Mechanistic Exploration of Aristolochic Acid I-Induced Hepatocellular Carcinoma: Insights from Network Toxicology, Machine Learning, Molecular Docking, and Molecular Dynamics Simulation
Abstract
1. Introduction
2. Results
2.1. Toxicity Prediction for AAI
2.2. Search of Targets for AAI and HCC
2.3. Recognition Core Gene from GEO
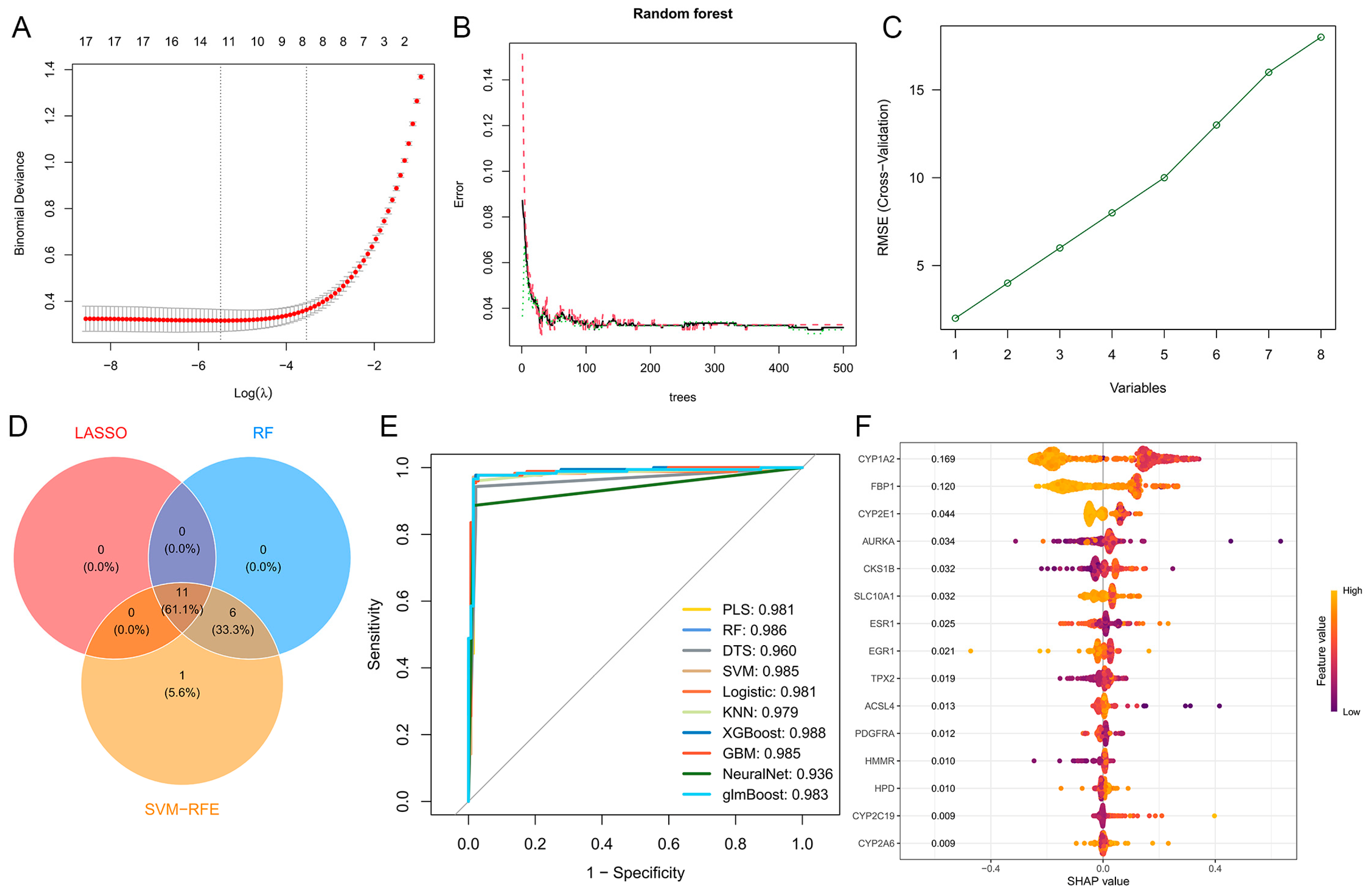
2.4. Genetic Screening Based on Machine Learning
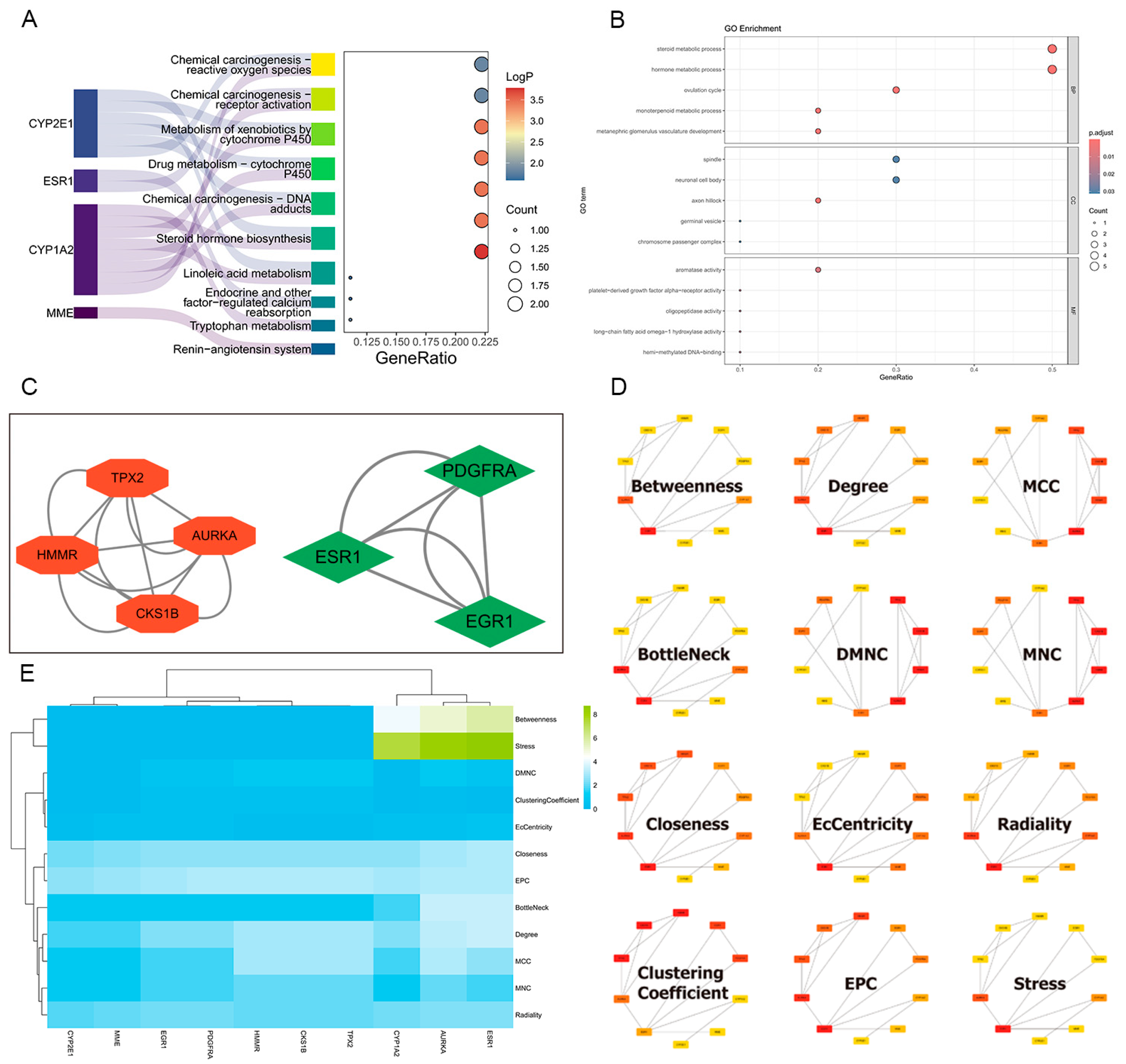
2.5. Construction of Protein–Protein Interaction Network and Hub Targets Screening
2.6. GO and KEGG Enrichment Analysis
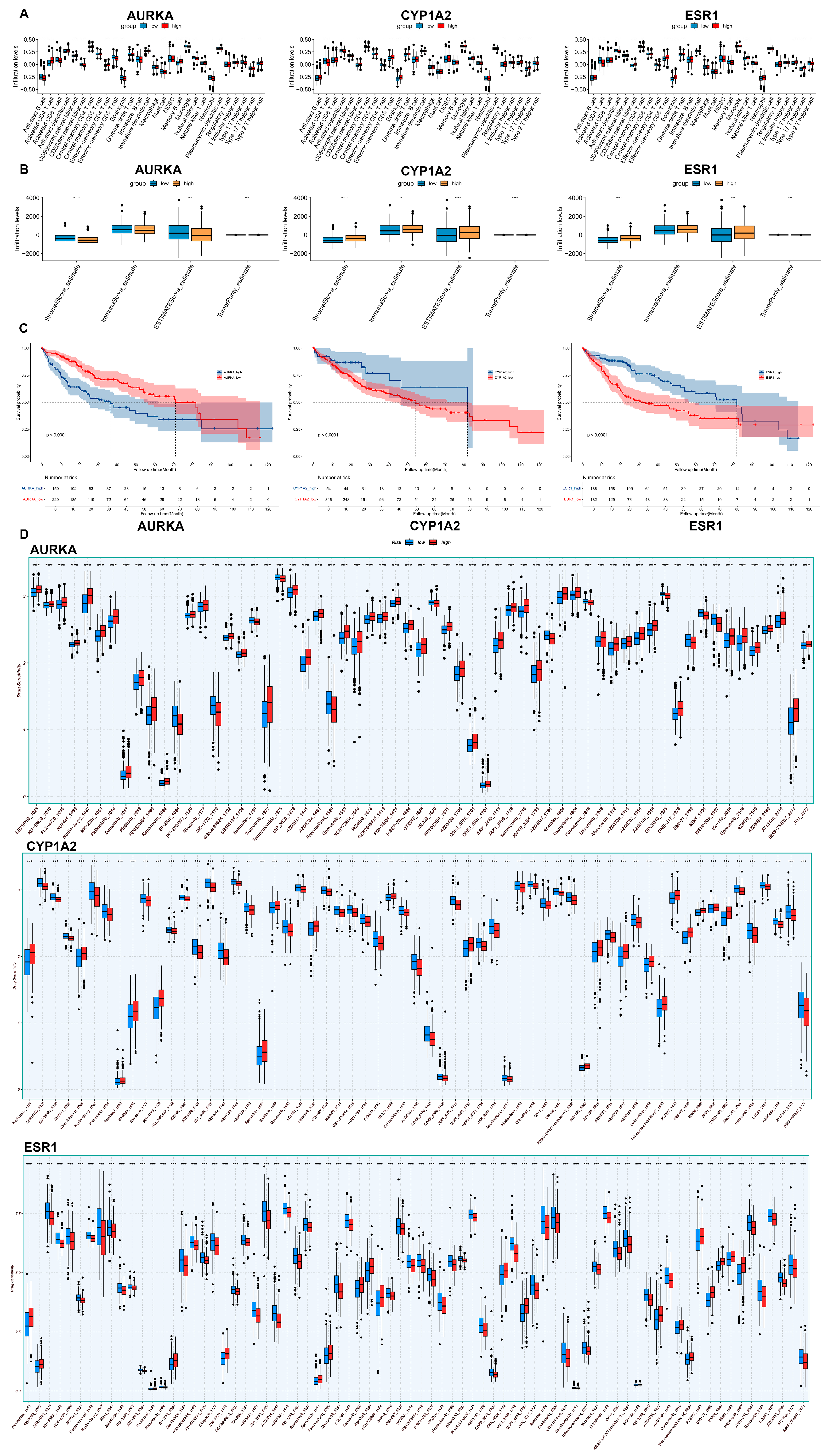
2.7. Immunoinfiltration and Drug Sensitivity Analysis
2.8. Survival Analysis
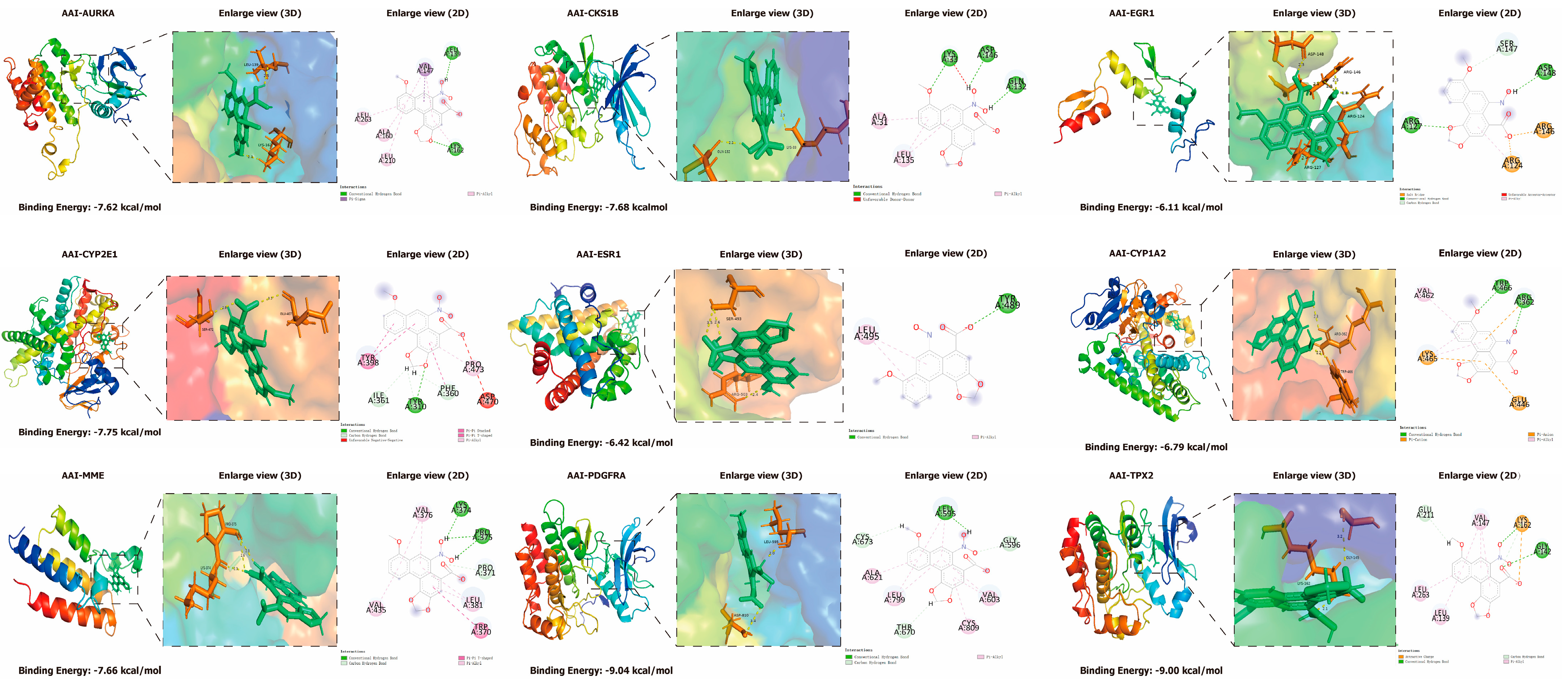
2.9. Molecular Docking
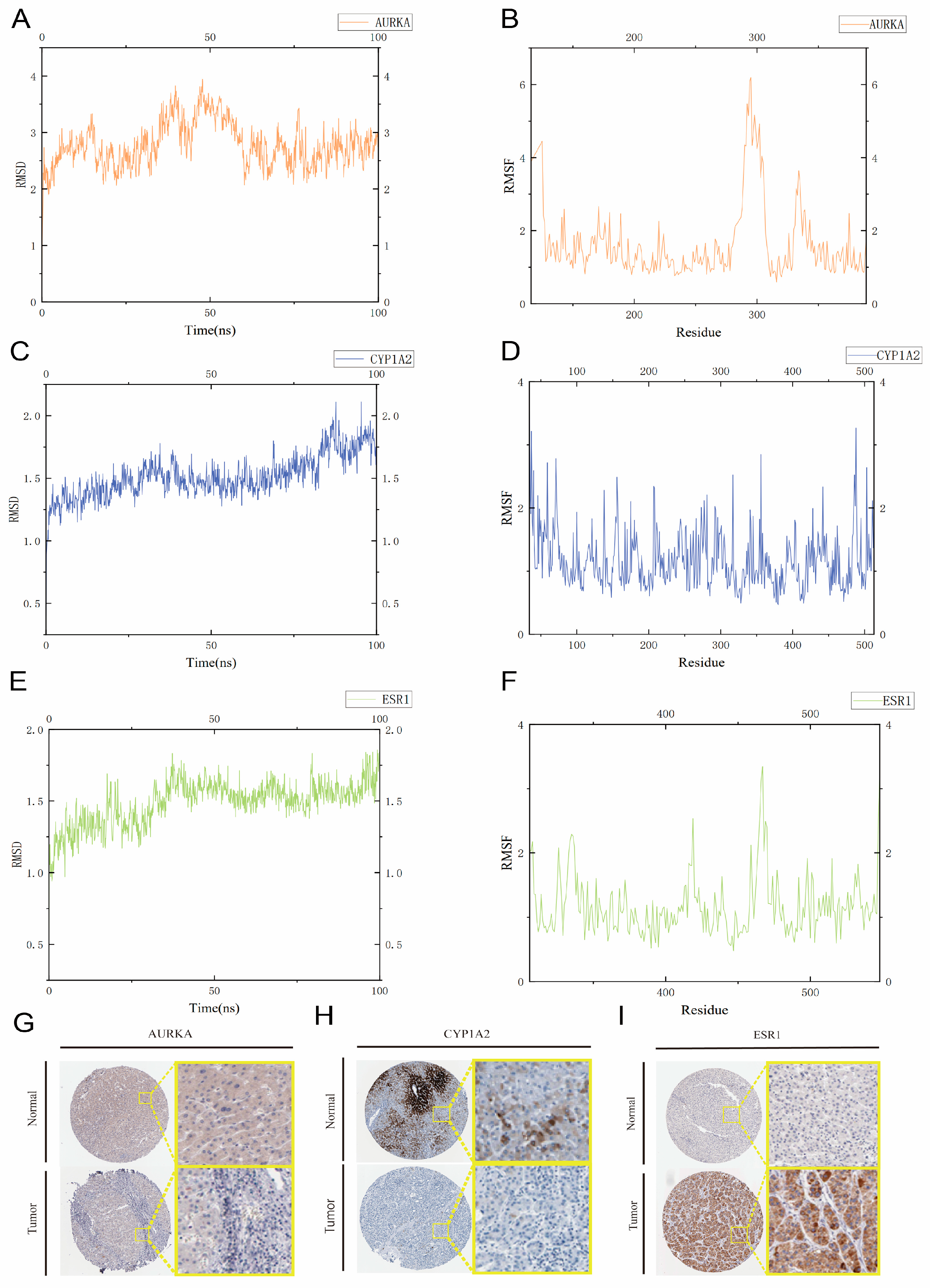
2.10. Molecular Dynamics Simulations
2.11. Immunohistochemistry Based on the HPA Database
3. Discussion
4. Conclusions
5. Methods and Materials
5.1. Study Design
5.2. Forecasting Toxicity Effects of AAI
5.3. Search for AAI Targets
5.4. Identification of Targets Associated with HCC
5.5. Recognition of Core Genes from GEO
5.6. Machine Learning Powered Genetic Assessment
5.7. Construction of Protein–Protein Interaction Network
5.8. Hub Target Screening
5.9. Functional Enrichment Profiling via GO and KEGG
5.10. Immuneinfiltration and Drug Sensitivity Analyses
5.11. Survival Analysis Using Samples from the TCGA Database
5.12. Molecular Docking of AAI and Its Core Targets
5.13. Molecular Dynamics Simulations for Core Genes
5.14. Immunohistochemistry Validation Form HPA Database
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- do Prado Schneidewind, F.C.C.; de Castilho, P.F.; Galvão, F.; de Andrade Dos Santos, J.V.; da Silva Dantas, F.G.; Negri, M.; da Silva Pinto, L.; Moraes, C.A.F.; Freitas, J.; de Souza, P.R.B.; et al. Effects of bioconversion by Battus polydamas on the chemical composition of Aristolochia spp. and evaluation of antimicrobial activity and biocompatibility. Fitoterapia 2024, 175, 105949. [Google Scholar] [CrossRef]
- Chan, C.K.; Liu, Y.; Pavlović, N.M.; Chan, W. Aristolochic Acids: Newly Identified Exposure Pathways of this Class of Environmental and Food-Borne Contaminants and its Potential Link to Chronic Kidney Diseases. Toxics 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Luan, Y.; Ma, S.; Mao, Y. Is it really the “dark side” of herbal medicine? Sci. China Life Sci. 2018, 61, 1118–1119. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhang, Q.; Jiang, W.; Wang, X.; Guo, X.; Chen, L.; Cheng, S.; Ying, J.; Ye, J.; Zhang, L. Aristolochic acid I induced mitochondrial Ca(2+) accumulation triggers the production of MitoROS and activates Src/FAK pathway in hepatocellular carcinoma cells. Chem. Biol. Interact. 2025, 405, 111269. [Google Scholar] [CrossRef]
- Drăghia, L.P.; Lukinich-Gruia, A.T.; Oprean, C.; Pavlović, N.M.; Păunescu, V.; Tatu, C.A. Aristolochic acid I: An investigation into the role of food crops contamination, as a potential natural exposure pathway. Environ. Geochem. Health 2021, 43, 4163–4178. [Google Scholar] [CrossRef]
