Screening a Strain of Aspergillus niger and Optimization of Fermentation Conditions for Degradation of Aflatoxin B1 †
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation of Microorganisms
2.2. Determination of Aflatoxin B1 Degradation
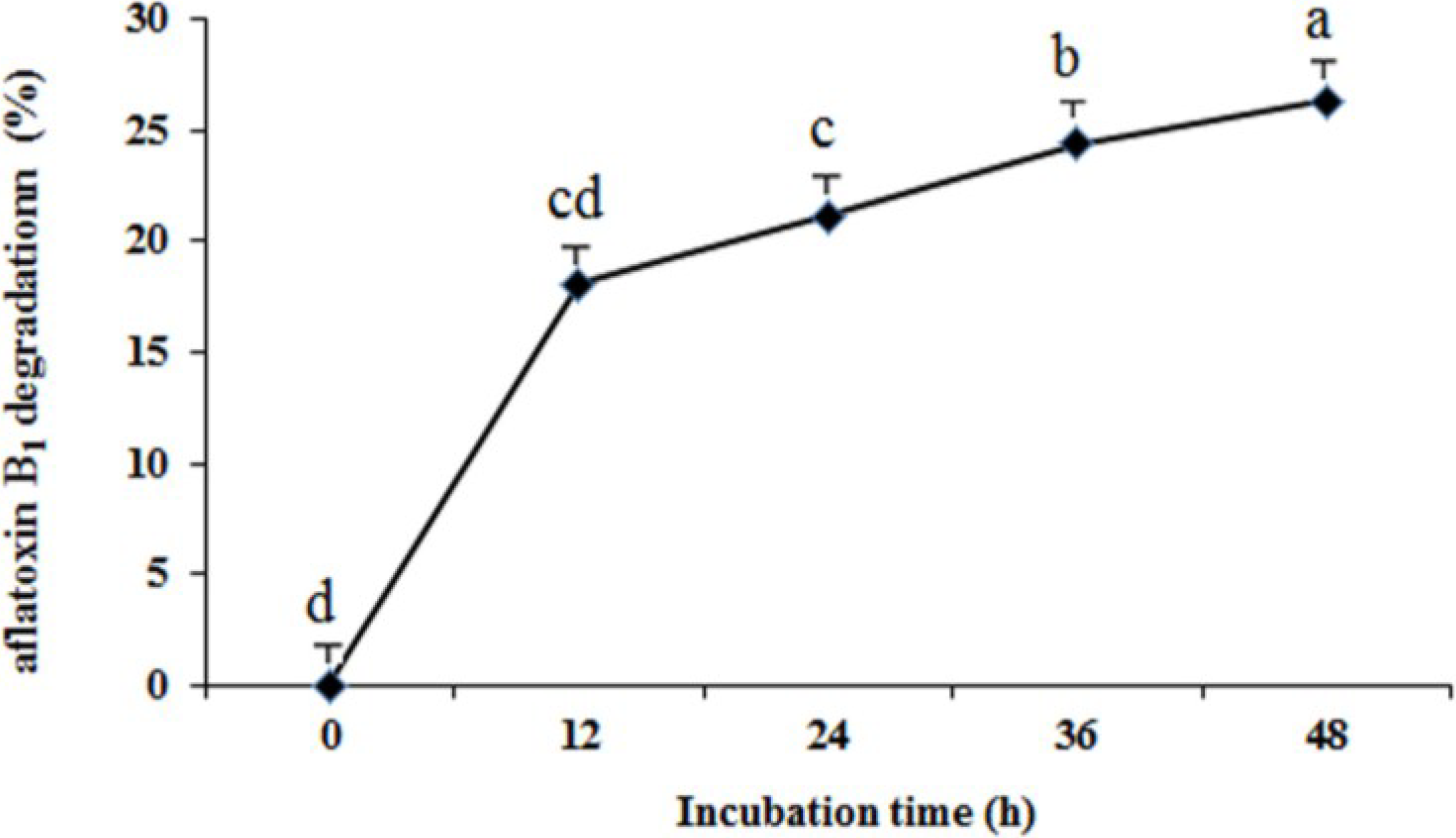
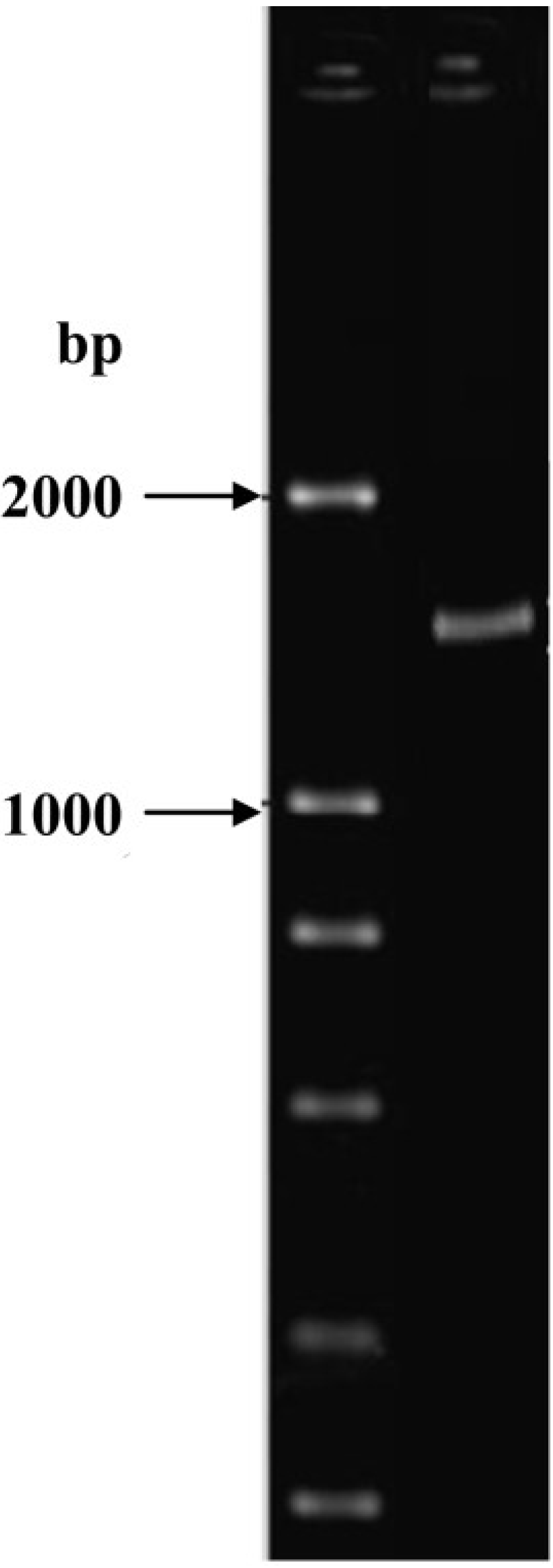
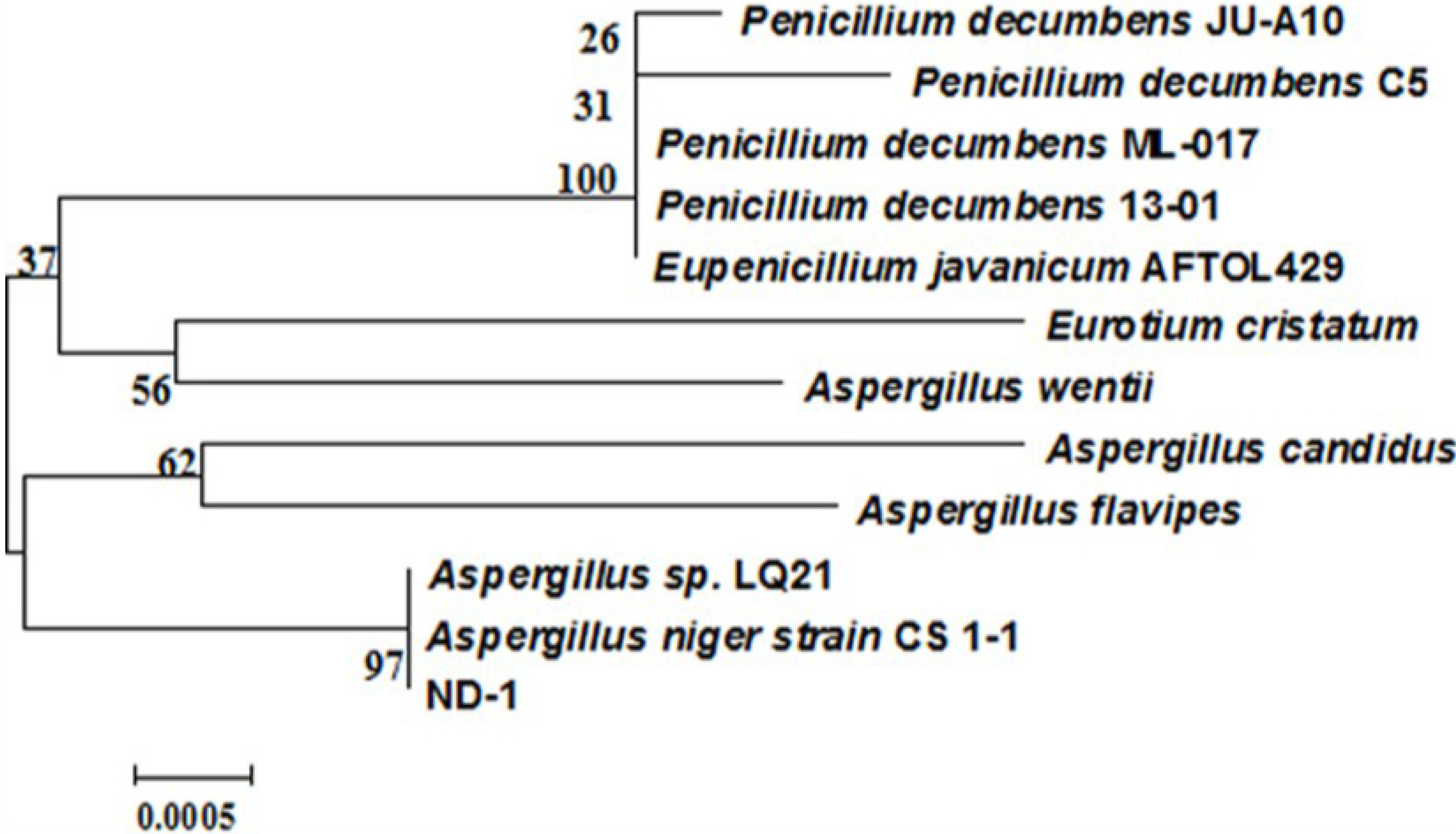
2.3. Identification of Isolate ND-1
