Development of Plate Reader and On-Line Microfluidic Screening to Identify Ligands of the 5-Hydroxytryptamine Binding Protein in Venoms
Abstract
:1. Introduction
2. Results and Discussion
2.1. Selection of a Suitable Tracer Ligand
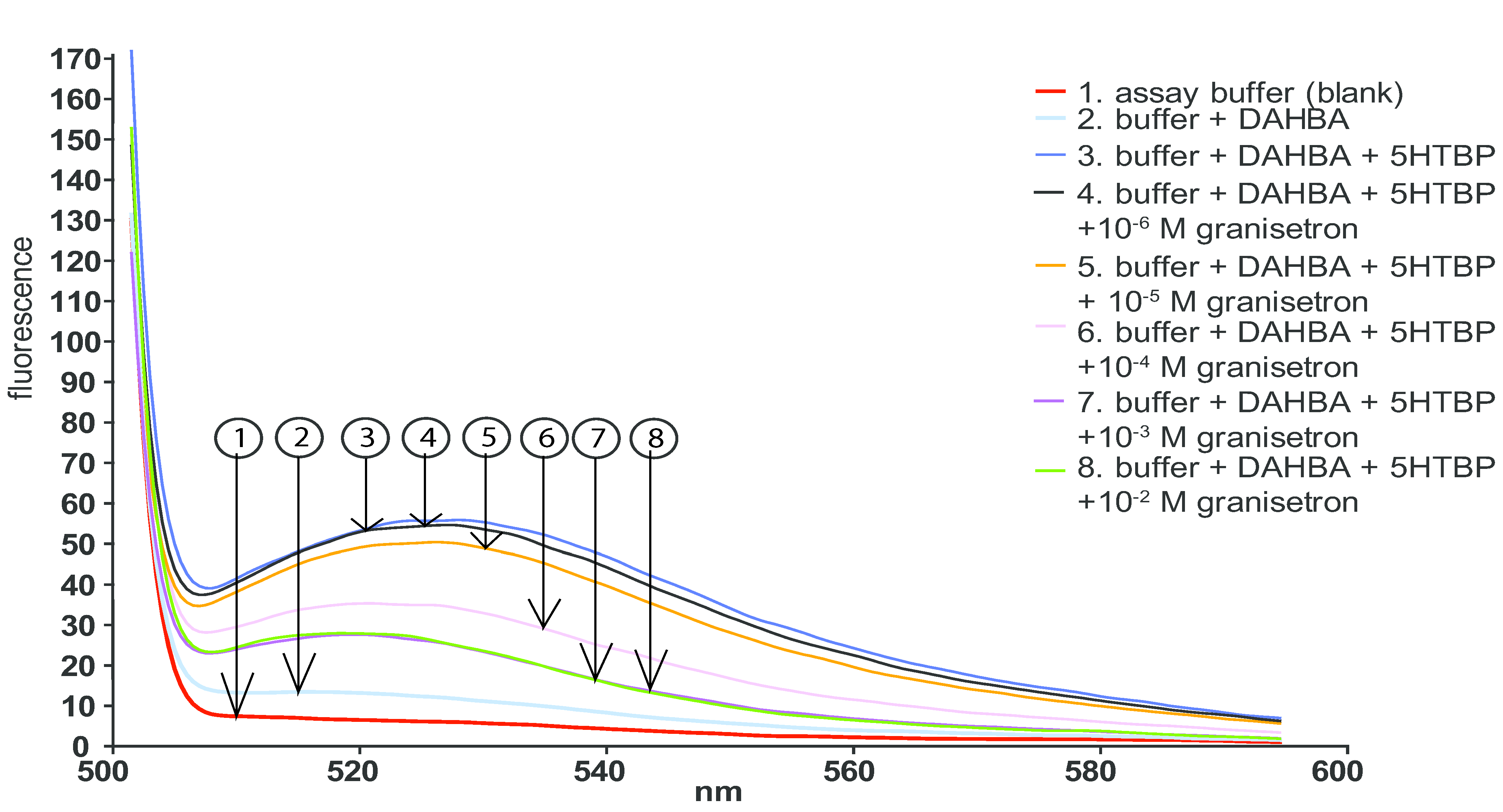
2.2. Fluorescence Enhancement Assay in Microplates
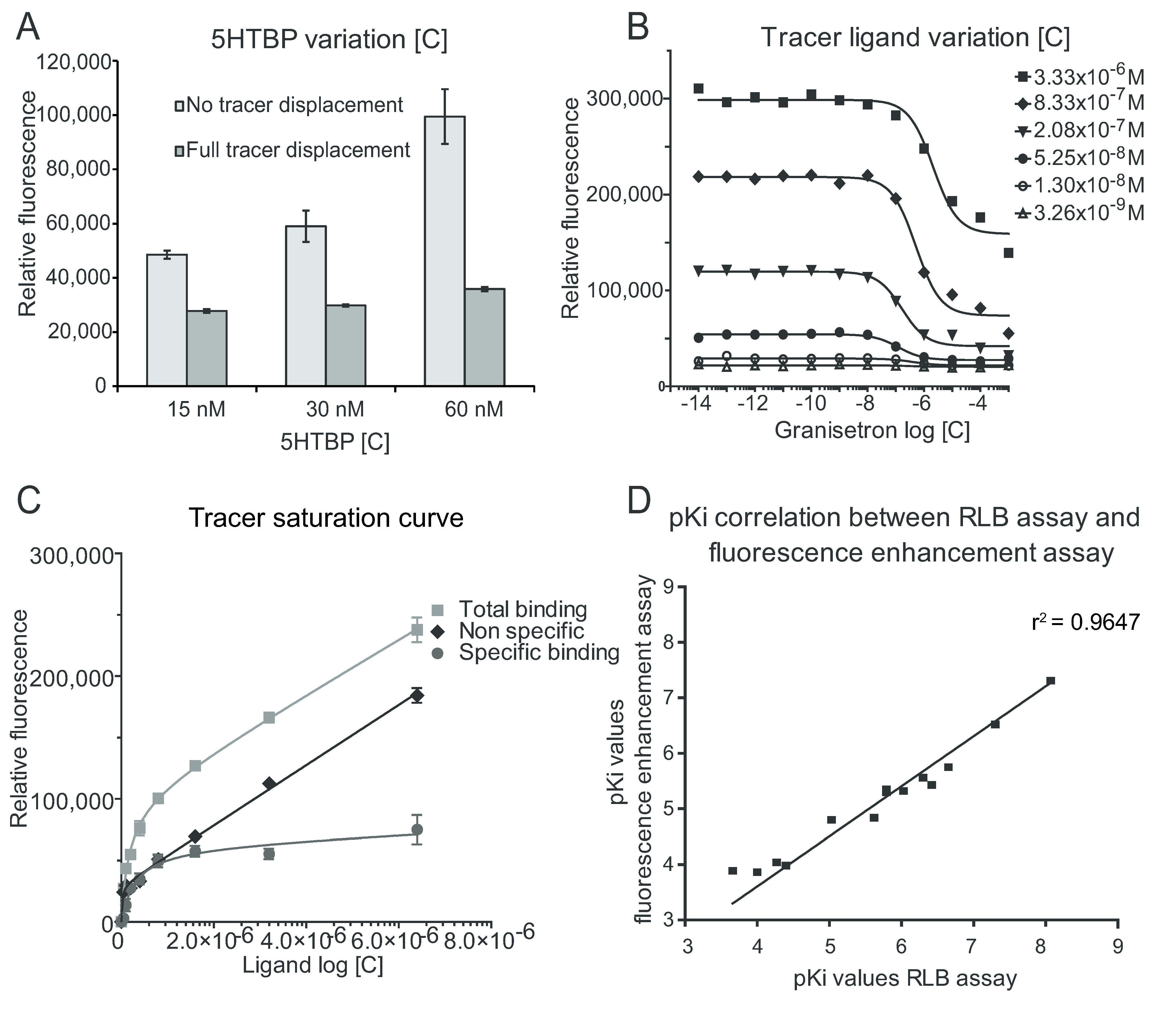
| Tracer c (M) | Z’-factor | Dynamic range | S/B | S/N | SW |
|---|---|---|---|---|---|
| 3.33 × 10−6 | 0.83 | 157302 | 2.16 | 18.8 | 4.1 |
| 8.33 × 10−7 | 0.77 | 136610 | 3.36 | 61.1 | 1.5 |
| 2.08 × 10−7 | 0.76 | 76749 | 3.26 | 38.0 | 1.4 |
| 5.25 × 10−8 | 0.87 | 23738 | 1.84 | 13.7 | 9.9 |
| 1.3 × 10−8 | −0.11 | 7513 | 1.30 | 1.8 | −4.3 |
| 3.26 × 10−9 | −0.99 | 3084 | 1.13 | 1.01 | −5.0 |
| 8.14 × 10−10 | ~0 | 45 | 1.00 | 0.01 | −5.3 |
2.3. Microfluidic On-Line Assay Optimization in Nano-LC Flow-injection Analysis Mode

2.4. Microfluidic On-Line HRS of Snake Venoms
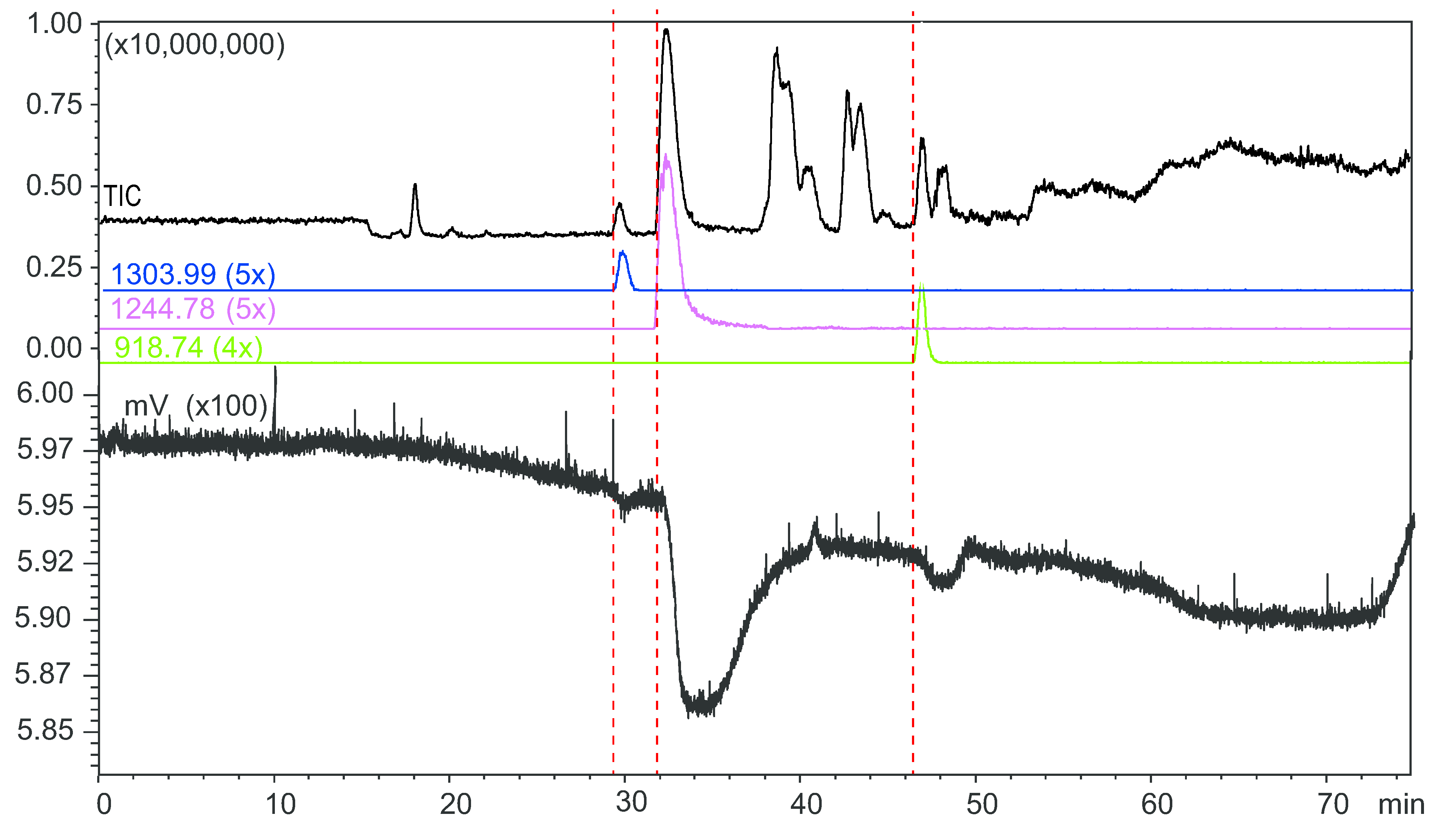

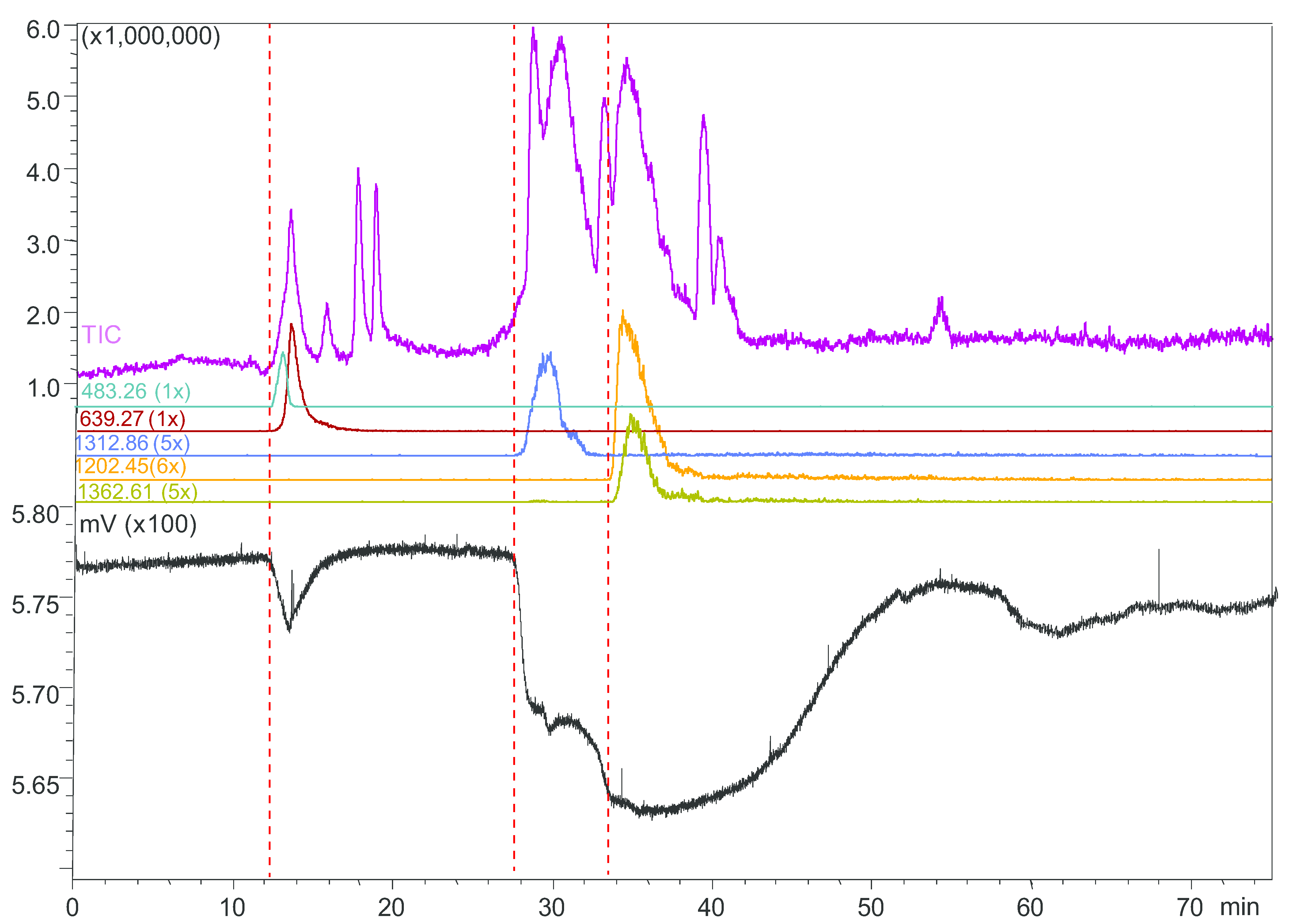
2.5. Protein Identification
3. Experimental Section
3.1. Chemicals and Biological Reagents
3.2. Expression and Purification of 5HTBPs
3.3. Snake Venom Samples
3.4. Fluorescence Enhancement by Anabaseine Derivatives with 5HTBP Constructs A1B2D1R and A1B2D1W
3.5. Fluorescence Enhancement Microplate Reader Assay
3.6. Radioligand Binding Assay with [3H]-granisetron
3.7. Microfluidic On-Line Assay Optimization in Nano-LC Flow-Injection Mode
3.8. Microfluidic On-Line HRS with Snake Venoms
3.9. Purification of Bioactives from Snake Venoms