- Tian, J.Z.; Liu, S.Y.; Gao, Y.; Zhang, B.L.; Liang, A.H. Risk assessment, safe medication and scientific supervision of traditional Chinese medicine containing aristolochic acids--toxicity is different among aristolochic acids, and detection and control of aristolochic acid Ⅰ/Ⅱ is critical. Zhongguo Zhong Yao Za Zhi 2022, 47, 3693–3700. [Google Scholar] [CrossRef] [PubMed]
- Stiborová, M.; Arlt, V.M.; Schmeiser, H.H. Balkan endemic nephropathy: An update on its aetiology. Arch. Toxicol. 2016, 90, 2595–2615. [Google Scholar] [CrossRef]
- Zhou, G.; Zhao, X. Carcinogens that induce the A:T > T:A nucleotide substitutions in the genome. Front. Med. 2018, 12, 236–238. [Google Scholar] [CrossRef]
- Chevalier, A.; Guo, T.; Gurevich, N.Q.; Xu, J.; Yajima, M.; Campbell, J.D. Characterization of highly active mutational signatures in tumors from a large Chinese population. medRxiv 2023. [Google Scholar] [CrossRef]
- Nault, J.C.; Letouzé, E. Mutational Processes in Hepatocellular Carcinoma: The Story of Aristolochic Acid. Semin. Liver Dis. 2019, 39, 334–340. [Google Scholar] [CrossRef]
- Luo, P.; Chen, J.; Zhang, Q.; Xia, F.; Wang, C.; Bai, Y.; Tang, H.; Liu, D.; Gu, L.; Du, Q.; et al. Dissection of cellular and molecular mechanisms of aristolochic acid-induced hepatotoxicity via single-cell transcriptomics. Precis. Clin. Med. 2022, 5, pbac023. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.M.; Zhao, X.H.; Sun, Z.H.; Li, G.C.; Liu, G.C.; Sun, L.R.; Hou, J.Q.; Zhou, W. Recognition of the toxicity of aristolochic acid. J. Clin. Pharm. Ther. 2019, 44, 157–162. [Google Scholar] [CrossRef]
- Rebhan, K.; Ertl, I.E.; Shariat, S.F.; Grollman, A.P.; Rosenquist, T. Aristolochic acid and its effect on different cancers in uro-oncology. Curr. Opin. Urol. 2020, 30, 689–695. [Google Scholar] [CrossRef]
- Kamaraju, S.; Conroy, M.; Harris, A.; Georgen, M.; Min, H.; Powell, M.; Kurzrock, R. Challenges to genetic testing for germline mutations associated with breast cancer among African Americans. Cancer Treat. Rev. 2024, 124, 102695. [Google Scholar] [CrossRef]
- Li, X.L.; Guo, X.Q.; Wang, H.R.; Chen, T.; Mei, N. Aristolochic Acid-Induced Genotoxicity and Toxicogenomic Changes in Rodents. World J. Tradit. Chin. Med. 2020, 6, 12–25. [Google Scholar] [CrossRef]
- Lu, Z.N.; Luo, Q.; Zhao, L.N.; Shi, Y.; Wang, N.; Wang, L.; Han, Z.G. The Mutational Features of Aristolochic Acid-Induced Mouse and Human Liver Cancers. Hepatology 2020, 71, 929–942. [Google Scholar] [CrossRef]
- Kristanc, L.; Kreft, S. European medicinal and edible plants associated with subacute and chronic toxicity part II: Plants with hepato-, neuro-, nephro- and immunotoxic effects. Food Chem. Toxicol. 2016, 92, 38–49. [Google Scholar] [CrossRef]
- Chen, S.; Dong, Y.; Qi, X.; Cao, Q.; Luo, T.; Bai, Z.; He, H.; Fan, Z.; Xu, L.; Xing, G.; et al. Aristolochic acids exposure was not the main cause of liver tumorigenesis in adulthood. Acta Pharm. Sin. B 2022, 12, 2252–2267. [Google Scholar] [CrossRef]
