; Wolf, M. 5.8S-28S rRNA interaction and HMM-based ITS2 annotation. Gene 2009, 430, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Leaw, C.-P.; Lim, P.-T.; Cheng, K.-W.; Ng, B.-K.; Usup, G. Morphology and molecular characterization of a new species of thecate benthic dinoflagellate, Cooliamalayensis sp. nov. (Dinophyceae). J. Phycol. 2010, 46, 162–171. [Google Scholar] [CrossRef]
- Mai, J.C.; Coleman, A.W. The internal transcribed spacer 2 exhibits a common secondary structure in green algae and flowering plants. J. Mol. Evol. 1997, 44, 258–271. [Google Scholar] [CrossRef] [PubMed]
- Joseph, N.; Krauskopf, E.; Vera, M.I.; Michot, B. Ribosomal internal transcribed spacer 2 (ITS2) exhibits a common core of secondary structure in vertebrates and yeast. Nucleic Acids Res. 1999, 27, 4533–4540. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Maisel, S.; Gerlach, D.; Muller, T.; Wolf, M. A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA 2005, 11, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Koetschan, C.; Hackl, T.; Muller, T.; Wolf, M.; Förster, F.; Schultz, J. ITS2 database IV: Interactive taxon sampling for internal transcribed spacer 2 based phylogenies. Mol. Phylogenet. Evol. 2012, 63, 585–588. [Google Scholar] [CrossRef] [PubMed]
- Van Hannen, E.J.; Fink, P.; Lurling, M. A revised secondary structure model for the internal transcribed spacer 2 of the green algae Scenedesmus and Desmodesmus and its implication for the phylogeny of these algae. Eur. J. Phycol. 2002, 37, 203–208. [Google Scholar] [CrossRef]
- Hershkovitz, M.A.; Zimmer, E.A. Conservation patterns in angiosperm rDNA ITS2 sequences. Nucleic Acids Res. 1996, 24, 2857–2867. [Google Scholar] [CrossRef] [PubMed]
- Salvi, D.; Mariottini, P. Molecular phylogenetics in 2D: ITS2 rRNA evolution and sequence-structure barcode from Veneridae to Bivalvia. Mol. Phylogenet. Evol. 2012, 65, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Parsons, M.L.; Aligizaki, K.; Bottein, M.Y.D.; Fraga, S.; Morton, S.L.; Penna, A.; Rhodes, L. Gambierdiscus and Ostreopsis: Reassessment of the state of knowledge of their taxonomy, geography, ecophysiology, and toxicology. Harmful Algae 2012, 14, 107–129. [Google Scholar] [CrossRef]
- Lund, J.W.G.; Kipling, C.; LeCren, E.D. The inverted microscope method of estimating algal numbers and the statistical basis of estimations by counting. Hydrobiologia 1958, 11, 143–170. [Google Scholar] [CrossRef]
- Komárek, J.; Anagnostidis, K. Cyanoprokaryota 1. Teil: Chroococcales. In Süsswasserflora von Mitteleuropa 19/1; Ettl, H., Gärtner, G., Heynig, H., Mollenhauer, D., Eds.; Gustav Fischer: Jena, Germany, 1988; Volume 1, pp. 1–548. [Google Scholar]
- Komárek, J.; Anagnostidis, K. Cyanoprokaryota 2. Teil: Oscillatoriales. In Süsswasserflora von Mitteleuropa 19/2; Büdel, B., Krienitz, L., Gärtner, G., Schagerl, M., Eds.; Elsevier/Spektrum: Heidelberg, Germany, 2005; Volume 2, pp. 1–759. [Google Scholar]
- Tomas, C.R. Identifying Marine Phytoplankton; Academic Press: San Diego, CA, USA, 1997; p. 858. [Google Scholar]
- Huber, T.; Faulkner, G.; Hugenholtz, P. Bellerophon: A program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 2004, 20, 2317–2319. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, S.; Correa, E.; Menezes, M.; Varela, D.; Paredes, J.; Morris, S. Growth and toxin profile of Ostreopsis cf. ovata (Dinophyta) from Rio de Janeiro, Brazil. Harmful Algae 2012, 13, 1–9. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.A.; Strimmer, K.; Vingron, M.; von Haeseler, A. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 2002, 18, 502–504. [Google Scholar] [CrossRef] [PubMed]