3.10. Tryptic Digestion
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Berger, M.; Gray, J.A.; Roth, B.L. The expanded biology of serotonin. Annu. Rev. Med. 2009, 60, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Davies, P.A.; Pistis, M.; Hanna, M.C.; Peters, J.A.; Lambert, J.J.; Hales, T.G.; Kirkness, E.F. The 5-HT3B subunit is a major determinant of serotonin-receptor function. Nature 1999, 397, 359–363. [Google Scholar] [CrossRef] [PubMed]
- Filip, M.; Bader, M. Overview on 5-HT receptors and their role in physiology and pathology of the central nervous system. Pharmacol. Rep. 2009, 61, 761–777. [Google Scholar] [CrossRef]
- Hoyer, D.; Clarke, D.E.; Fozard, J.R.; Hartig, P.R.; Martin, G.R.; Mylecharane, E.J.; Saxena, P.R.; Humphrey, P.P. International Union of Pharmacology classification of receptors for 5-hydroxytryptamine (Serotonin). Pharmacol. Rev. 1994, 46, 157–203. [Google Scholar] [PubMed]
- Thompson, A.J.; Lester, H.A.; Lummis, S.C. The structural basis of function in Cys-loop receptors. Q. Rev. Biophys. 2010, 43, 449–499. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; Lummis, S.C. The 5-HT3 receptor as a therapeutic target. Expert Opin. Ther. Targets 2007, 11, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Walstab, J.; Rappold, G.; Niesler, B. 5-HT3 receptors: Role in disease and target of drugs. Pharmacol. Ther. 2010, 128, 146–169. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Spencer, R.H.; Kiss, L. High throughput assay technologies for ion channel drug discovery. Assay Drug Dev. Technol. 