- Han, J.; Xian, Z.; Zhang, Y.; Liu, J.; Liang, A. Systematic Overview of Aristolochic Acids: Nephrotoxicity, Carcinogenicity, and Underlying Mechanisms. Front. Pharmacol. 2019, 10, 648. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.W.T.; Poon, S.L.; Huang, M.N.; Lim, J.Q.; Boot, A.; Yu, W.; Suzuki, Y.; Thangaraju, S.; Ng, C.C.Y.; Tan, P.; et al. Aristolochic acids and their derivatives are widely implicated in liver cancers in Taiwan and throughout Asia. Sci. Transl. Med. 2017, 9, eaan6446. [Google Scholar] [CrossRef]
- Wang, X.; Song, J.; Dong, Y.; Hu, L.; Chen, X.; Yang, F.; Qi, H.; Qi, X.; Wen, W.; Chen, S.; et al. Qualitative and quantitative bioanalytical methods validation of aristolochic acid DNA adducts and application in human formalin-fixed paraffin-embedded hepatocellular carcinoma tissues. bioRxiv 2020. [Google Scholar] [CrossRef]
- Li, X.-Y.; Jin, X.; Li, Y.-Z.; Gao, D.-D.; Liu, R.; Liu, C.-X. Network toxicology and LC-MS-based metabolomics:New approaches for mechanism of action of toxic components in traditional Chinese medicines. Chin. Herbal. Med. 2019, 11, 357–363. [Google Scholar] [CrossRef]
- Bai, C.; Wu, L.; Li, R.; Cao, Y.; He, S.; Bo, X. Machine Learning-Enabled Drug-Induced Toxicity Prediction. Adv Sci. 2025, 12, e2413405. [Google Scholar] [CrossRef]
- Kong, J.; Tian, Y.; Liu, C.X.; Huang, J.M. Mechanism of hepatotoxicity induced by ethanol extract of Dysosma versipellis based on "quantity-weight-evidence" network toxicology. Zhongguo Zhong Yao Za Zhi 2022, 47, 511–527. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Ding, Y.; Yu, L.; Xiang, C.; Yang, M. Exploring the mechanism of Alisma orientale for the treatment of pregnancy induced hypertension and potential hepato-nephrotoxicity by using network pharmacology, network toxicology, molecular docking and molecular dynamics simulation. Front. Pharmacol. 2022, 13, 1027112. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Liu, X.; Yu, S. Utilizing machine learning algorithms to identify biomarkers associated with diabetic nephropathy: A review. Medicine 2024, 103, e37235. [Google Scholar] [CrossRef] [PubMed]
- Salazar, R.M.; Nair, S.S.; Leone, A.O.; Xu, T.; Mumme, R.P.; Duryea, J.D.; De, B.; Corrigan, K.L.; Rooney, M.K.; Ning, M.S.; et al. Performance Comparison of 10 State-of-the-Art Machine Learning Algorithms for Outcome Prediction Modeling of Radiation-Induced Toxicity. Adv. Radiat. Oncol. 2025, 10, 101675. [Google Scholar] [CrossRef]
- Ponce-Bobadilla, A.V.; Schmitt, V.; Maier, C.S.; Mensing, S.; Stodtmann, S. Practical guide to SHAP analysis: Explaining supervised machine learning model predictions in drug development. Clin. Transl. Sci. 2024, 17, e70056. [Google Scholar] [CrossRef]
- Ye, W.L.; Shen, C.; Xiong, G.L.; Ding, J.J.; Lu, A.P.; Hou, T.J.; Cao, D.S. Improving Docking-Based Virtual Screening Ability by Integrating Multiple Energy Auxiliary Terms from Molecular Docking Scoring. J. Chem. Inf. Model. 2020, 60, 4216–4230. [Google Scholar] [CrossRef]
- Vidal-Limon, A.; Aguilar-Toalá, J.E.; Liceaga, A.M. Integration of Molecular Docking Analysis and Molecular Dynamics Simulations for Studying Food Proteins and Bioactive Peptides. J. Agric. Food Chem. 2022, 70, 934–943. [Google Scholar] [CrossRef]
