Geographic Variation in Venom Proteome and Toxicity Profiles of Chinese Naja atra: Implications for Antivenom Optimization
Abstract
1. Introduction
2. Results
2.1. Regional Differences in Venom LD50
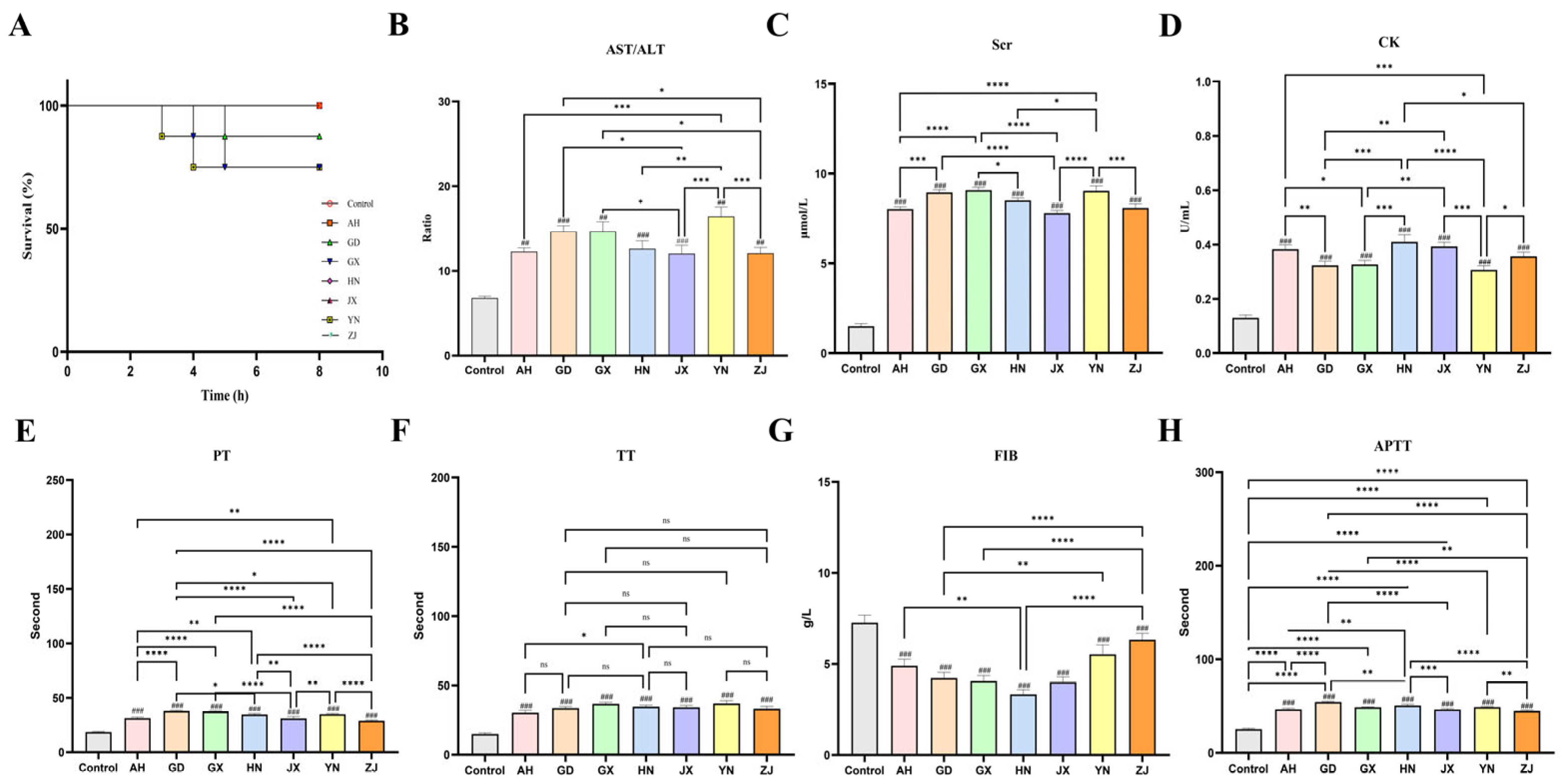
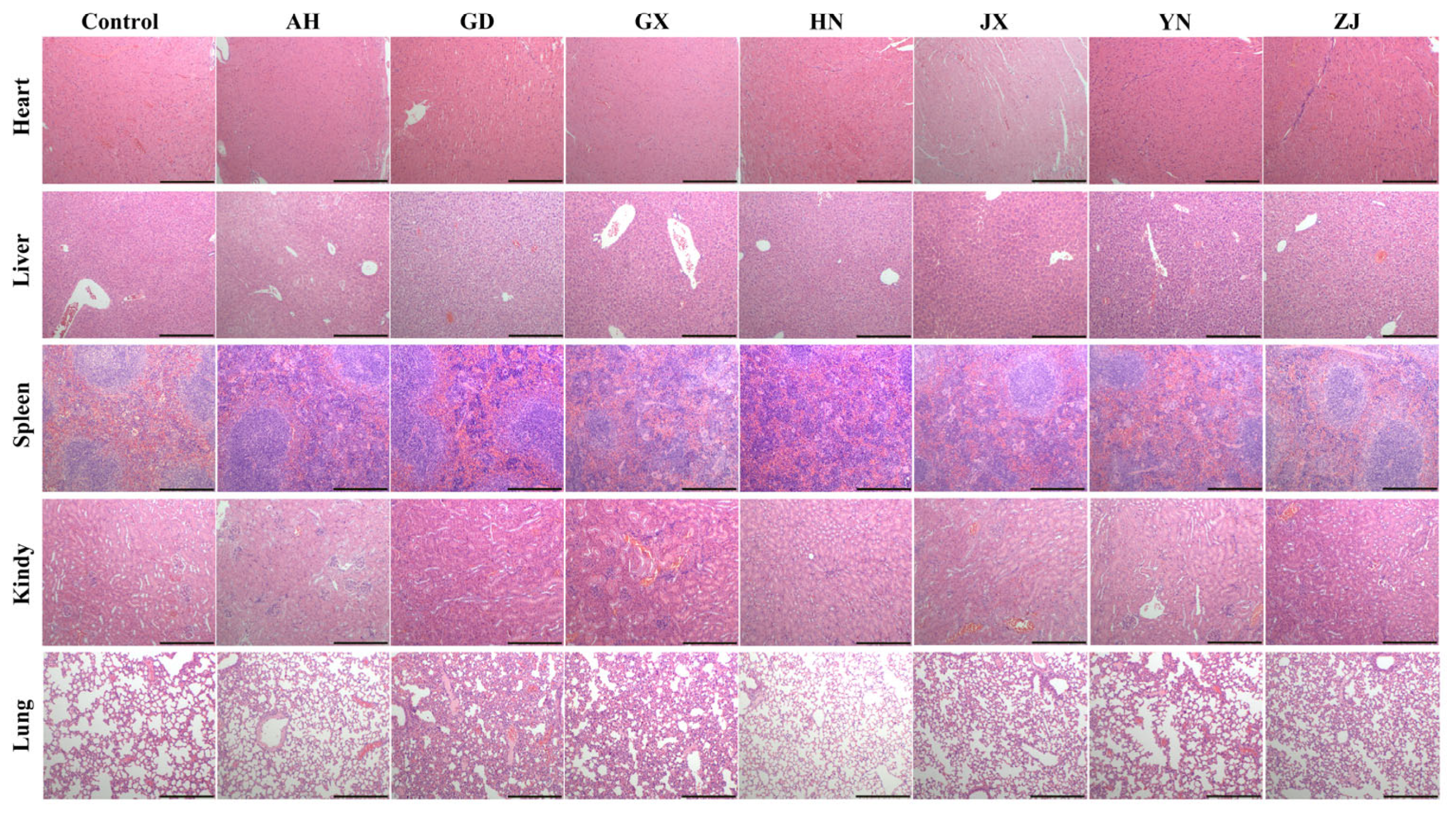
2.2. Geographic Variation in Systemic Toxicity and Multiorgan Damage Induced by Venom
2.3. Proteomic Profiling Reveals Geographical Variation in Naja atra Venom Composition
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Snake Venom
5.2. Animals Model and Ethics
5.3. LD50 Test
5.4. Determination of Neutralizing Efficacy of Antivenom Serum
5.5. Ser-Enzyme Assays
5.6. Evaluation of Hemostatic Parameters
5.7. Histological Analysis
5.8. Sample Preparation for LC-MS/MS Analysis
5.9. LC-MS/MS Analysis
5.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Calvete, J.J. Venomics: Integrative venom proteomics and beyond. Biochem. J. 2017, 474, 611–634. [Google Scholar] [CrossRef]
- Alfaro-Chinchilla, A.; Lomonte, B.; Zúniga, L.; Acevedo, M.; Neri-Castro, E.; Alagón, A.; Bonilla, F.; Diaz, C.; Sasa, M. Venom composition, toxicity and cross-neutralization by PoliVal-ICP antivenom, of Mesoamerican jumping pitvipers genus Metlapilcoatlus (Viperidae: Crotalinae). Trans. R. Soc. Trop. Med. Hyg. 2025, 119, 954–970. [Google Scholar] [CrossRef] [PubMed]
- Chong, H.P.; Tan, K.Y.; Liu, B.S.; Sung, W.C.; Tan, C.H. Cytotoxicity of Venoms and Cytotoxins from Asiatic Cobras (Naja kaouthia, Naja sumatrana, Naja atra) and Neutralization by Antivenoms from Thailand, Vietnam, and Taiwan. Toxins 2022, 14, 334. [Google Scholar] [CrossRef]
- Liu, C.C.; You, C.H.; Wang, P.J.; Yu, J.S.; Huang, G.J.; Liu, C.H.; Hsieh, W.C.; Lin, C.C. Analysis of the efficacy of Taiwanese freeze-dried neurotoxic antivenom against, and through proteomics and animal model approaches. PLoS Negl. Trop. Dis. 2017, 11, e0006138. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Zhang, Y.Q.; Qian, X.R.; Zhao, H.Y.; Lu, H.L.; Gao, J.F. First Look at the Venoms of Two Snakes: Differences in Yield, Proteomic Profiles, and Immunorecognition by Commercial Antivenoms. Toxins 2025, 17, 19. [Google Scholar] [CrossRef] [PubMed]
- Boldrini-França, J.; Corrêa-Netto, C.; Silva, M.M.S.; Rodrigues, R.S.; De La Torre, P.; Pérez, A.; Soares, A.M.; Zingali, R.B.; Nogueira, R.A.; Rodrigues, V.M.; et al. Snake venomics and antivenomics of subspecies from Brazil: Assessment of geographic variation and its implication on snakebite management. J. Proteom. 2010, 73, 1758–1776. [Google Scholar] [CrossRef]
- Calvete, J.J.; Sanz, L.; Pérez, A.; Borges, A.; Vargas, A.M.; Lomonte, B.; Angulo, Y.; Gutiérrez, J.M.; Chalkidis, H.M.; Mourao, R.H.V.; et al. Snake population venomics and antivenomics of: Paedomorphism along its transamazonian dispersal and implications of geographic venom variability on snakebite management. J. Proteom. 2011, 74, 510–527. [Google Scholar] [CrossRef]
- Sunagar, K.; Undheim, E.A.B.; Scheib, H.; Gren, E.C.K.; Cochran, C.; Person, C.E.; Koludarov, I.; Kelln, W.; Hayes, W.K.; King, G.F.; et al. Intraspecific venom variation in the medically significant Southern Pacific Rattlesnake: Biodiscovery, clinical and evolutionary implications. J. Proteom. 2014, 99, 68–83. [Google Scholar] [CrossRef]