2004, 2, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; Verheij, M.H.; Leurs, R.; de Esch, I.J.; Lummis, S.C. An efficient and information-rich biochemical method design for fragment library screening on ion channels. Biotechniques 2010, 49, 822–829. [Google Scholar] [CrossRef] [PubMed]
- Kesters, D.; Thompson, A.J.; Brams, M.; van Elk, R.; Spurny, R.; Geitmann, M.; Villalgordo, J.M.; Guskov, A.; Danielson, U.H.; Lummis, S.C.; et al. Structural basis of ligand recognition in 5-HT3 receptors. EMBO Rep. 2013, 14, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.B.; Syed, N.I.; Schaap, D.; van Minnen, J.; Klumperman, J.; Kits, K.S.; Lodder, H.; van der Schors, R.C.; van Elk, R.; Sorgedrager, B.; et al. A glia-derived acetylcholine-binding protein that modulates synaptic transmission. Nature 2001, 411, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Werner, P.; Kawashima, E.; Reid, J.; Hussy, N.; Lundstrom, K.; Buell, G.; Humbert, Y.; Jones, K.A. Organization of the mouse 5-HT3 receptor gene and functional expression of two splice variants. Brain Res. Mol. Brain Res. 1994, 26, 233–241. [Google Scholar] [CrossRef]
- Lummis, S.C.; Thompson, A.J.; Bencherif, M.; Lester, H.A. Varenicline is a potent agonist of the human 5-hydroxytryptamine3 receptor. J. Pharmacol. Exp. Ther. 2011, 339, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Drisdel, R.C.; Sharp, D.; Henderson, T.; Hales, T.G.; Green, W.N. High affinity binding of epibatidine to serotonin type 3 receptors. J. Biol. Chem. 2008, 283, 9659–9665. [Google Scholar] [CrossRef] [PubMed]