- Cao, F.; Guo, C.; Guo, J. Deciphering CSU pathogenesis: Network toxicologyand molecular dynamics of DOTP exposure. Ecotoxicol. Environ. Saf. 2025, 291, 117864. [Google Scholar] [CrossRef]
- Naqvi, A.A.T.; Mohammad, T.; Hasan, G.M.; Hassan, M.I. Advancements in Docking and Molecular Dynamics Simulations Towards Ligand-receptor Interactions and Structure-function Relationships. Curr. Top. Med. Chem. 2018, 18, 1755–1768. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, S.; Ren, T.; Zhang, Y.; Li, B.; Geng, X. Mechanism of emodin in treating hepatitis B virus-associated hepatocellular carcinoma: Network pharmacology and cell experiments. Front. Cell Infect. Microbiol. 2024, 14, 1458913. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.Q.; Ke, J.J.; Xu, Q.S.; Wu, W.Q.; Wan, Y.Y. Integrated network analysis to identify the key genes, transcription factors, and microRNAs involved in hepatocellular carcinoma. Neoplasma 2018, 65, 66–74. [Google Scholar] [CrossRef]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C.; et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef]
- Cao, L.; Zhu, L.; Cheng, L. ncRNA-Regulated LAYN Serves as a Prognostic Biomarker and Correlates with Immune Cell Infiltration in Hepatocellular Carcinoma: A Bioinformatics Analysis. Biomed. Res. Int. 2022, 2022, 5357114. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Chen, B.; Zeng, Y.; Wang, H. UHRF1 Could Be a Prognostic Biomarker and Correlated with Immune Cell Infiltration in Hepatocellular Carcinoma. Int. J. Gen. Med. 2021, 14, 6769–6776. [Google Scholar] [CrossRef]
- Li, X.; Gao, Z.; Chen, J.; Feng, S.; Luo, X.; Shi, Y.; Tang, Z.; Liu, W.; Zhang, X.; Huang, A.; et al. Integrated single cell and bulk sequencing analysis identifies tumor reactive CXCR6(+) CD8 T cells as a predictor of immune infiltration and immunotherapy outcomes in hepatocellular carcinoma. Front. Oncol. 2023, 13, 1099385. [Google Scholar] [CrossRef]
- Sciarra, A.; Pintea, B.; Nahm, J.H.; Donadon, M.; Morenghi, E.; Maggioni, M.; Blanc, J.F.; Torzilli, G.; Yeh, M.; Bioulac-Sage, P.; et al. CYP1A2 is a predictor of HCC recurrence in HCV-related chronic liver disease: A retrospective multicentric validation study. Dig. Liver Dis. 2017, 49, 434–439. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, J.; Shu, J.; Ying, X.; Khan, S.; Sarfaraz, S.; Mirzaeiebrahimabadi, R.; Alhomrani, M.; Alamri, A.S.; ALSuhaymi, N. Exploring potential key genes and pathways associatedwith hepatocellular carcinoma prognosis through bioinformatics analysis, followed by experimental validation. Am. J. Transl. Res. 2024, 16, 7286–7302. [Google Scholar] [CrossRef] [PubMed]
- Grisetti, L.; Garcia, C.J.C.; Saponaro, A.A.; Tiribelli, C.; Pascut, D. The role of Aurora kinase A in hepatocellular carcinoma: Unveiling the intriguing functions of a key but still underexplored factor in liver cancer. Cell Prolif. 2024, 57, e13641. [Google Scholar] [CrossRef]
- Sundberg, C.D.; Hankinson, O. A CRISPR/Cas9 Whole-Genome Screen Identifies Genes Required for Aryl Hydrocarbon Receptor-Dependent Induction of Functional CYP1A1. Toxicol. Sci. 2019, 170, 310–319. [Google Scholar] [CrossRef] [PubMed]