2.4. Optimization of Fermentation Conditions for Aflatoxin B1 Degradation
| Carbon sources | Aflatoxin B1 degradation (%) | Concentrations of starch (%) | Aflatoxin B1 degradation (%) |
|---|---|---|---|
| Starch | 33.5 ± 1.5 A | 4.0 | 35.0 ± 2.5 A |
| Lactose | 14.9 ± 1.1 B | 3.0 | 34.0 ± 1.5 A |
| Galactose | 11.1 ± 1.6 B,C | 2.0 | 33.1 ± 1.6 A |
| Sucrose | 8.5 ± 0.9 B,C | 1.0 | 19.5 ± 2.2 B |
| Maltose | 8.0 ± 0.7 B,C | 0.5 | 14.0 ± 0.5 B,C |
| Glucose | 5.74 ± 0.4 B,C | 0.2 | 9.0 ± 1.0 C |
| Mannitol | 4.9 ± 0.5 C | - | - |
| Nitrogen sources | Aflatoxin B1 degradation (%) | Concentrations of tryptone (%) | Aflatoxin B1 degradation (%) | Initial pH value | Aflatoxin B1 degradation (%) |
|---|---|---|---|---|---|
| tryptone | 50.7 ± 0.6 a | 0.5 | 50.7 ± 0.6 A | 6.0 | 58.2 ± 0.9 A |