- Kalita, B.; Mackessy, S.P.; Mukherjee, A.K. Proteomic analysis reveals geographic variation in venom composition of Russell’s Viper in the Indian subcontinent: Implications for clinical manifestations post-envenomation and antivenom treatment. Expert. Rev. Proteomic 2018, 15, 837–849. [Google Scholar] [CrossRef]
- Oliveira, A.L.; Viegas, M.F.; da Silva, S.L.; Soares, A.M.; Ramos, M.J.; Fernandes, P.A. The chemistry of snake venom and its medicinal potential. Nat. Rev. Chem. 2022, 6, 451–469. [Google Scholar] [CrossRef]
- Puschhof, J.; Post, Y.; Beumer, J.; Kerkkamp, H.M.; Bittenbinder, M.; Vonk, F.J.; Casewell, N.R.; Richardson, M.K.; Clevers, H. Derivation of snake venom gland organoids for in vitro venom production. Nat. Protoc. 2021, 16, 1494–1510. [Google Scholar] [CrossRef]
- Tasoulis, T.; Isbister, G.K. A current perspective on snake venom composition and constituent protein families. Arch. Toxicol. 2023, 97, 133–153. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.C.; Liu, P.Y.; Chiang, L.C.; Lai, C.S.; Lai, K.L.; Ho, C.H.; Wang, T.H.; Yang, C.C. Snakebite in Taiwan. Clin. Toxicol. 2018, 56, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Fu, K.P.; Zhao, J.Q.; Zhong, L.P.; Xu, H.Y.; Yu, X.H.; Bi, X.W.; Huang, C.H. Dual therapy with phospholipase and metalloproteinase inhibitors from alleviated acute kidney and liver injury caused by multiple snake venoms. Biomed. Pharmacother. 2024, 177, 116967. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.Q.; Liu, G.Y.; Shi, X.; Huang, C.H. Combination of Rhamnetin and RXP03 Mitigates Venom-Induced Toxicity in Murine Models: Preclinical Insights into Dual-Target Antivenom Therapy. Toxins 2025, 17, 280. [Google Scholar] [CrossRef]
- Huang, H.W.; Liu, B.S.; Chien, K.Y.; Chiang, L.C.; Huang, S.Y.; Sung, W.C.; Wu, W.G. Cobra venom proteome and glycome determined from individual snakes of reveal medically important dynamic range and systematic geographic variation. J. Proteom. 2015, 128, 92–104. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, Y.; Lee, W.H.; Xu, X.; Zhang, Y.; Zhao, R.P.; Zhang, Y.; Wang, W. Venom gland transcriptomes of two elapid snakes (Bungarus multicinctus and Naja atra) and evolution of toxin genes. BMC Genom. 2011, 12, 1. [Google Scholar] [CrossRef]
- Shan, L.L.; Gao, J.F.; Zhang, Y.X.; Shen, S.S.; He, Y.; Wang, J.; Ma, X.M.; Ji, X. Proteomic characterization and comparison of venoms from two elapid snakes from China. J. Proteom. 2016, 138, 83–94. [Google Scholar] [CrossRef]
- Xu, N.; Zhao, H.Y.; Yin, Y.; Shen, S.S.; Shan, L.L.; Chen, C.X.; Zhang, Y.X.; Gao, J.F.; Ji, X. Combined venomics, antivenomics and venom gland transcriptome analysis of the monocoled cobra from China. J. Proteom. 2017, 159, 19–31. [Google Scholar] [CrossRef]
- Marcussi, S.; Oliveira, C.Z.; Sant’Ana, C.D.; Quintero, A.; Menaldo, D.L.; Beleboni, R.O.; Stabeli, R.G.; Giglio, J.R.; Fontes, M.R.M.; Soares, A.M. Snake venom phospholipase A inhibitors: Medicinal chemistry and therapeutic potential. Curr. Top. Med. Chem. 2007, 7, 743–756. [Google Scholar] [CrossRef]
- Vulfius, C.A.; Kasheverov, I.E.; Kryukova, E.V.; Spirova, E.N.; Shelukhina, I.V.; Starkov, V.G.; Andreeva, T.V.; Faure, G.; Zouridakis, M.; Tsetlin, V.I.; et al. Pancreatic and snake venom presynaptically active phospholipases A inhibit nicotinic acetylcholine receptors. PLoS ONE 2017, 12, e0186206. [Google Scholar] [CrossRef] [PubMed]