- Macor, J.E.; Gurley, D.; Lanthorn, T.; Loch, J.; Mack, R.A.; Mullen, G.; Tran, O.; Wright, N.; Gordon, J.C. The 5-HT3 antagonist tropisetron (ICS 205-930) is a potent and selective alpha7 nicotinic receptor partial agonist. Bioorg. Med. Chem. Lett. 2001, 11, 319–321. [Google Scholar] [CrossRef]
- Dastmalchi, K.; Ollilainen, V.; Lackman, P.; Boije af Gennas, G.; Dorman, H.J.; Jarvinen, P.P.; Yli-Kauhaluoma, J.; Hiltunen, R. Acetylcholinesterase inhibitory guided fractionation of Melissa officinalis L. Bioorg. Med. Chem. 2009, 17, 867–871. [Google Scholar] [CrossRef] [PubMed]
- Phillipson, D.W.; Milgram, K.E.; Yanovsky, A.I.; Rusnak, L.S.; Haggerty, D.A.; Farrell, W.P.; Greig, M.J.; Xiong, X.; Proefke, M.L. High-throughput bioassay-guided fractionation: A technique for rapidly assigning observed activity to individual components of combinatorial libraries, screened in HTS bioassays. J. Comb. Chem. 2002, 4, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Kool, J.; Giera, M.; Irth, H.; Niessen, W.M. Advances in mass spectrometry-based post-column bioaffinity profiling of mixtures. Anal. Bioanal. Chem. 2011, 399, 2655–2668. [Google Scholar] [CrossRef] [PubMed]
- Przyjazny, A.; Kjellstrom, T.L.; Bachas, L.G. High-performance liquid chromatographic postcolumn reaction detection based on a competitive binding system. Anal. Chem. 1990, 62, 2536–2540. [Google Scholar] [CrossRef] [PubMed]