| Acidicase peptone | 47.0 ± 1.0 b | 0.7 | 50.4 ± 0.4 A | 6.5 | 50.7 ± 0.6 B |
| Proteose peptone | 45.1 ± 1.5 b | 0.9 | 49.8 ± 0.3 A | 7.0 | 49.2 ± 1.0 B |
| Ammonium nitrate | 39.1 ± 1.1 c | 0.3 | 38.9 ± 1.6 B | 7.5 | 47.9 ± 1.6 B |
| Mixed ammonium salt | 37.0 ± 1.0 c | 0.1 | 14.1 ± 0.9 C | 8.0 | 47.1 ± 1.1 B |
| Beef extract peptone | 33.3 ± 0.8 d | - | - | 5.5 | 35.2 ± 0.9 C |
| Peptone | 33.1 ± 1.6 d | - | - | 5.0 | 27.4 ± 1.8 D |
| Beef extract | 32.6 ± 1.1 d | - | - | - | - |
2.5. Degradation of Aflatoxin B1 by the Supernatant, Cells and Cell Extracts of ND-1
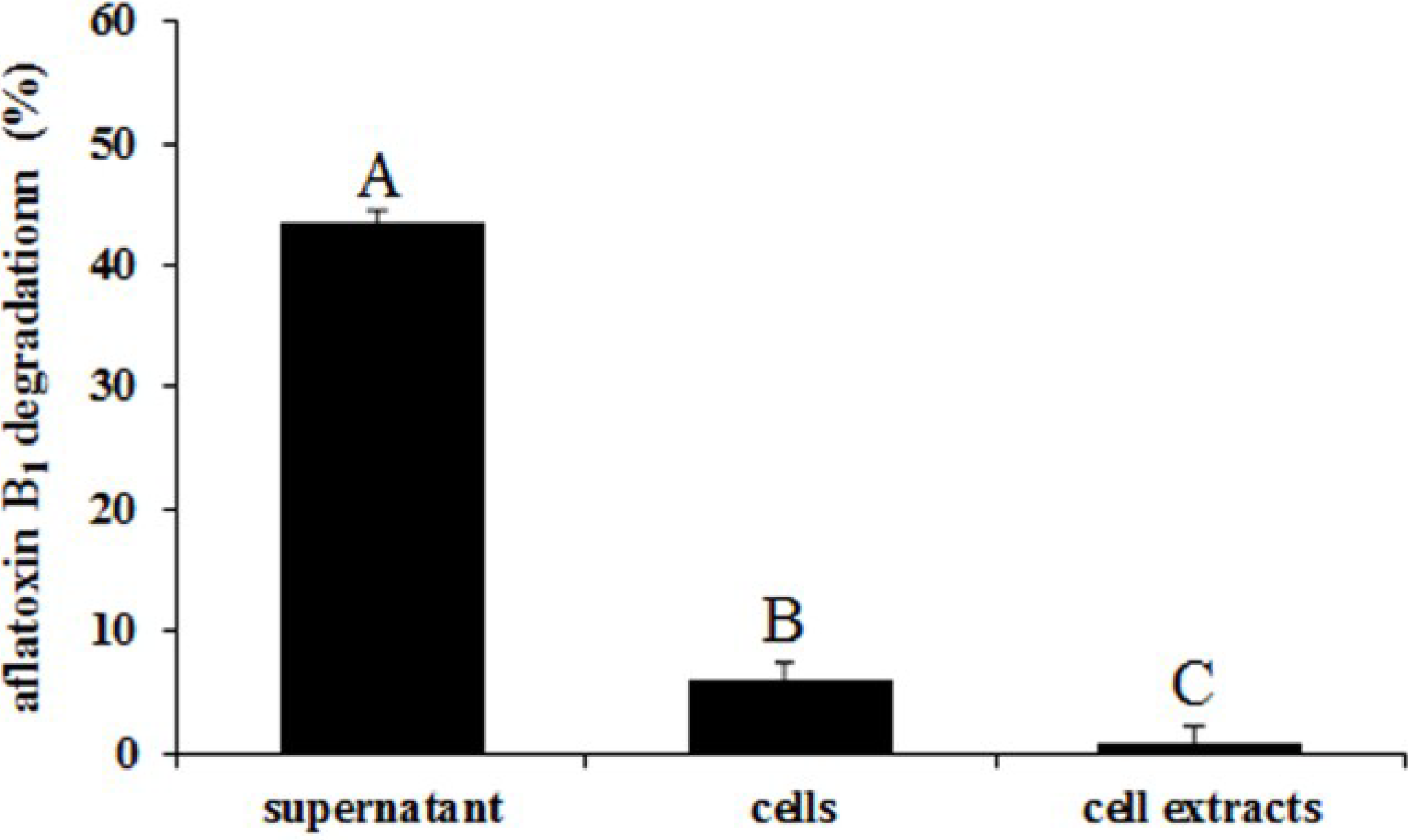
2.6. Effects of Heat Treatment, Temperature, pH, and Metal Ions on Aflatoxin B1 Degradation by the Supernatant
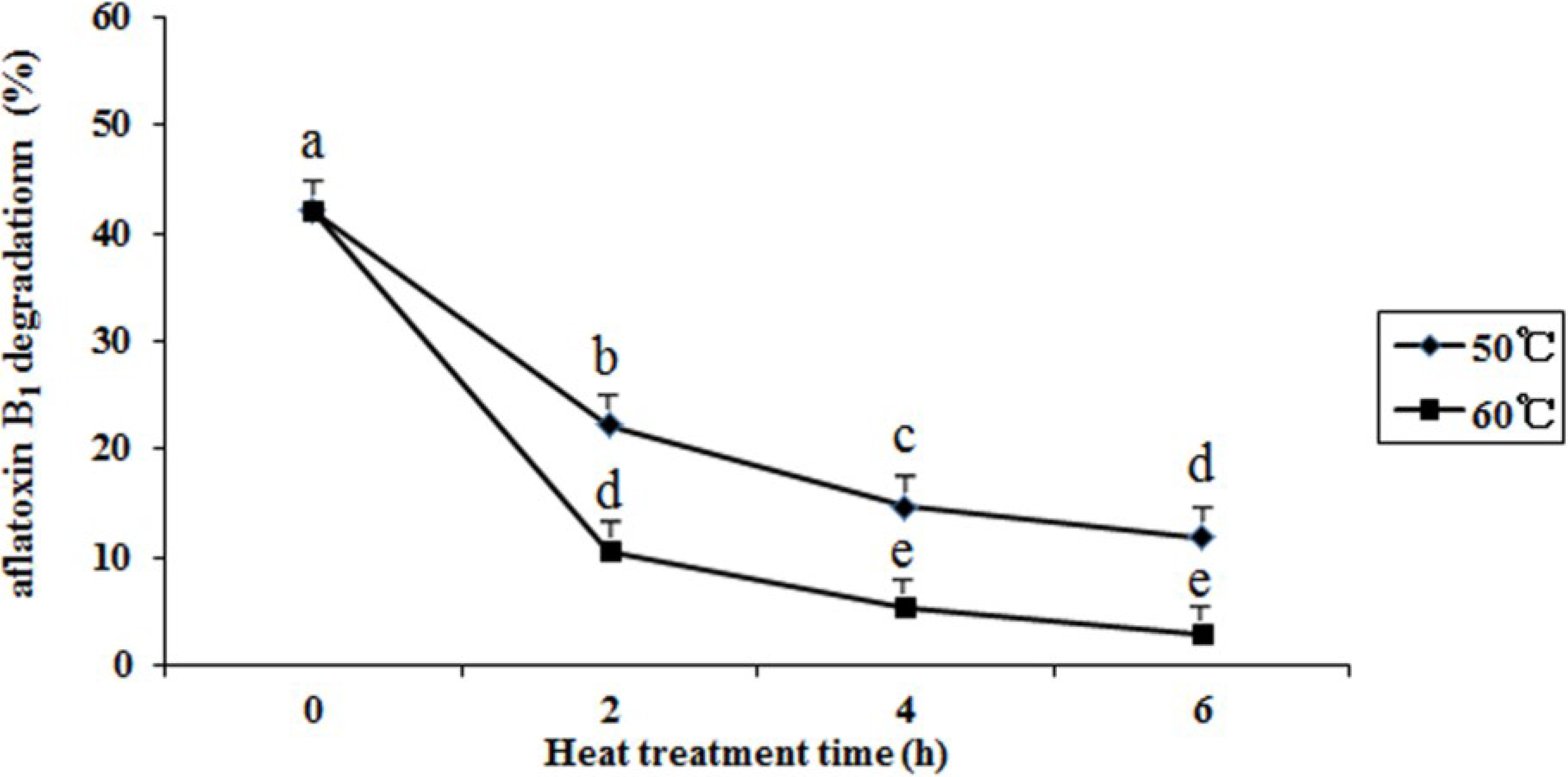
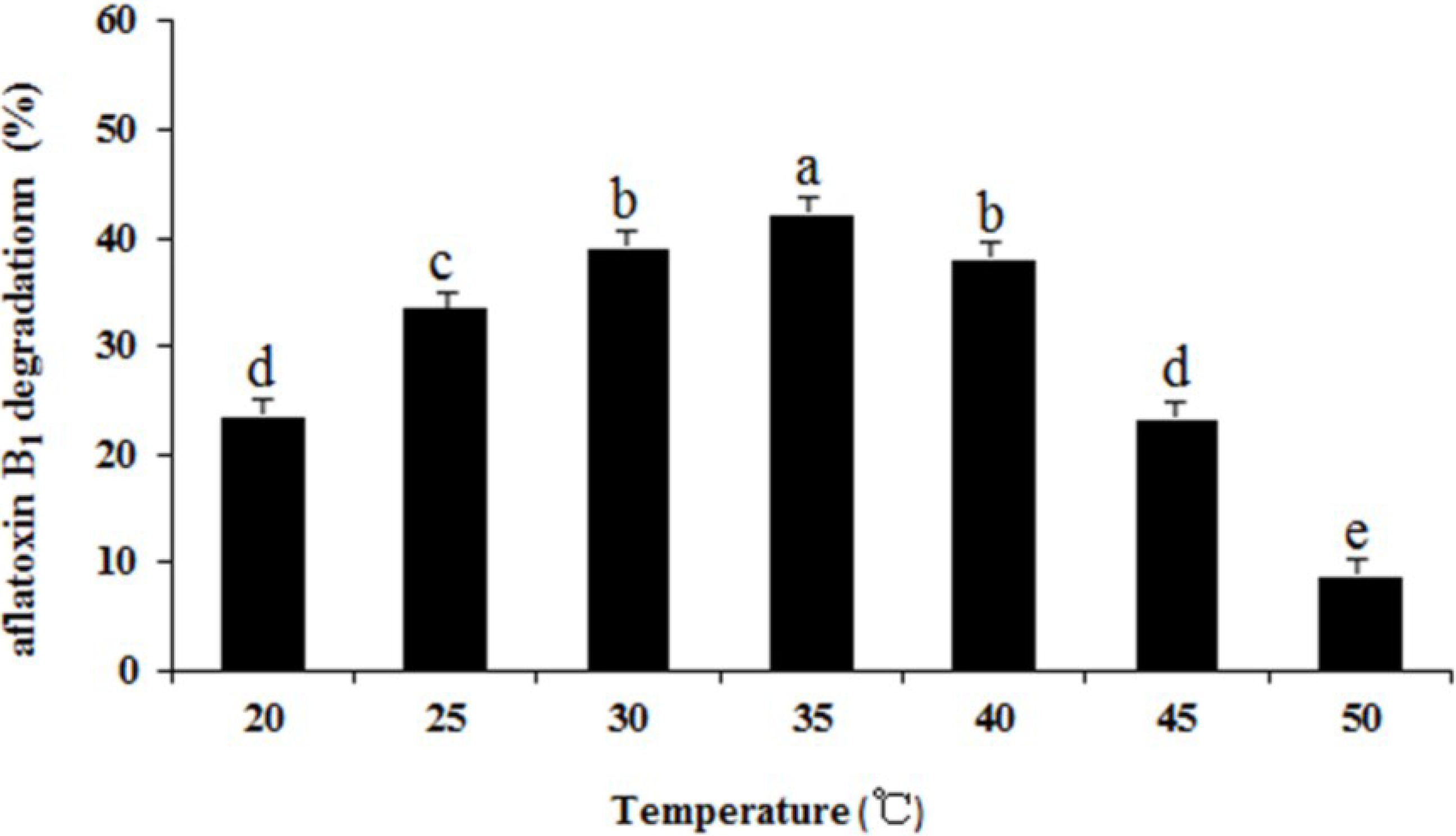