- Sampat, G.H.; Hiremath, K.; Dodakallanavar, J.; Patil, V.S.; Harish, D.R.; Biradar, P.; Mahadevamurthy, R.K.; Barvaliya, M.; Roy, S. Unraveling snake venom phospholipase A: An overview of its structure, pharmacology, and inhibitors. Pharmacol. Rep. 2023, 75, 1454–1473. [Google Scholar] [CrossRef] [PubMed]
- Kumar, T.K.S.; Yang, P.W.; Lin, S.H.; Wu, C.Y.; Lei, B.; Lo, S.J.; Tu, S.C.; Yu, C. Cloning, direct expression, and purification of a snake venom cardiotoxin in Escherichia coli. Biochem. Biophys. Res. Commun. 1996, 219, 450–456. [Google Scholar] [CrossRef]
- Das, D.; Sharma, M.; Das, H.K.; Sahu, P.P.; Doley, R. Purification and Characterization of Nk-3FTx: A Three Finger Toxin from the Venom of North East Indian Monocled Cobra. J. Biochem. Mol. Toxic. 2016, 30, 59–70. [Google Scholar] [CrossRef]
- Averin, A.S.; Nenov, M.N.; Starkov, V.G.; Tsetlin, V.I.; Utkin, Y.N. Effects of Cardiotoxins from Cobra Venom on Rat Heart Muscle and Aorta: A Comparative Study of Toxin-Induced Contraction Mechanisms. Toxins 2022, 14, 88. [Google Scholar] [CrossRef]
- Sonavane, M.; Almeida, J.R.; Rajan, E.; Williams, H.F.; Townsend, F.; Cornish, E.; Mitchell, R.D.; Patel, K.; Vaiyapuri, S. Intramuscular Bleeding and Formation of Microthrombi during Skeletal Muscle Damage Caused by a Snake Venom Metalloprotease and a Cardiotoxin. Toxins 2023, 15, 530. [Google Scholar] [CrossRef]
- Fox, J.W.; Serrano, S.M.T. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008, 275, 3016–3030. [Google Scholar] [CrossRef]
- Markland, F.S.; Swenson, S. Snake venom metalloproteinases. Toxicon 2013, 62, 3–18. [Google Scholar] [CrossRef]
- Glenn, J.L.; Straight, R.C.; Wolfe, M.C.; Hardy, D.L. Geographical Variation in Crotalus-Scutulatus Scutulatus (Mojave Rattlesnake) Venom Properties. Toxicon 1983, 21, 119–130. [Google Scholar] [CrossRef]
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef]
- Qian, H.; Wang, X.L. Global relationships between beta diversity and latitude after accounting for regional diversity. Ecol. Inform. 2015, 25, 10–13. [Google Scholar] [CrossRef]
- Lin, L.H.; Hua, L.; Qu, Y.F.; Gao, J.F.; Ji, X. The Phylogeographical Pattern and Conservation of the Chinese Cobra across Its Range Based on Mitochondrial Control Region Sequences. PLoS ONE 2014, 9, e106944. [Google Scholar] [CrossRef]
- Lomonte, B.; Rey-Suárez, P.; Fernández, J.; Sasa, M.; Pla, D.; Vargas, N.; Bénard-Valle, M.; Sanz, L.; Corrêa-Netto, C.; Núñez, V.; et al. Venoms of coral snakes: Evolutionary trends in compositional patterns emerging from proteomic analyses. Toxicon 2016, 122, 7–25. [Google Scholar] [CrossRef]
- Laustsen, A.H. Guiding recombinant antivenom development by omics technologies. New Biotechnol. 2018, 45, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.; Chanda, A.; Mukherjee, A.K. Quantitative proteomic analysis of venom from Southern India common krait and identification of poorly immunogenic toxins by immune-profiling against commercial antivenom. Expert. Rev. Proteomic 2019, 16, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Glanville, J.; Bellin, M.; Pletnev, S.; Zhang, B.S.; Andrade, J.C.; Kim, S.; Tsao, D.; Verardi, R.; Bedi, R.; Liao, S.; et al. Snake venom protection by a cocktail of varespladib and broadly neutralizing human antibodies. Cell 2025, 188, 3117–3134.e11. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.F.; Slagboom, J.; Albulescu, L.O.; Somsen, G.W.; Vonk, F.J.; Casewell, N.R.; Kool, J. Neutralising effects of small molecule toxin inhibitors on nanofractionated coagulopathic Crotalinae snake venoms. Acta Pharmacol. Sin. B 2020, 10, 1835–1845. [Google Scholar] [CrossRef]