- Du, R.; Li, K.; Zhou, Z.; Huang, Y.; Guo, K.; Zhang, H.; Chen, Z.; Zhao, X.; Han, L.; Bian, H. Bioinformatics and experimental validation of an AURKA/TPX2 axis as a potential target in esophageal squamous cell carcinoma. Oncol. Rep. 2023, 49, 116. [Google Scholar] [CrossRef]
- Bonomo, S.; Jørgensen, F.S.; Olsen, L. Dissecting the Cytochrome P450 1A2- and 3A4-Mediated Metabolism of Aflatoxin B1 in Ligand and Protein Contributions. Chemistry 2017, 23, 2884–2893. [Google Scholar] [CrossRef]
- Chen, X.; Wang, H.; Xie, W.; Liang, R.; Wei, Z.; Zhi, L.; Zhang, X.; Hao, B.; Zhong, S.; Zhou, G.; et al. Association of CYP1A2 genetic polymorphisms with hepatocellular carcinoma susceptibility: A case-control study in a high-risk region of China. Pharmacogenet Genom. 2006, 16, 219–227. [Google Scholar] [CrossRef]
- Deng, J.; Zhao, L.; Zhang, N.Y.; Karrow, N.A.; Krumm, C.S.; Qi, D.S.; Sun, L.H. Aflatoxin B(1) metabolism: Regulation by phase I and II metabolizing enzymes and chemoprotective agents. Mutat. Res. Rev. Mutat. Res. 2018, 778, 79–89. [Google Scholar] [CrossRef]
- Dohnal, V.; Wu, Q.; Kuča, K. Metabolism of aflatoxins: Key enzymes and interindividual as well as interspecies differences. Arch. Toxicol. 2014, 88, 1635–1644. [Google Scholar] [CrossRef]
- Daly, A.K. Polymorphic Variants of Cytochrome P450: Relevance to Cancer and Other Diseases. Adv. Pharmacol. 2015, 74, 85–111. [Google Scholar] [CrossRef]
- Wojnowski, L.; Turner, P.C.; Pedersen, B.; Hustert, E.; Brockmöller, J.; Mendy, M.; Whittle, H.C.; Kirk, G.; Wild, C.P. Increased levels of aflatoxin-albumin adducts are associated with CYP3A5 polymorphisms in The Gambia, West Africa. Pharmacogenetics 2004, 14, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; He, L.; Zeng, H.; Fu, W.; Wang, J.; Tan, Y.; Zheng, C.; Qiu, Z.; Luo, J.; Lv, C.; et al. Low-dose microcystin-LR antagonizes aflatoxin B1 induced hepatocarcinogenesis through decreasing cytochrome P450 1A2 expression and aflatoxin B1-DNA adduct generation. Chemosphere 2020, 248, 126036. [Google Scholar] [CrossRef]
- Dračínská, H.; Bárta, F.; Levová, K.; Hudecová, A.; Moserová, M.; Schmeiser, H.H.; Kopka, K.; Frei, E.; Arlt, V.M.; Stiborová, M. Induction of cytochromes P450 1A1 and 1A2 suppresses formation of DNA adducts by carcinogenic aristolochic acid I in rats in vivo. Toxicology 2016, 344–346, 7–18. [Google Scholar] [CrossRef]
- Li, C.X.; Xiao, X.H.; Li, X.Y.; Xiao, D.K.; Wang, Y.K.; Wang, X.L.; Zhang, P.; Li, Y.R.; Niu, M.; Bai, Z.F. Stir-fried Semen Armeniacae Amarum Suppresses Aristolochic Acid I-Induced Nephrotoxicity and DNA Adducts. Chin. J. Integr. Med. 2025, 31, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Valentín López, J.C.; Lange, C.A.; Dehm, S.M. Androgen receptor and estrogen receptor variants in prostate and breast cancers. J. Steroid Biochem. Mol. Biol. 2024, 241, 106522. [Google Scholar] [CrossRef]