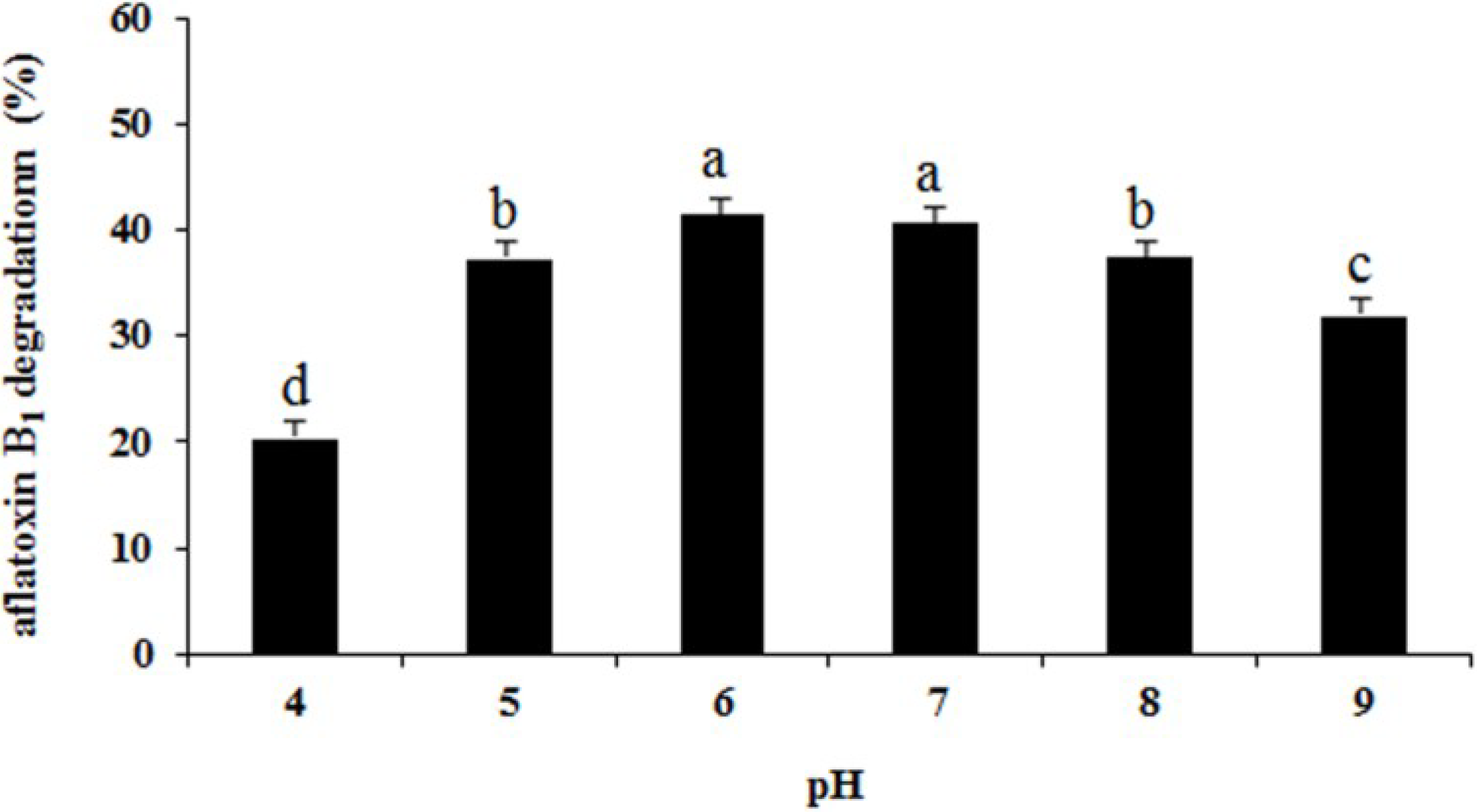
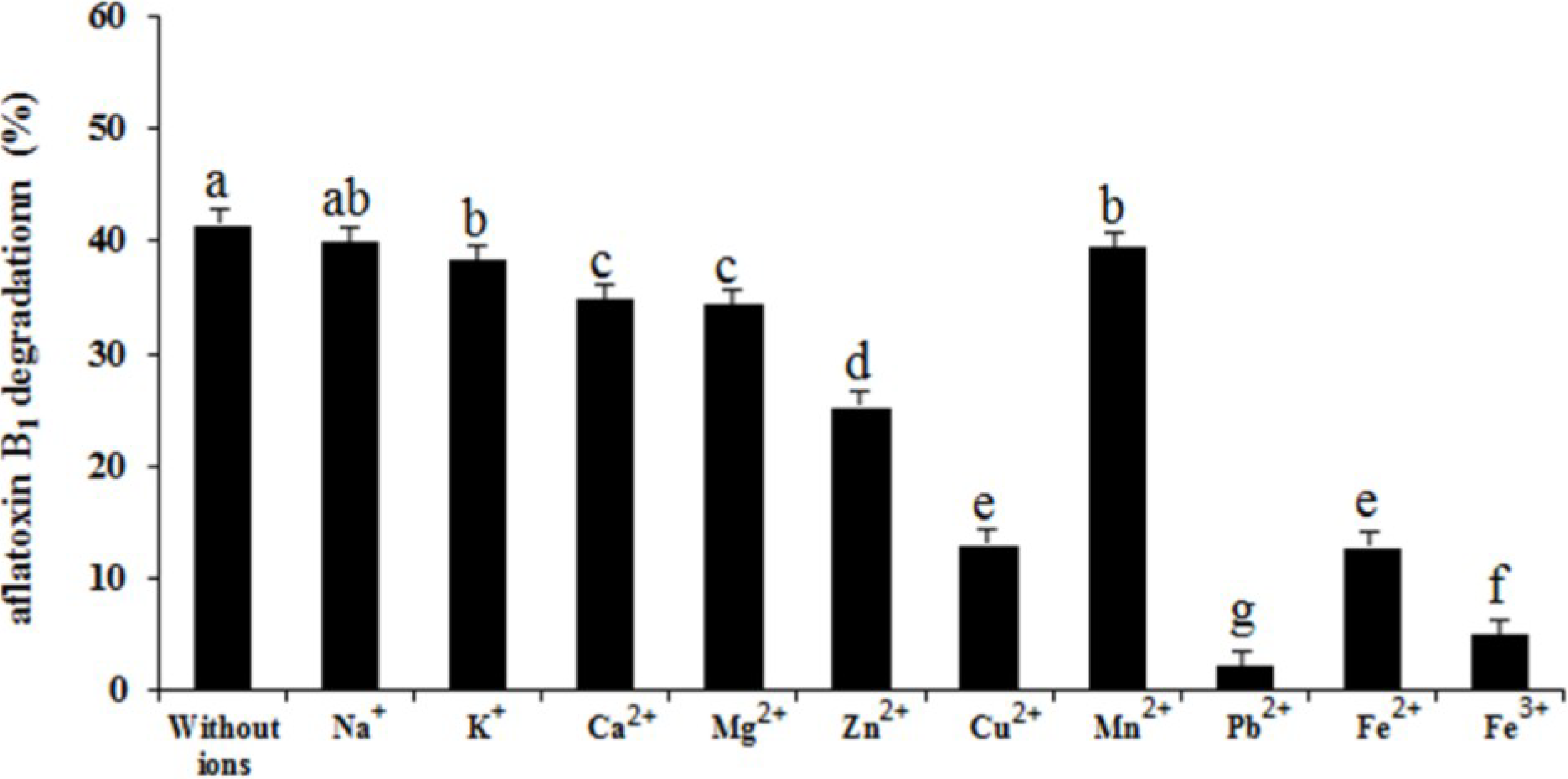
3. Materials and Methods
3.1. Culture Media
3.2. Isolation of Microorganisms
3.3. Determination of Aflatoxin B1 Degradation
- X1: The concentration of aflatoxin B1 in the control after fermentation;
- X2: The concentration of aflatoxin B1 in treatment after fermentation;
- X3: The initial concentration of aflatoxin B1 before fermentation.