- Albulescu, L.O.; Xie, C.F.; Ainsworth, S.; Alsolaiss, J.; Crittenden, E.; Dawson, C.A.; Softley, R.; Bartlett, K.E.; Harrison, R.A.; Kool, J.; et al. A therapeutic combination of two small molecule toxin inhibitors provides broad preclinical efficacy against viper snakebite. Nat. Commun. 2021, 12, 6094. [Google Scholar] [CrossRef]
- Du, T.Y.; Hall, S.R.; Chung, F.L.C.Y.; Kurdyukov, S.; Crittenden, E.; Patel, K.; Dawson, C.A.; Westhorpe, A.P.; Bartlett, K.E.; Rasmussen, S.A.; et al. Molecular dissection of cobra venom highlights heparinoids as an antidote for spitting cobra envenoming. Sci. Transl. Med. 2024, 16, eadk4802. [Google Scholar] [CrossRef]
- Sanz, L.; Quesada-Bernat, S.; Ramos, T.; Casais-E-Silva, L.L.; Corrêa-Netto, C.; Silva-Haad, J.J.; Sasa, M.; Lomonte, B.; Calvete, J.J. New insights into the phylogeographic distribution of the 3FTx/PLA venom dichotomy across genus in South America. J. Proteom. 2019, 200, 90–101. [Google Scholar] [CrossRef]
- Yang, C.Y.; Ding, L.; He, Q.Y.; Chen, X.Y.; Zhu, H.T.; Chen, F.; Yang, W.Z.; Pan, Y.X.; Tai, Z.Y.; Zhang, W.H.; et al. Proteomic Profiling of Venoms from Bungarus suzhenae and B. bungaroides: Enzymatic Activities and Toxicity Assessment. Toxins 2024, 16, 494. [Google Scholar] [CrossRef]

| AH | GD | GX | HN | JX | YN | ZJ |
|---|---|---|---|---|---|---|
| 0.51 mg/kg | 0.31 mg/kg | 0.27 mg/kg | 0.48 mg/kg | 0.45 mg/kg | 0.23 mg/kg | 0.63 mg/kg |
| Category | Protein ID | Protein Name | Relative Quantification (Mean) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| AH | GD | GX | HN | JX | YN | ZJ | |||
| Metalloprotease | |||||||||
| A0A024AXX7 | p-III snake venom metalloprotease | / | 0.063% | / | 0.070% | / | 0.054% | 0.094% | |
| A0A194ARL7 | Metalloproteinase Type III | / | 0.028% | 0.028% | 0.021% | / | 0.019% | 0.013% | |
| D3TTC2 | Zinc metalloproteinase-disintegrin-like atragin | 2.801% | 4.830% | 4.221% | 3.882% | 3.364% | 3.258% | 3.258% | |
| D5LMJ3 | Zinc metalloproteinase-disintegrin-like atrase-A | 0.405% | 0.666% | 0.575% | 0.424% | 0.696% | 0.709% | 0.345% | |
| D6PXE8 | Zinc metalloproteinase-disintegrin-like atrase-B | 0.917% | 2.003% | 2.096% | 1.359% | 1.904% | 1.578% | 1.412% | |
| E9JG34 | Snake venom metalloproteinase | 0.044% | 0.053% | 0.073% | 0.040% | 0.065% | 0.109% | 0.042% | |
| P0DJ43 | Zinc metalloproteinase-disintegrin-like mikarin | / | 0.030% | / | / | / | 0.045% | / | |
| P82942 | Hemorrhagic metalloproteinase-disintegrin-like kaouthiagin | 2.159% | 2.846% | 2.689% | 1.801% | 1.788% | 2.031% | 1.862% | |
| Q10749 | Snake venom metalloproteinase-disintegrin-like mocarhagin | 1.894% | 1.617% | 2.040% | 1.229% | 1.599% | 1.175% | 1.255% | |
| Q2EI26 | Snake venom metalloproteinase AaPA | / | 0.005% | / | / | / | / | / | |
| Q7LZS9 | Snake venom metalloproteinase Ac1 | / | 0.019% | / | / | / | 0.003% | / | |
| Q9PVK7 | Zinc metalloproteinase-disintegrin-like cobrin | / | 0.009% | / | / | / | 0.012% | 0.011% | |
| R4G2I1 | Zinc metalloproteinase-Hop-23 | / | 0.003% | / | / | / | 0.002% | / | |
| Phospholipase | |||||||||
