Response Surface Methodology Optimization of Time-Resolved Fluorescence Immunoassay for Rapid Detection of AflatoxinB1 in Yellow Rice Wine
Abstract
:1. Introduction
2. Results and Discussion
2.1. Method Detection Line and Linear Range
2.2. Results of the Single-Factor Experiment
2.3. Screening of Important Variables Using Plackett-Burman Design
2.4. Path of Steepest Ascent
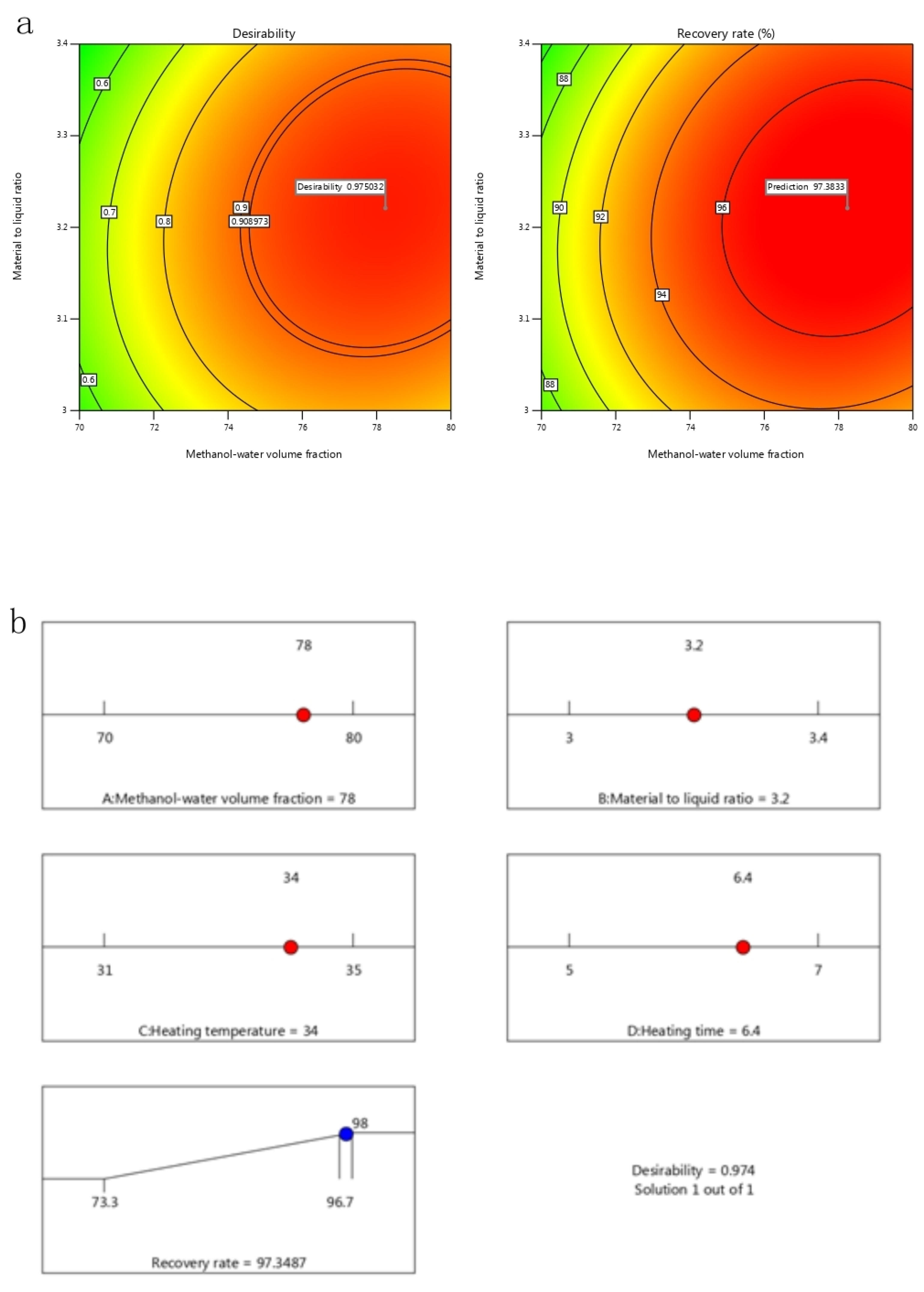
2.5. Optimization by Response Surface Methodology
2.5.1. RSM Regression Equation and Model Analysis
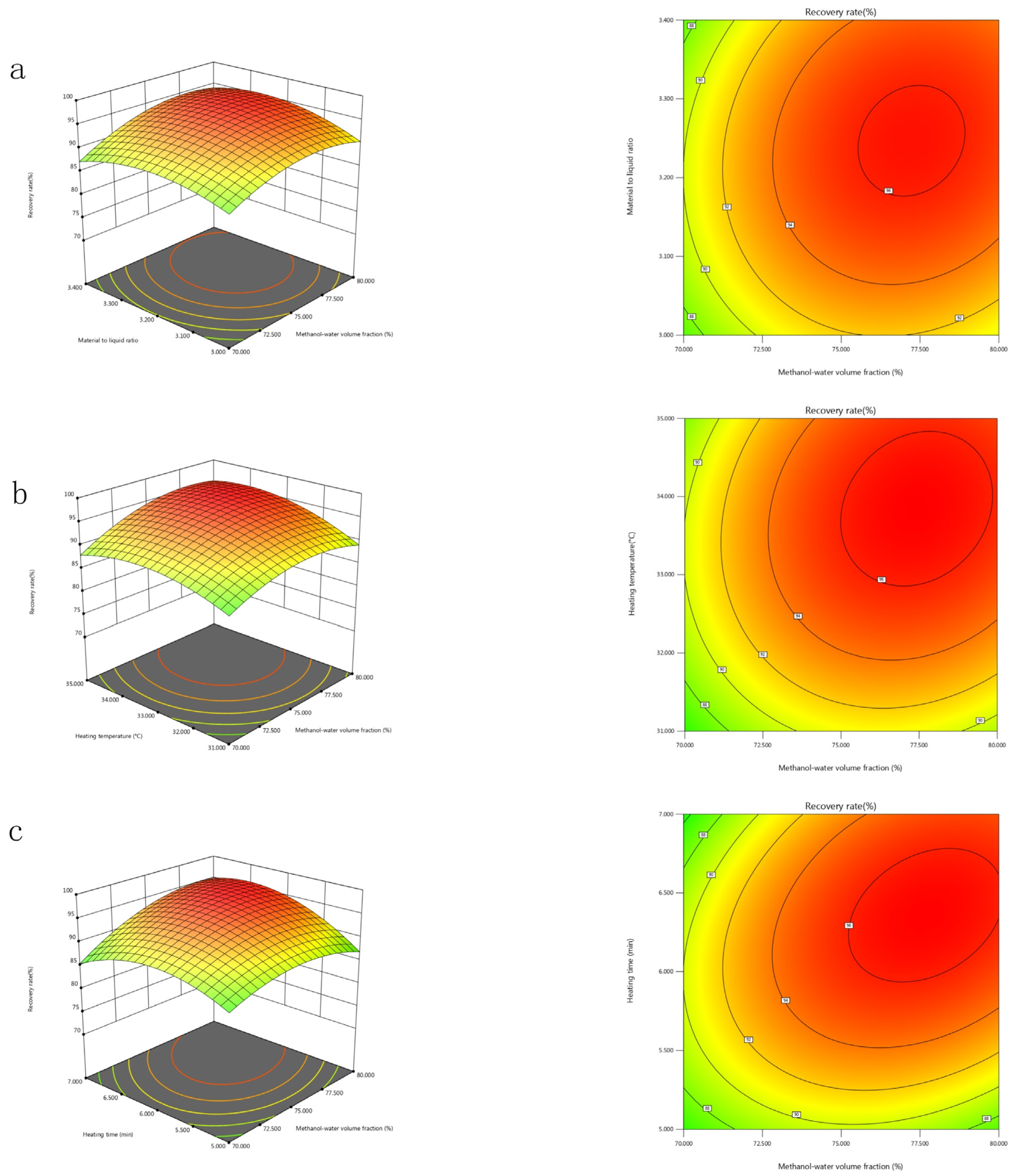
2.5.2. Mutual Interactions Between the Significant Factors
2.6. Validation of the Model
2.6.1. Intra-Assay Precision of the Method
2.6.2. Inter-Assay Precision of the Method
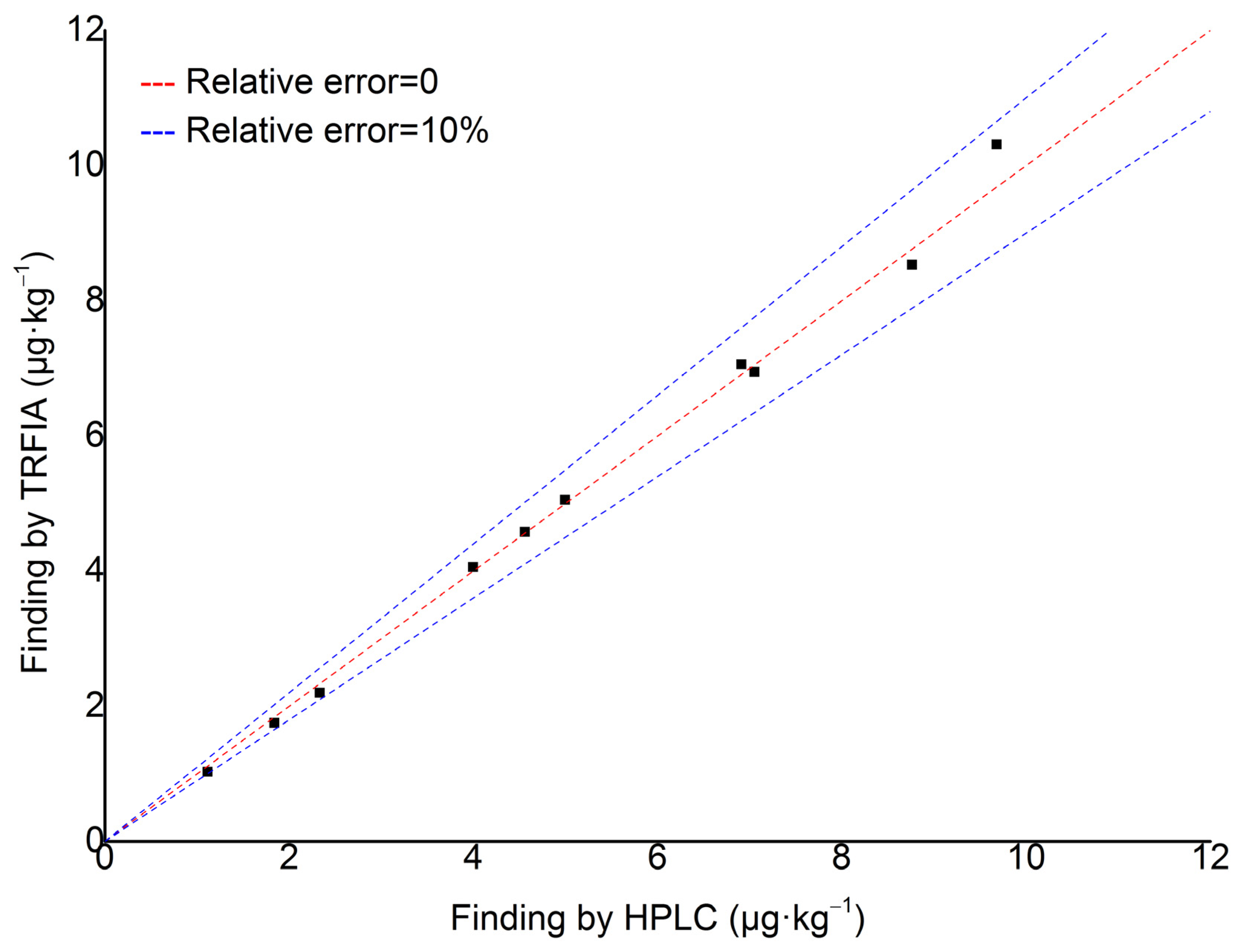
2.6.3. Comparison of Result Between TRFIA and HPLC
2.6.4. Performance Comparison of TRFIA, ELISA, HPLC, and LC-MS/MS
3. Conclusions
4. Materials and Methods
4.1. Establish a Standard Curve
4.1.1. Preparation of Sample Diluent
4.1.2. Selection of Blank Matrix Solution
4.1.3. Standard Working Solution Preparation
4.1.4. Detection Limit and Linear Range
4.2. Sample Pretreatment and Determination of AFB1 Content
4.2.1. Sample Pretreatment
4.2.2. Determination of AFB1 Content
4.3. Experimental Designs
4.3.1. Single-Factor Experiment
4.3.2. Plackett-Burman Design
4.3.3. Path of Steepest Ascent
4.3.4. Response Surface Methodology
4.4. Validation of the Model
4.4.1. Intra-Assay and Inter-Assay Precision of the Method
4.4.2. Comparison of Result Between TRFIA and HPLC
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jin, Z.; Cai, G.; Wu, C.; Hu, Z.; Xu, X.; Xie, G.; Wu, D.; Lu, J. Profiling the key metabolites produced during the modern brewing process of Chinese rice wine. Food Res. Int. 2021, 139, 109955. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.F.; Ruan, H.; Jin, J.J.F.F.; Jiang, Y. Research progress on healthy efficacy and related bioactive substances of Huangjiu. China Brew. 2021, 40, 7–11. [Google Scholar]