3.4. Identification of the Aflatoxin B1 Degradation Strains
3.5. Optimization of Fermentation Conditions
3.5.1. Optimization of Fermentation Medium
3.5.2. Optimization of Incubation Temperature, Period, Amount of Inoculum and pH
| Factors | A (°C) | B (h) | C (%) |
|---|---|---|---|
| Level 1 | 28 | 12 | 1 |
| Level 2 | 32 | 24 | 3 |
| Level 3 | 36 | 36 | 5 |
| Level 4 | 40 | 48 | 7 |
3.6. Degradation of Aflatoxin B1 by the Supernatant, Cells and Cell Extracts of Strains
3.7. Effects of Heat Treatment, Temperature, pH and Metal Ions on Aflatoxin B1 Degradation by the Supernatant
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Diener, U.L.; Cole, R.J.; Sanders, T.H.; Payne, G.A.; Lee, L.S.; Klich, M.A. Epidemiology of aflatoxin formation by Aspergillus flavus. Annu. Rev. Phytopathol. 1987, 25, 240–270. [Google Scholar] [CrossRef]
- Leontopoulos, D.; Siafaka, A.; Markaki, P. Black olives as substrate for Aspergillus parasiticus growth and aflatoxin B1 production. Food Microbiol. 2003, 20, 119–126. [Google Scholar]
- Ito, Y.; Peterson, S.W.; Wicklow, D.T.; Goto, T. Aspergillus pseudotamarii, a new aflatoxin-producing species in Aspergillus section Flavi. Mycol. Res. 2001, 105, 233–239. [Google Scholar] [CrossRef]
- Peterson, S.W.; Ito, Y.; Horn, B.W.; Goto, T. Aspergillus bombycis, a new aflatoxigenic species and genetic variation in its sibling species, A. nomius. Mycologia 2001, 93, 689–703. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Horn, B.W.; Hesseltine, C.W. Aspergillus nomius, a new aflatoxin-producing species related to Aspergillus flavus and Aspergillus tamari. Antonie Van Leeuwenhoek 1987, 53, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Richard, J.L.; Payne, G.A.; Desjardins, A.E. Mycotoxins: Risks in plant, animal and human systems. CAST Task Force Rep. 2003, 139, 101–103. [Google Scholar]
- Hussein, S.H.; Brasel, J.M. Toxicity, metabolism, and impact of mycotoxins on humans and animals. Toxicology 2001, 167, 101–134. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Albores, A.; Arámbula-Villa, G.; Loarca-Piña, M.G.F.; Castaño-Tostado, E.; Moreno-Martínez, E. Safety and efficacy evaluation of aqueous citric acid to degrade B-aflatoxins in maize. Food Chem. Toxicol. 2005, 43, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Gowda, N.K.S.; Suganthi, R.U.; Malathi, V.; Raghavendra, A. Efficacy of heat treatment and sun drying of aflatoxin-contaminated feed for reducing the harmful biological effects in sheep. Anim. Feed Sci. Technol. 2007, 133, 167–175. [Google Scholar] [CrossRef]
- Basappa, S.C.; Shantha, T. Methods for detoxification of Aflatoxins in foods and feeds—A critical appraisal. J. Food Sci. Technol. 1996, 33, 95–107. [Google Scholar]
- Henry, S.H.; Bosch, F.X.; Troxell, T.C. Reducing liver cancer—Global control of aflatoxin. Science 1999, 286, 2453–2454. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P.; Johnson, W.W.; Ueng, Y.F.; Yamazaki, H.; Shimada, T. Involvement of cytochrome P450, glutathione S-transferase, and epoxide hydrolase in the metabolism of aflatoxin B1 and relevance to risk of human liver cancer. Environ. Health Perspect. 1996, 104, 557–562. [Google Scholar] [PubMed]
- Laciaková, A.; Ciconova, P.; Máté, D.; Laciak, V. Aflatoxins and possibilities for their biological detoxification. Med. Weter 2008, 64, 276–279. [Google Scholar]
- Farzaneh, M.; Shi, Z.Q.; Ghassempour, A.; Sedaghat, N.; Ahmadzadeh, M.; Mirabolfathy, M.; Javan-Nikkhah, M. Aflatoxin B1 degradation by Bacillus subtilis UTBSP1 isolated from pistachio nuts of Iran. Food Control 2012, 23, 100–106. [Google Scholar] [CrossRef]
- Khanafari, A.; Soudi, H.; Miraboulfathi, M.; Osboo, R.K. An in vitro investigation of Aflatoxin B1 biological control by Lactobacillus plantarum. J. Biol. Sci. 2007, 4, 2553–2556. [Google Scholar]
- Cao, H.; Liu, D.L.; Mo, X.M.; Xie, C.F.; Yao, D.S. A fungal enzyme with the ability of aflatoxin B1 conversion: Purification and ESI-MS/MS identification. Microbiol. Res. 2011, 166, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Hormisch, D.; Brost, I.; Kohring, G.W.; Giffhorn, F.; Kroppensted, R.M.; Stackebrandt, E.; Farber, P.; Holzapfel, W.H. Mycobacterium fluoranthenivorans sp. nov., a fluoranthene and aflatoxin B1 degrading bacterium from contaminated soil of a former coal gas plant. J. Appl. Microbiol. 2004, 27, 653–660. [Google Scholar]
- Alberts, J.F.; Engelbrecht, Y.; Steyn, P.S.; Holzapfel, W.H.; Vanzyl, W.H. Biological degradation of aflatoxin B1 by Rhodococcus erythropolis cultures. Int. J. Food Microbiol. 2006, 109, 121–126. [Google Scholar] [PubMed]
- Guan, S.; Ji, C.; Zhou, T.; Li, J.; Ma, Q.; Niu, T. Aflatoxin B1 degradation by Stenotrophomonas maltophilia and other microbes selected using coumarin medium. Int. J. Mol. Sci. 2008, 9, 1489–1503. [Google Scholar] [PubMed]