| A0A098LWY9 | Phospholipase B-like | / | 0.003% | / | / | / | 0.006% | / | |
| A0A898INR6 | Phospholipase A2 | 0.017% | 0.021% | 0.035% | 0.023% | 0.024% | 0.040% | 0.033% | |
| P00595 | Basic phospholipase A2 | / | 0.044% | / | / | 0.143% | 0.070% | 0.082% | |
| P00596 | Acidic phospholipase A2 | 3.027% | 3.572% | 3.507% | 11.574% | 4.284% | 5.862% | 3.931% | |
| P00617 | Basic phospholipase A2 beta-bungarotoxin A1 chain | / | / | / | / | / | 0.000% | / | |
| P00618 | Basic phospholipase A2 beta-bungarotoxin A2 chain | / | / | / | / | / | 0.000% | / | |
| Q92084 | Neutral phospholipase A2 muscarinic inhibitor | 19.128% | 20.183% | 20.376% | 25.987% | 22.966% | 41.201% | 24.994% | |
| Oxidase | |||||||||
| A0A0B8RQ82 | Methanethiol oxidase | / | / | / | / | / | 0.000% | / | |
| A0A098LX00 | Amine oxidase | / | 0.006% | / | 0.008% | / | 0.009% | 0.005% | |
| A8QL58 | L-amino-acid oxidase | 0.645% | 0.506% | 0.577% | 0.481% | 0.529% | 0.901% | 0.512% | |
| V8P7T9 | Sulfhydryl oxidase | / | 0.006% | / | / | / | 0.006% | / | |
| Cysteine-rich secretory protein | |||||||||
| A0A0F7Z2U7 | Cysteine-rich secretory protein 1 | 0.181% | 0.061% | / | 0.122% | / | 0.068% | 0.093% | |
| P0DL16 | Cysteine-rich venom protein mossambin | 0.602% | 0.262% | 0.266% | 0.279% | 0.321% | 0.099% | 0.138% | |
| P84805 | Cysteine-rich venom protein kaouthin-1 | 4.103% | 3.623% | 3.171% | 2.594% | 2.358% | 3.744% | 2.639% | |
| P84808 | Cysteine-rich venom protein kaouthin-2 | 0.138% | 0.089% | 0.134% | 0.104% | 0.105% | 0.093% | 0.088% | |
| Q2XXP4 | Cysteine-rich venom protein TRI1 | 0.135% | 0.077% | 0.092% | 0.057% | 0.069% | 0.094% | 0.060% | |
| Q3SB03 | Cysteine-rich venom protein pseudechetoxin-like | / | 0.051% | 0.030% | / | / | / | 0.034% | |
| E3P6P4 | Cystatin | 0.053% | 0.050% | 0.064% | 0.070% | 0.044% | 0.065% | 0.070% | |
| Muscarinic toxin-like protein | |||||||||
| P0DQQ3 | Muscarinic toxin-like protein Tx-NM3-2 | / | 0.108% | / | 0.205% | 0.226% | 0.212% | 0.101% | |
| P82463 | Muscarinic toxin-like protein 2 | 0.057% | 0.108% | 0.094% | 0.111% | 0.068% | 0.132% | 0.087% | |
| P82464 | Muscarinic toxin-like protein 3 | / | 1.586% | / | 1.721% | / | 2.068% | 1.392% | |
| RNA processing and regulatory proteins | |||||||||
| A0A0B8RTT7 | 40S ribosomal protein S10 | / | / | / | / | / | 0.008% | / | |
| J3S9G0 | 60S ribosomal protein L18a | / | / | / | / | / | 0.003% | / | |
| V8NRR7 | 39S ribosomal protein L16, mitochondrial | / | / | / | / | / | 0.000% | / | |
| V8NTR1 | Small ribosomal subunit protein uS5 | / | / | / | / | / | 0.001% | / | |
| V8P8H9 | Heterogeneous nuclear ribonucleoprotein A1 (Fragment) | / | 0.003% | / | / | / | 0.075% | 0.010% | |
| A0A0B8RX21 | U6 snRNA-associated Sm-like protein LSm8 | / | / | / | / | / | 0.004% | / | |
| A0A2D4G7G6 | RRM domain-containing protein | / | 0.004% | / | / | / | 0.004% | 0.023% | |
| V8NZ75 | Putative RNA-binding protein Luc7-like 2 | / | / | / | / | / | 0.002% | / | |
| J3SC47 | Elongation factor 2 | / | / | / | / | / | 0.005% | / | |
| Histone | |||||||||
| V8NDF2 | Histone H2A | 0.008% | 0.012% | / | / | / | 0.002% | / | |
| V8N8S2 | Histone H4 | / | 0.007% | / | / | / | 0.002% | / | |
| Metabolic enzymes | |||||||||
| A0A0B8RST8 | 6-phosphogluconate dehydrogenase | / | / | / | / | / | 0.002% | / | |
| J3S119 | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex | / | 0.001% | / | / | / | 0.004% | / | |