- Schobel, U.; Frenay, M.; van Elswijk, D.A.; McAndrews, J.M.; Long, K.R.; Olson, L.M.; Bobzin, S.C.; Irth, H. High resolution screening of plant natural product extracts for estrogen receptor alpha and beta binding activity using an online HPLC-MS biochemical detection system. J. Biomol. Screen 2001, 6, 291–303. [Google Scholar] [PubMed]
- Heus, F.; Vonk, F.; Otvos, R.A.; Bruyneel, B.; Smit, A.B.; Lingeman, H.; Richardson, M.; Niessen, W.M.; Kool, J. An efficient analytical platform for on-line microfluidic profiling of neuroactive snake venoms towards nicotinic receptor affinity. Toxicon 2013, 61, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Chung, B.G.; Langer, R.; Khademhosseini, A. Microfluidics for drug discovery and development: From target selection to product lifecycle management. Drug Discov. Today 2008, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A. Strategies for discovering drugs from previously unexplored natural products. Drug Discov. Today 2000, 5, 294–300. [Google Scholar] [CrossRef]
- Vetter, I.; Davis, J.L.; Rash, L.D.; Anangi, R.; Mobli, M.; Alewood, P.F.; Lewis, R.J.; King, G.F. Venomics: A new paradigm for natural products-based drug discovery. Amino Acids 2011, 40, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Kool, J.; de Kloe, G.E.; Bruyneel, B.; de Vlieger, J.S.; Retra, K.; Wijtmans, M.; van Elk, R.; Smit, A.B.; Leurs, R.; Lingeman, H.; et al. Online fluorescence enhancement assay for the acetylcholine binding protein with parallel mass spectrometric identification. J. Med. Chem. 2010, 53, 4720–4730. [Google Scholar] [CrossRef] [PubMed]