- Samuel, M.S.; Sivaramakrishna, A.; Mehta, A. Degradation and detoxification of aflatoxin B1 by Pseudomonas putida. Int. Biodeterior. Biodegrad. 2014, 86, 202–209. [Google Scholar] [CrossRef]
- Abrunhosa, L.; Santos, L.; Venâncio, A. Degradation of ochratoxin A by proteases and by a crude enzyme of Aspergillus niger. Food Biotechnol. 2006, 20, 231–242. [Google Scholar] [CrossRef]
- Sun, X.; He, X.; Li, Y.; Xu, D.; Qian, H. Biological detoxification of zearalenone by Aspergillus niger strain FS10. Food Chem. Toxicol. 2014, 72, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Jonathan, S.G.; Fasidi, I.O. Effect of carbon, nitrogen and mineral sources on growth of Psathyerella atroumbonata (Pegler), a Nigerian edible mushroom. Food Chem. 2001, 72, 479–483. [Google Scholar]
- Brzonkalik, K.; Herrling, T.; Syldatk, C.; Neumann, A. The influence of different nitrogen and carbon sources on mycotoxin production in Alternaria alternate. Int. J. Food Microbiol. 2011, 147, 120–126. [Google Scholar] [PubMed]
- Nancib, A.; Nancib, N.; Meziane-Cherif, D.; Boubendir, A.; Fick, M.; Boudrant, J. Joint effect of nitrogen sources and B vitamin supplementation of date juice on lactic acid production by Lactobacillus casei subsp. Rhamnosus. Bioresour. Technol. 2005, 96, 63–67. [Google Scholar] [CrossRef]
- Kohut, G.; Ádám, A.L.; Fazekas, B.; Hornok, L. N-starvation stress induced FUM gene expression and fumonisin production is mediated via the HOG-type MAPK pathway in Fusarium proliferatum. Int. J. Food microbiol. 2009, 130, 65–69. [Google Scholar] [CrossRef]
- Sohail, M.; Siddiqi, R.; Ahmad, A.; Khan, S.A. Cellulase production from Aspergillus niger MS82: Effect of temperature and pH. New Biotechnol. 2009, 25, 437–441. [Google Scholar] [CrossRef]
- Sharma, A.; Vivekanand, V.; Singh, R.P. Solid-state fermentation for gluconic acid production from sugarcane molasses by Aspergillus niger ARNU-4 employing tea waste as the novel solid support. Bioresour. Technol. 2008, 99, 3444–3450. [Google Scholar] [PubMed]
- Sabu, A.; Augur, C.; Swati, C.; Pandey, A. Tannase production by Lactobacillus sp. ASR-S1 under solid-state fermentation. Process Biochem. 2006, 41, 575–580. [Google Scholar]
- Shu, C.H.; Lung, M.Y. Effect of pH on the production and molecular weight distribution of exopolysaccharide by Antrodia camphorate in batch cultures. Process Biochem. 2004, 39, 931–937. [Google Scholar] [CrossRef]
- Wang, Y.; McNeil, B. pH effects on exopolysaccharide and oxalic acid production in cultures of Sclerotium glucanicum. Enzym. Microb. Technol. 1995, 17, 124–130. [Google Scholar] [CrossRef]
- D’Souza, D.H.; Brackett, R.E. The role of trace metal ions in aflatoxin B1 degradation by Flavobacterium aurantiacum. J. Food Prot. 1998, 61, 1666–1669. [Google Scholar] [PubMed]
- D’Souza, D.H.; Brackett, R.E. The influence of divalent cations and chelators on aflatoxin B1 degradation by Flavobacterium aurantiacum. J. Food Prot. 2000, 63, 102–105. [Google Scholar] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial disk susceptibility tests; Approved Standard—Tenth Edition (M02-A10); CLSI: Wayne, PA, USA, 2009. [Google Scholar]
- Sivakumaran, S.; Bridge, P.; Roberts, P. Genetic relatedness among Filobasidiella species. Mycopathologia 2002, 156, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Teniola, O.D.; Addo, P.A.; Brost, I.M.; Färber, P.; Jany, K.D.; Alberts, J.F.; van Zyi, W.H.; Steyn, P.S.; Holzapfel, W.H. Degradation of aflatoxin B1 by cell-free extracts of Rhodococcus erythropolis and Mycobacterium fluoranthenivorans sp. nov. DSM44556T. Int. J. Food Microbiol. 2005, 105, 111–117. [Google Scholar] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Xue, B.; Li, M.; Mu, Y.; Chen, Z.; Li, J.; Shan, A. Screening a Strain of Aspergillus niger and Optimization of Fermentation Conditions for Degradation of Aflatoxin B1 . Toxins 2014, 6, 3157-3172. https://doi.org/10.3390/toxins6113157
Zhang W, Xue B, Li M, Mu Y, Chen Z, Li J, Shan A. Screening a Strain of Aspergillus niger and Optimization of Fermentation Conditions for Degradation of Aflatoxin B1 . Toxins. 2014; 6(11):3157-3172. https://doi.org/10.3390/toxins6113157
Chicago/Turabian StyleZhang, Wei, Beibei Xue, Mengmeng Li, Yang Mu, Zhihui Chen, Jianping Li, and Anshan Shan. 2014. "Screening a Strain of Aspergillus niger and Optimization of Fermentation Conditions for Degradation of Aflatoxin B1 " Toxins 6, no. 11: 3157-3172. https://doi.org/10.3390/toxins6113157
APA StyleZhang, W., Xue, B., Li, M., Mu, Y., Chen, Z., Li, J., & Shan, A. (2014). Screening a Strain of Aspergillus niger and Optimization of Fermentation Conditions for Degradation of Aflatoxin B1 . Toxins, 6(11), 3157-3172. https://doi.org/10.3390/toxins6113157