| V8NIF8 | L-lactate dehydrogenase | / | 0.001% | / | / | / | 0.000% | / | |
| Resultant protein | |||||||||
| A0A0B8RPH0 | Vimentin | 0.044% | 0.023% | 0.053% | 0.041% | 0.040% | 0.020% | 0.057% | |
| A0A0F7Z8W5 | Tubulin alpha chain | / | / | / | / | / | 0.005% | / | |
| J3SFJ0 | Tubulin beta chain | / | / | / | / | / | 0.026% | / | |
| V8N9Y8 | Tubulin alpha-8 chain | / | / | / | / | / | 0.001% | / | |
| A0A1W7RJI5 | Alpha-actinin-1-like protein | / | / | / | / | / | 0.000% | / | |
| A0A0B8RYW7 | Ezrin-like protein | / | 0.012% | / | / | / | 0.007% | / | |
| A0A0B8RUG1 | Katanin p60 ATPase-containing subunit A1 | / | 0.003% | / | / | / | 0.001% | / | |
| V8NBM9 | Keratin, type II cytoskeletal 5 | / | 0.017% | / | / | / | 0.002% | / | |
| V8NYV8 | Keratin, type II cytoskeletal 1 | 0.105% | 0.082% | / | 0.044% | / | 0.019% | 0.031% | |
| V8P8L1 | Keratin, type I cytoskeletal 19 | 0.234% | 0.210% | 0.204% | 0.157% | 0.195% | 0.084% | 0.131% | |
| V8NL73 | Extracellular matrix protein 1 | / | 0.021% | 0.029% | / | 0.015% | 0.008% | 0.025% | |
| V8NRX1 | IF rod domain-containing protein | 0.187% | 0.343% | 0.136% | 0.111% | 0.157% | 0.065% | 0.082% | |
| Molecular chaperones and stress response proteins | |||||||||
| V8N9M0 | Heat shock protein HSP 90-alpha | / | / | / | / | / | 0.026% | / | |
| A0A0B8RUJ6 | Glucose-regulated protein | / | 0.033% | 0.033% | / | / | 0.003% | / | |
| A0A1W7REY2 | Protein disulfide-isomerase A6 | / | 0.040% | 0.041% | / | / | 0.003% | / | |
| V8NBS9 | Endoplasmic reticulum resident protein 44 | / | 0.005% | / | / | / | 0.001% | / | |
| J3SFD9 | T-complex protein 1 subunit beta | / | / | / | / | / | 0.001% | / | |
| V8NWK2 | T-complex protein 1 subunit epsilon | / | / | / | / | / | 0.003% | / | |
| Immune and complement-related proteins | |||||||||
| V8NCP4 | Complement C3 | 0.379% | 0.851% | 0.473% | / | 0.442% | 0.538% | / | |
| A0A1W7RH78 | Gamma-interferon-inducible lysosomal thiol reductase | / | 0.013% | / | / | / | 0.011% | 0.016% | |
| D2YVI2 | C-type lectin galactose-binding isoform | / | 0.160% | / | 0.190% | / | 0.313% | 0.132% | |
| 3FTX | |||||||||
| P01400 | Weak toxin S4C11 | 0.436% | 0.479% | 0.607% | 0.267% | / | 0.288% | 0.215% | |
| P01401 | Weak toxin CM-11 | 1.351% | 1.516% | 2.145% | 0.817% | 1.017% | 1.646% | 1.046% | |
| P29181 | Weak neurotoxin 7 | 0.731% | 0.488% | 0.548% | 0.814% | 0.707% | 0.865% | 0.524% | |
| O42256 | Weak neurotoxin 6 | 1.008% | 1.073% | 1.431% | 0.556% | 0.722% | 1.052% | 0.567% | |
| P01424 | Short neurotoxin 1 | / | 0.002% | / | / | / | 0.002% | / | |
| Q9YGI4 | Probable weak neurotoxin NNAM2 | 2.112% | 5.167% | 2.550% | 4.336% | 3.549% | 3.002% | 3.128% | |
| D5J9Q0 | Non-conventional three finger toxin isoform 6 | / | 0.002% | / | / | / | 0.002% | / | |
| Q9W717 | Neurotoxin-like protein NTL2 | / | 0.047% | / | / | / | 0.056% | / | |
| C0HJW9 | Neurotoxin Nk-3FTx (Fragment) | 0.037% | 0.051% | 0.050% | 0.070% | 0.073% | 0.039% | 0.069% | |
| Q9DEQ3 | Neurotoxin homolog NL1 | / | / | / | / | / | 0.000% | / | |
| P34074 | Long neurotoxin 1 | 0.086% | 0.530% | 0.594% | 0.506% | 0.691% | 0.649% | 0.467% | |
| P60308 | Cytotoxin SP15c | 0.494% | 1.784% | 0.664% | 1.180% | 0.890% | 0.506% | 0.419% | |
| Q91135 | Cytotoxin I-like P-15 | 0.288% | 0.464% | 0.308% | 0.150% | 0.105% | 0.090% | 0.083% | |