- Ren, Q.; Xu, J.L.; Liu, J.H. Safety risk and its control of yellow rice wine. J. Food Sci. Technol. 2016, 34, 12–19. [Google Scholar]
- Zhuang, Z.H.; Zheng, C.Q.; Wang, S.H. Optimization of Aspergillus flvaus Culture Conditions and Extraction of Aflatoxin B1. Chin. J. Appl. Environ. Biol. 2010, 16, 724–729. [Google Scholar]
- Wilson, C.L.; Droby, S. Microbial Food Contamination; CRC Press: New York, NY, USA, 2001. [Google Scholar]
- Guo, X.; Wen, F.; Zheng, N.; Luo, Q.; Wang, H.; Wang, H.; Li, S.; Wang, J. Development of an ultrasensitive aptsensor for the detection of aflatoxin B1. Biosens. Bioelectron. 2014, 56, 340–344. [Google Scholar] [CrossRef]
- Mao, Q.Z.; Lu, W.G.; Chen, B.L.; Yu, G.S.; Wu, B.Y. Study and analysis on safety of rice wine. Liquor. Mak. 2006, 3, 60–65. [Google Scholar]
- Yang, X.D.; Luo, H.B.; Ye, G.B.; Li, D.Y.; Wang, C.H.; Wang, Y. Verificat-ion of the Safety of Aflatoxin B1 in Nong-flavor Liquor Production by Enzyme-linked Immunosorbent Assay(ELISA). Liquor-Mak. Sci. Technol. 2013, 4, 92–94. [Google Scholar]
- Han, X.W. Analysis Methods Development for Three Mycotoxins and Application in Liquor Production. Master’sThesis, Tianjin University of Science and Technology, Tianjin, China, 2014. [Google Scholar]
- Nagatomi, Y.; Inoue, T.; Uyama, A.; Mochizuki, N. The fate of mycotoxins during the distillation process of barley shochu, a distilled alcoholic beverage. Biosci. Biotechnol. Biochem. 2012, 76, 202–204. [Google Scholar] [CrossRef]