- Falck, D.; de Vlieger, J.S.; Niessen, W.M.; Kool, J.; Honing, M.; Giera, M.; Irth, H. Development of an online p38alpha mitogen-activated protein kinase binding assay and integration of LC-HR-MS. Anal. Bioanal. Chem. 2010, 398, 1771–1780. [Google Scholar] [CrossRef] [PubMed]
- Tyler, M.I.; Spence, I.; Barnett, D.; Howden, M.E. Pseudonajatoxin b: Unusual amino acid sequence of a lethal neurotoxin from the venom of the Australian common brown snake, Pseudonaja textilis. Eur. J. Biochem. 1987, 166, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; Verheij, M.H.; de Esch, I.J.; Lummis, S.C. VUF10166, a novel compound with differing activities at 5-HT(3)A and 5-HT(3)AB receptors. J. Pharmacol. Exp. Ther. 2012, 341, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999, 4, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Heus, F.; Giera, M.; de Kloe, G.E.; van Iperen, D.; Buijs, J.; Nahar, T.T.; Smit, A.B.; Lingeman, H.; de Esch, I.J.; Niessen, W.M.; et al. Development of a microfluidic confocal fluorescence detection system for the hyphenation of nano-LC to on-line biochemical assays. Anal. Bioanal. Chem. 2010, 398, 3023–3032. [Google Scholar] [CrossRef] [PubMed]
- Schebb, N.H.; Heus, F.; Saenger, T.; Karst, U.; Irth, H.; Kool, J. Development of a countergradient parking system for gradient liquid chromatography with online biochemical detection of serine protease inhibitors. Anal. Chem. 2008, 80, 6764–6772. [Google Scholar] [CrossRef] [PubMed]