| P49122 | Cytotoxin 7 | / | 0.249% | / | 0.384% | 0.166% | 0.140% | 0.343% | |
| P80245 | Cytotoxin 6 | 3.321% | 0.063% | / | 0.214% | 0.160% | 0.006% | 0.041% | |
| Q9W6W9 | Cytotoxin 4N | 0.476% | 1.456% | 0.543% | 1.174% | 0.879% | 1.025% | 0.686% | |
| Q98962 | Cytotoxin 3d | 0.071% | 0.025% | 0.036% | 0.018% | / | 0.010% | 0.010% | |
| P01470 | Cytotoxin 3 | 29.251% | 20.677% | 24.175% | 15.939% | 26.952% | 6.192% | 30.163% | |
| P01440 | Cytotoxin 2 | 4.710% | 2.753% | 3.893% | 4.753% | 5.856% | 1.761% | 3.813% | |
| Q98956 | Cytotoxin 1b | 0.404% | 0.088% | 0.410% | 0.201% | 0.240% | 0.054% | 0.364% | |
| P86541 | Cytotoxin 10 | 0.196% | 0.163% | / | 0.168% | 0.194% | 0.120% | 0.077% | |
| P0CH80 | Cytotoxin 1 | 1.567% | 0.635% | 1.487% | 0.474% | 0.483% | 0.118% | 0.432% | |
| P59276 | Cobrotoxin-c | / | 0.034% | 0.046% | 0.008% | / | 0.008% | 0.011% | |
| P59275 | Cobrotoxin-b | 0.132% | 0.276% | 0.330% | 0.194% | 0.108% | 0.181% | 0.194% | |
| Q91126 | Cardiotoxin 7a | 0.532% | 0.515% | 0.408% | 0.333% | 0.439% | 0.460% | 0.239% | |
| O57326 | Alpha-neurotoxin NTX-3 | 0.025% | 0.029% | 0.029% | 0.013% | 0.017% | 0.023% | 0.012% | |
| C0HM08 | Alpha-elapitoxin-Nn2a | / | 0.001% | / | / | / | 0.000% | / | |
| E2ITZ3 | Alpha-elapitoxin-Na1a | 1.318% | 0.974% | 1.032% | 1.070% | 0.781% | 0.300% | 0.631% | |
| Others | |||||||||
| A0A2D0TC04 | Venom phosphodiesterase | 0.779% | 0.467% | 0.587% | 0.726% | 0.678% | 0.712% | 0.738% | |
| V8NX10 | WD repeat and FYVE domain-containing protein 1 | / | 0.013% | / | / | / | 0.015% | 0.017% | |
| A0A098LYI7 | Vespryn | / | / | / | / | / | 0.012% | / | |
| Q5YF89 | Venom nerve growth factor | 0.531% | 0.497% | 0.485% | 0.515% | 0.423% | 0.767% | 0.431% | |
| A0A2D4Q7C6 | Uncharacterized protein | / | / | 1.022% | 0.519% | / | 0.753% | 0.549% | |
| A0A194AS98 | Snake venom 5′-nucleotidase | 0.752% | 0.697% | 0.897% | 0.866% | 0.707% | 1.779% | 0.954% | |
| Q9DEF9 | Snaclec anticoagulant protein subunit A | / | 0.001% | / | / | / | / | / | |
| B0FXL8 | Siamenotoxin I | 1.824% | 1.750% | 2.185% | 0.853% | 1.007% | 0.693% | 0.871% | |
| V8NKT2 | ShKT domain-containing protein | / | / | / | / | / | 0.059% | / | |
| A0A2D4HD83 | SH3 domain-containing protein | / | 0.003% | / | / | / | / | / | |
| A0A2D4G403 | SCP domain-containing protein | / | / | 0.152% | / | / | / | / | |
| C1IC50 | Protease inhibitor 1 | 0.361% | 0.195% | 0.288% | 0.266% | 0.391% | 0.063% | 0.209% | |
| A0A8C6VK71 | Plasminogen activator | 1.274% | 0.929% | 1.063% | 1.133% | 0.936% | 1.145% | 0.951% | |
| A0A0B8RU52 | Peptidyl-glycine alpha-amidating monooxygenase | / | 0.003% | / | / | / | 0.003% | / | |
| A0A8C6Y6B8 | Peptidase S1 domain-containing protein | / | 0.006% | / | / | / | 0.005% | / | |
| I2C090 | Ophiophagus venom factor | 0.589% | 1.238% | 1.240% | 0.892% | 0.872% | 1.341% | 0.906% | |
| V8P0W2 | Neuroserpin | / | / | / | / | / | 0.005% | 0.004% | |
| V8P1Y2 | Neuroendocrine convertase 1 | / | 0.009% | / | / | / | 0.006% | / | |
| V8N4D8 | Nerve growth factor-related domain-containing protein | 2.396% | 3.436% | 2.331% | 3.013% | 2.043% | 3.290% | 2.833% | |
| V8NQ76 | Neprilysin | / | 0.006% | / | / | / | 0.004% | / | |
| A0A2D4GN98 | Multiple inositol polyphosphate phosphatase 1 | 0.046% | 0.066% | 0.072% | 0.028% | 0.042% | 0.048% | 0.018% | |