- Xu, Z.; Tan, S.M.; Jiao, Y.C.; Zhang, Y.F. Determination of AflatoxinB1 in Dry Capsicum by Enzyme-linked Immunosorbent Assay (ELISA). Food Sci. 2009, 30, 245–247. [Google Scholar]
- Ji, X.F.; Chen, X.Y.; Wei, W. Determination of aflatoxin B1 in bottled Chinese rice wine by enzyme-linked immunosorbent assay (ELISA). Acta Agric. Zhejiangensis 2015, 27, 2006–2010. [Google Scholar]
- Liu, J.; Xiong, N.; Liu, L.; Yu, D. Verification of rapid detection of aflatoxin b1 in grain by immunochromatographic test paper method. J. Henan Univ. Technol. 2011, 32, 51–57. [Google Scholar]
- Li, P.W.; Ma, L.; Yang, J.E.; Zhang, W.; Yang, C.H.; Yang, M. A reiew on analyticalmethods for aflatox in B1 in grains and oilseeds products. ChineseJournal Oil Crop Sci. 2005, 27, 77–81. [Google Scholar]
- Hu, Z.; Chen, J.; Li, Y.; Wang, Y.; Zhang, Q.; Hussain, E.; Yang, M.; Shahzad, S.A.; Yu, D.; Yu, C. Nucleic acid-controlled quantum dots aggregation: A label-free fluorescence turn-on strategy for alkaline phosphatase detection. Talanta 2017, 169, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Gao, Y.; Li, Y.; Zhang, Q.; Hu, Z.; Zhang, Y.; Hussain, E.; Yang, X.; Yu, D.; Yu, C. Polyphosphoric acid-induced perylene probe self-assembly and label-free fluorescence turn-on detection of alkaline phosphatase. Anal. Bioanal. Chem. 2017, 409, 1031–1036. [Google Scholar] [CrossRef]
- Cusati, T.; Granucci, G.; Persico, M. Photodynamics and time-resolved fluorescence of azobenzene in solution: A mixed quantum-classical simulation. J. Am. Chem. Soc. 2011, 133, 5109–5123. [Google Scholar] [CrossRef]
- Mizukami, S.; Tonai, K.; Kaneko, M.; Kikuchi, K. Lanthanide-based protease activity sensors for time-resolved fluorescence measurements. J. Am. Chem. Soc. 2008, 130, 14376–14377. [Google Scholar] [CrossRef]