- Martínez-Galán, J.; Torres-Torres, B.; Núñez, M.I.; López-Peñalver, J.; Del Moral, R.; Ruiz De Almodóvar, J.M.; Menjón, S.; Concha, A.; Chamorro, C.; Ríos, S.; et al. ESR1 gene promoter region methylation in free circulating DNA and its correlation with estrogen receptor protein expression in tumor tissue in breast cancer patients. BMC Cancer 2014, 14, 59. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Hou, J.; Shi, W.; Zhang, L. Estrogen receptor 1 (ESR1) genetic variations in cancer risk: A systematic review and meta-analysis. Clin. Res. Hepatol. Gastroenterol. 2015, 39, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Deng, Q.; Pan, Y.; He, B.; Ying, H.; Chen, J.; Liu, X.; Wang, S. Association between estrogen receptor 1 (ESR1) genetic variations and cancer risk: A meta-analysis. J. Buon 2015, 20, 296–308. [Google Scholar]
- Liu, X.; Huang, J.; Lin, H.; Xiong, L.; Ma, Y.; Lao, H. ESR1 PvuII (rs2234693 T>C) polymorphism and cancer susceptibility: Evidence from 80 studies. J. Cancer 2018, 9, 2963–2972. [Google Scholar] [CrossRef]
- Ding, X.; Cui, F.M.; Xu, S.T.; Pu, J.X.; Huang, Y.H.; Zhang, J.L.; Wei, X.D.; Hou, J.Q.; Yan, C.Y. Variants on ESR1 and their association with prostate cancer risk: A meta-analysis. Asian Pac. J. Cancer Prev. 2012, 13, 3931–3936. [Google Scholar] [CrossRef][Green Version]
- Garcia, C.J.C.; Grisetti, L.; Tiribelli, C.; Pascut, D. The ncRNA-AURKA Interaction in Hepatocellular Carcinoma: Insights into Oncogenic Pathways, Therapeutic Opportunities, and Future Challenges. Life 2024, 14, 1430. [Google Scholar] [CrossRef]
- Liu, R.; Jiang, Z.; Kong, W.; Zheng, S.; Dai, T.; Wang, G. A Novel Nine-Gene Signature Associated With Immune Infiltration for Predicting Prognosis in Hepatocellular Carcinoma. Front. Genet. 2021, 12, 730732. [Google Scholar] [CrossRef]
- Zhang, K.; Chen, J.; Chen, D.; Huang, J.; Feng, B.; Han, S.; Chen, Y.; Song, H.; De, W.; Zhu, Z.; et al. Aurora-A promotes chemoresistance in hepatocelluar carcinoma by targeting NF-kappaB/microRNA-21/PTEN signaling pathway. Oncotarget 2014, 5, 12916–12935. [Google Scholar] [CrossRef]
- Du, R.; Huang, C.; Liu, K.; Li, X.; Dong, Z. Targeting AURKA in Cancer: Molecular mechanisms and opportunities for Cancer therapy. Mol. Cancer 2021, 20, 15. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Tóth, F.; Polyák, H.; Szabó, Á.; Mándi, Y.; Vécsei, L. Immune Influencers in Action: Metabolites and Enzymes of the Tryptophan-Kynurenine Metabolic Pathway. Biomedicines 2021, 9, 734. [Google Scholar] [CrossRef] [PubMed]
- Teunis, C.; Nieuwdorp, M.; Hanssen, N. Interactions between Tryptophan Metabolism, the Gut Microbiome and the Immune System as Potential Drivers of Non-Alcoholic Fatty Liver Disease (NAFLD) and Metabolic Diseases. Metabolites 2022, 12, 514. [Google Scholar] [CrossRef]
- Niu, B.; Pan, T.; Xiao, Y.; Wang, H.; Zhu, J.; Tian, F.; Lu, W.; Chen, W. The therapeutic potential of dietary intervention: Based on the mechanism of a tryptophan derivative-indole propionic acid on metabolic disorders. Crit. Rev. Food Sci. Nutr. 2025, 65, 1729–1748. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, S.; Ding, Y.; Saedi, N.; Choi, M.; Sridharan, G.V.; Sherr, D.H.; Yarmush, M.L.; Alaniz, R.C.; Jayaraman, A.; Lee, K. Gut Microbiota-Derived Tryptophan Metabolites Modulate Inflammatory Response in Hepatocytes and Macrophages. Cell Rep. 2018, 23, 1099–1111. [Google Scholar] [CrossRef]