| A0A898INP5 | Kunitz peptide | / | / | / | / | 0.035% | 0.046% | 0.036% | |
| V8NNL9 | Insulin-like growth factor-binding protein 3 | 0.011% | 0.019% | 0.017% | 0.008% | / | 0.010% | 0.007% | |
| A0A2D4LE84 | Ig-like domain-containing protein | 0.036% | 0.061% | 0.069% | 0.079% | / | 0.019% | 0.164% | |
| A0A898INC5 | Hyaluronidase | / | 0.003% | / | / | / | 0.001% | / | |
| A0A6J1VMA6 | Hepatocyte growth factor activator | / | 0.001% | / | / | / | 0.002% | / | |
| A0A2D4IPJ7 | Granulins domain-containing protein | 0.010% | 0.010% | 0.013% | 0.009% | 0.009% | 0.011% | 0.008% | |
| V8P395 | Glutathione peroxidase | 0.676% | 0.666% | 0.775% | 0.685% | 0.559% | 0.788% | 0.786% | |
| A0A2D4FFX4 | GH18 domain-containing protein | / | 0.005% | / | / | / | 0.006% | / | |
| U3FCT9 | Endonuclease domain-containing 1 protein | / | 0.256% | / | / | / | 0.215% | 0.072% | |
| V8NG26 | EH domain-containing protein 4 | / | / | / | / | / | 0.007% | / | |
| A6MJH5 | Dipeptidyl peptidase 4 | / | 0.004% | / | / | / | 0.004% | 0.006% | |
| A0A346CI96 | Cobra venom factor | / | / | 0.143% | 0.091% | / | 0.084% | 0.083% | |
| A0A670ZPJ2 | Coagulation factor VII | / | / | / | / | / | 0.000% | / | |
| J3SE58 | Chromobox protein 3 like | / | / | / | / | / | 0.015% | / | |
| A0A0B8RRA8 | Chitotriosidase | / | 0.007% | / | / | / | 0.006% | 0.012% | |
| U3FD65 | Cathepsin B | / | 0.004% | / | / | / | 0.006% | / | |
| P83346 | Bucain | / | / | / | / | / | 0.000% | / | |
| A0A2D4GU19 | BPTI/Kunitz inhibitor domain-containing protein | / | 0.025% | / | 0.038% | / | 0.025% | 0.032% | |
| A0A6J1VTT9 | BPTI/Kunitz domain-containing protein-like | 0.032% | 0.024% | 0.023% | 0.052% | 0.038% | 0.025% | 0.038% | |
| V8NEU2 | B30.2/SPRY domain-containing protein | 3.455% | 2.526% | 2.739% | 1.919% | 4.169% | 1.970% | 1.187% | |
| A0A0B8RR92 | ATP-dependent Clp protease ATP-binding subunit clpX-likeserine-threonine-like protein | 0.005% | 0.004% | 0.006% | 0.004% | / | 0.004% | 0.006% | |
| U3FZS8 | Aminopeptidase | / | 0.017% | 0.007% | / | / | 0.017% | 0.013% | |
| V8ND09 | Alpha-fetoprotein | / | 0.013% | / | 0.074% | / | 0.027% | 0.108% | |
| A0A0B8RVP6 | ADP/ATP translocase | / | / | / | / | / | 0.001% | / | |
| Q0ZZJ6 | A.superbus venom factor 1 | 1.411% | 3.127% | 3.564% | 1.888% | 2.662% | 2.504% | 2.371% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; Shi, X.; Liu, G.; Yang, Y.; Huang, C. Geographic Variation in Venom Proteome and Toxicity Profiles of Chinese Naja atra: Implications for Antivenom Optimization. Toxins 2025, 17, 404. https://doi.org/10.3390/toxins17080404
Zhao J, Shi X, Liu G, Yang Y, Huang C. Geographic Variation in Venom Proteome and Toxicity Profiles of Chinese Naja atra: Implications for Antivenom Optimization. Toxins. 2025; 17(8):404. https://doi.org/10.3390/toxins17080404
Chicago/Turabian StyleZhao, Jianqi, Xiao Shi, Guangyao Liu, Yang Yang, and Chunhong Huang. 2025. "Geographic Variation in Venom Proteome and Toxicity Profiles of Chinese Naja atra: Implications for Antivenom Optimization" Toxins 17, no. 8: 404. https://doi.org/10.3390/toxins17080404
APA StyleZhao, J., Shi, X., Liu, G., Yang, Y., & Huang, C. (2025). Geographic Variation in Venom Proteome and Toxicity Profiles of Chinese Naja atra: Implications for Antivenom Optimization. Toxins, 17(8), 404. https://doi.org/10.3390/toxins17080404