- Bunzli, J.C.G.; Piguet, C. Taking advantage of luminescent lanthanide ions. Chem. Soc. Rev. 2005, 34, 1048–1077. [Google Scholar] [CrossRef]
- Shi, J.; Huang, B.; Zhang, L.-F.; Mi, X.-L.; Xu, G.-R.; Jin, J. Use of ultrasensitive time-resolved fluoroimmunoassay for rapid detection of clenbuterol hydrochloride in urine. J. Hyg. Res. 2006, 35, 798–801. [Google Scholar]
- Kennedy, M.; Krouse, D. Strategies for improving fermentation mediumperformance:Areview. J. Ind. Microbiol. Biotechnol. 1999, 23, 456–475. [Google Scholar] [CrossRef]
- Abdel; Fattah, Y.R.; Saeed, H.M.; Gohar, Y.M.; El-Baz, M.A. 5. Improved production of Pseudomonas aeruginosa uricase by optimization of process parameters through statistical experimental designs. Process Biochem. 2004, 5, 1707–1714. [Google Scholar]
- Gheshlaghi, R.; Scharer, J.M.; Moo-Young, M.; Douglas, P.L. Medium optimization for hen cgg white lysozyme production by recombinant Aspergillus niger using statistical methods. Biotechnol. Bioeng. 2005, 90, 754–760. [Google Scholar] [CrossRef] [PubMed]
- GB 5009.22—2016; Determination of AFB and AFG Groups in Food. National Food Safety Standards: Beijing, China, 2016; pp. 11–17.
- Zhang, Z.; Tang, X.; Wang, D.; Zhang, Q.; Li, P.; Ding, X. Rapid on-site sensing aflatoxinB1 in food and feed via a chromatographic time-resolved fluoroimmunoassay. PLoS ONE 2015, 10, 0123266. [Google Scholar]
- Zhao, Z.; Yang, X.; Zhao, X.; Bai, B.; Yao, C.; Liu, N.; Wang, J.; Zhou, C. Vortex-assisted dispersive liquid-liquid microextraction for the analysis of major Aspergillus and Penicillium mycotoxins in rice wine by liquid chromatography-tandem mass spectrometry. Food Control 2017, 73, 862–868. [Google Scholar] [CrossRef]
- Wang, A.F. Research on Analysis and Confirmed System of Typical Toxins During the Production of Chinese Rice Wine. Ph.D. Thesis, Fujian Agriculture and Forestry University, Fuzhou, China, April 2023. [Google Scholar]