- Ding, Y.; Yanagi, K.; Yang, F.; Callaway, E.; Cheng, C.; Hensel, M.E.; Menon, R.; Alaniz, R.C.; Lee, K.; Jayaraman, A. Oral supplementation of gut microbial metabolite indole-3-acetate alleviates diet-induced steatosis and inflammation in mice. elife 2024, 12, RP87458. [Google Scholar] [CrossRef]
- Newman, A.C.; Falcone, M.; Huerta Uribe, A.; Zhang, T.; Athineos, D.; Pietzke, M.; Vazquez, A.; Blyth, K.; Maddocks, O.D.K. Immune-regulated IDO1-dependent tryptophan metabolism is source of one-carbon units for pancreatic cancer and stellate cells. Mol. Cell 2021, 81, 2290–2302.e2297. [Google Scholar] [CrossRef]
- Dang, S.; Jain, A.; Dhanda, G.; Bhattacharya, N.; Bhattacharya, A.; Senapati, S. One carbon metabolism and its implication in health and immune functions. Cell Biochem. Funct. 2024, 42, e3926. [Google Scholar] [CrossRef]
- Chiang, J.Y.L.; Ferrell, J.M. Bile acid receptors FXR and TGR5 signaling in fatty liver diseases and therapy. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G554–G573. [Google Scholar] [CrossRef]
- Song, J.; Zhong, R.L.; Xia, Z.; Wu, H.; Zhong, Q.X.; Zhang, Z.H.; Wei, Y.J.; Shi, Z.Q.; Feng, L.; Jia, X.B. Research and application of hepatotoxicity evaluation technique of traditional Chinese medicine. Zhongguo Zhong Yao Za Zhi 2017, 42, 41–48. [Google Scholar] [CrossRef]
- Choudhuri, S.; Klaassen, C.D. Molecular Regulation of Bile Acid Homeostasis. Drug Metab. Dispos. 2022, 50, 425–455. [Google Scholar] [CrossRef] [PubMed]
- Trabelsi, M.S.; Lestavel, S.; Staels, B.; Collet, X. Intestinal bile acid receptors are key regulators of glucose homeostasis. Proc. Nutr. Soc. 2017, 76, 192–202. [Google Scholar] [CrossRef]
- Kim, H.; Fang, S. Crosstalk between FXR and TGR5 controls glucagon-like peptide 1 secretion to maintain glycemic homeostasis. Lab. Anim. Res. 2018, 34, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, B.; Jones, S.A.; Price, R.R.; Watson, M.A.; McKee, D.D.; Moore, L.B.; Galardi, C.; Wilson, J.G.; Lewis, M.C.; Roth, M.E.; et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 2000, 6, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Anakk, S.; Watanabe, M.; Ochsner, S.A.; McKenna, N.J.; Finegold, M.J.; Moore, D.D. Combined deletion of Fxr and Shp in mice induces Cyp17a1 and results in juvenile onset cholestasis. J. Clin. Investig. 2011, 121, 86–95. [Google Scholar] [CrossRef]
- Li, T.; Matozel, M.; Boehme, S.; Kong, B.; Nilsson, L.M.; Guo, G.; Ellis, E.; Chiang, J.Y. Overexpression of cholesterol 7α-hydroxylase promotes hepatic bile acid synthesis and secretion and maintains cholesterol homeostasis. Hepatology 2011, 53, 996–1006. [Google Scholar] [CrossRef]
- Liu, H.; Pathak, P.; Boehme, S.; Chiang, J.L. Cholesterol 7α-hydroxylase protects the liver from inflammation and fibrosis by maintaining cholesterol homeostasis. J. Lipid Res. 2016, 57, 1831–1844. [Google Scholar] [CrossRef] [PubMed]
- Hafner, M.; Rezen, T.; Rozman, D. Regulation of hepatic cytochromes p450 by lipids and cholesterol. Curr. Drug Metab. 2011, 12, 173–185. [Google Scholar] [CrossRef]