- Stover, B.C.; Muller, K.F. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 2010, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Mathews, D.H. Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA 2004, 10, 1178–1190. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.J.; Gloor, J.W.; Mathews, D.H. Improved RNA secondary structure prediction by maximizing expected pair accuracy. RNA 2009, 15, 1805–1813. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.; Achtziger, M.; Schultz, J.; Dandekar, T.; Müller, T. Homology modeling revealed more than 20,000 rRNA internal transcribed spacer 2 (ITS2) secondary structures. RNA 2005, 11, 1616–1623. [Google Scholar] [CrossRef] [PubMed]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinformatics 2010, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Perini, F.; Casabianca, A.; Battocchi, C.; Accoroni, S.; Totti, C.; Penna, A. New approach using the real-time PCR method for estimation of the toxic marine dinoflagellate Ostreopsis cf. ovata in marine environment. PLoS ONE 2011, 6, e17699. [Google Scholar] [CrossRef] [PubMed]
- Battocchi, C.; Totti, C.; Vila, M.; Masó, M.; Capellacci, S.; Accoroni, S.; Reñé, A.; Scardi, M.; Penna, A. Monitoring toxic microalgae Ostreopsis (dinoflagellate) species in coastal waters of the Mediterranean Sea using molecular PCR-based assay combined with light microscopy. Mar. Pollut. Bull. 2010, 60, 1074–1084. [Google Scholar] [CrossRef] [PubMed]
- Casabianca, S.; Casabianca, A.; Riobó, P.; Franco, J.M.; Vila, M.; Penna, A. Quantification of the toxic dinoflagellate Ostreopsis spp. by qPCR assay in marine aerosol. Environ. Sci. Technol. 2013, 47, 3788–3795. [Google Scholar] [CrossRef] [PubMed]
- Penna, A.; Battocchi, C.; Capellacci, S.; Fraga, S.; Aligizaki, K.; Lemée, R.; Vernesi, C. Mitochondrial, but not rDNA, genes fail to discriminate dinoflagellate species in the genus Ostreopsis. Harmful Algae 2014, 40, 40–50. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramos, V.; Salvi, D.; Machado, J.P.; Vale, M.; Azevedo, J.; Vasconcelos, V. Culture-Independent Study of the Late-Stage of a Bloom of the Toxic Dinoflagellate Ostreopsis cf. ovata: Preliminary Findings Suggest Genetic Differences at the Sub-Species Level and Allow ITS2 Structure Characterization. Toxins 2015, 7, 2514-2533. https://doi.org/10.3390/toxins7072514
Ramos V, Salvi D, Machado JP, Vale M, Azevedo J, Vasconcelos V. Culture-Independent Study of the Late-Stage of a Bloom of the Toxic Dinoflagellate Ostreopsis cf. ovata: Preliminary Findings Suggest Genetic Differences at the Sub-Species Level and Allow ITS2 Structure Characterization. Toxins. 2015; 7(7):2514-2533. https://doi.org/10.3390/toxins7072514
Chicago/Turabian StyleRamos, Vitor, Daniele Salvi, João Paulo Machado, Micaela Vale, Joana Azevedo, and Vitor Vasconcelos. 2015. "Culture-Independent Study of the Late-Stage of a Bloom of the Toxic Dinoflagellate Ostreopsis cf. ovata: Preliminary Findings Suggest Genetic Differences at the Sub-Species Level and Allow ITS2 Structure Characterization" Toxins 7, no. 7: 2514-2533. https://doi.org/10.3390/toxins7072514
APA StyleRamos, V., Salvi, D., Machado, J. P., Vale, M., Azevedo, J., & Vasconcelos, V. (2015). Culture-Independent Study of the Late-Stage of a Bloom of the Toxic Dinoflagellate Ostreopsis cf. ovata: Preliminary Findings Suggest Genetic Differences at the Sub-Species Level and Allow ITS2 Structure Characterization. Toxins, 7(7), 2514-2533. https://doi.org/10.3390/toxins7072514