- Nijmeijer, S.; Vischer, H.F.; Rudebeck, A.F.; Fleurbaaij, F.; Falck, D.; Leurs, R.; Niessen, W.M.; Kool, J. Development of a profiling strategy for metabolic mixtures by combining chromatography and mass spectrometry with cell-based GPCR signaling. J. Biomol. Screen. 2012, 17, 1329–1338. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Otvos, R.A.; Iyer, J.K.; Van Elk, R.; Ulens, C.; Niessen, W.M.A.; Somsen, G.W.; Kini, R.M.; Smit, A.B.; Kool, J. Development of Plate Reader and On-Line Microfluidic Screening to Identify Ligands of the 5-Hydroxytryptamine Binding Protein in Venoms. Toxins 2015, 7, 2336-2353. https://doi.org/10.3390/toxins7072336
Otvos RA, Iyer JK, Van Elk R, Ulens C, Niessen WMA, Somsen GW, Kini RM, Smit AB, Kool J. Development of Plate Reader and On-Line Microfluidic Screening to Identify Ligands of the 5-Hydroxytryptamine Binding Protein in Venoms. Toxins. 2015; 7(7):2336-2353. https://doi.org/10.3390/toxins7072336
Chicago/Turabian StyleOtvos, Reka A., Janaki Krishnamoorthy Iyer, René Van Elk, Chris Ulens, Wilfried M. A. Niessen, Govert W. Somsen, R. Manjunatha Kini, August B. Smit, and Jeroen Kool. 2015. "Development of Plate Reader and On-Line Microfluidic Screening to Identify Ligands of the 5-Hydroxytryptamine Binding Protein in Venoms" Toxins 7, no. 7: 2336-2353. https://doi.org/10.3390/toxins7072336
APA StyleOtvos, R. A., Iyer, J. K., Van Elk, R., Ulens, C., Niessen, W. M. A., Somsen, G. W., Kini, R. M., Smit, A. B., & Kool, J. (2015). Development of Plate Reader and On-Line Microfluidic Screening to Identify Ligands of the 5-Hydroxytryptamine Binding Protein in Venoms. Toxins, 7(7), 2336-2353. https://doi.org/10.3390/toxins7072336