- Eloranta, J.J.; Kullak-Ublick, G.A. Coordinate transcriptional regulation of bile acid homeostasis and drug metabolism. Arch. Biochem. Biophys. 2005, 433, 397–412. [Google Scholar] [CrossRef] [PubMed]
- Li, P.L.; Huang, C.H.; Mao, L.; Li, J.; Sheng, Z.G.; Zhu, B.Z. An unprecedented free radical mechanism for the formation of DNA adducts by the carcinogenic N-sulfonated metabolite of aristolochic acids. Free Radic. Biol. Med. 2023, 205, 332–345. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, R.; Zhao, Y.; Chen, Z.; Zhai, H.; Si, H. Network Pharmacology, Molecular Docking, Molecular Dynamics to Explore the Mechanism of Danggui Shaoyao Powder for Hepatic Encephalopathy. Curr. Pharm. Des. 2025, 31, 1562–1582. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, X.; Wang, Y.; Zhong, H.; Liu, L.; Ye, Y. Network pharmacology integrated with molecular docking technology to reveal the potential mechanism of Shuganfang against drug-induced liver injury. Medicine 2023, 102, e36349. [Google Scholar] [CrossRef]
- Zhang, S.; Mo, Z.; Zhang, S.; Li, X. A Network Pharmacology Approach to Reveal the Underlying Mechanisms of Artemisia annua on the Treatment of Hepatocellular Carcinoma. Evid. Based Complement. Altern. Med. 2021, 2021, 8947304. [Google Scholar] [CrossRef]
- Wang, N.; Tang, L.; Duan, R.; Shu, Y. Investigating the Mechanism of Hedysarum Multijugum Maxim in the Treatment of Liver Cancer through Network Pharmacology and Molecular Docking Validation. Oncology 2024, 1–18. [Google Scholar] [CrossRef]
- Kim, S. Getting the most out of PubChem for virtual screening. Expert. Opin. Drug Discov. 2016, 11, 843–855. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tu, T.; Zheng, T.; Lin, H.; Cheng, P.; Yang, Y.; Liu, B.; Ying, X.; Xie, Q. Mechanistic Exploration of Aristolochic Acid I-Induced Hepatocellular Carcinoma: Insights from Network Toxicology, Machine Learning, Molecular Docking, and Molecular Dynamics Simulation. Toxins 2025, 17, 390. https://doi.org/10.3390/toxins17080390
Tu T, Zheng T, Lin H, Cheng P, Yang Y, Liu B, Ying X, Xie Q. Mechanistic Exploration of Aristolochic Acid I-Induced Hepatocellular Carcinoma: Insights from Network Toxicology, Machine Learning, Molecular Docking, and Molecular Dynamics Simulation. Toxins. 2025; 17(8):390. https://doi.org/10.3390/toxins17080390
Chicago/Turabian StyleTu, Tiantaixi, Tongtong Zheng, Hangqi Lin, Peifeng Cheng, Ye Yang, Bolin Liu, Xinwang Ying, and Qingfeng Xie. 2025. "Mechanistic Exploration of Aristolochic Acid I-Induced Hepatocellular Carcinoma: Insights from Network Toxicology, Machine Learning, Molecular Docking, and Molecular Dynamics Simulation" Toxins 17, no. 8: 390. https://doi.org/10.3390/toxins17080390
APA StyleTu, T., Zheng, T., Lin, H., Cheng, P., Yang, Y., Liu, B., Ying, X., & Xie, Q. (2025). Mechanistic Exploration of Aristolochic Acid I-Induced Hepatocellular Carcinoma: Insights from Network Toxicology, Machine Learning, Molecular Docking, and Molecular Dynamics Simulation. Toxins, 17(8), 390. https://doi.org/10.3390/toxins17080390