- Zhang, Z.W.; Li, P.W.; Zhang, Q.; Ding, X.X. Study on Time-Resolved Fluorescence Immunochromatography for Afaltoxin Determination in Agricultural Product. Sci. Agric. Sin. 2014, 47, 3668–3674. [Google Scholar]
- GB/T 27404—2008; Riterion on Quality Control of Laboratories-Chemical Testing of Food. Chinese National Standards: Beijing, China, 2008.
- Stanbury, P.F.; Whitaker, A.; Hall, S.J. 6. Media for industrial fermentations. In Principles of Fermentation Technology; Pergamon Press: Oxford, UK, 1989; pp. 93–122. [Google Scholar]
- Chen, X.C.; Bai, J.X.; Cao, J.M.; Li, Z.J.; Xiong, J.; Zhang, L.; Hong, Y.; Ying, H.J. Mediumoptimization for the production of cycfic adenosine 3′.5′-monophosphate by Microbacterium sp. no. 205 using response surface methodology. Bioresour. Technol. 2009, 100, 919–924. [Google Scholar] [CrossRef]
- He, Y.Q.; Tan, T.W. Use of response surface methodology to optiIllize culture medium for production of lipase with Candida sp. 99-l25. J. Mokcular Catal. B Enzym. 2006, 43, 9–14. [Google Scholar] [CrossRef]
- Tang, X.J.; He, G.Q.; Chen, Q.H.; Zhang, X.Y.; Ali, M.A.M. Medium optimization for the production of thermal stable β-glucanase by Bacillus subtilis ZJF-1A5 using response surface methodology. Bioresour. Technol. 2004, 93, 175–181. [Google Scholar] [CrossRef]



| Experimental Value | |||
|---|---|---|---|
| Variable | Symbol | Low (−1) | High (+1) |
| Methanol-water volume fraction (%) | A | 60 | 80 |
| Sample to extraction solvent ratio | B | 1:2 | 1:4 |
| Oscillation time (min) | C | 12 | 18 |
| Centrifugal speed (r/min) | D | 5000 | 7000 |
| Centrifugal time (min) | E | 3 | 5 |
| Heating temperature (°C) | F | 30 | 37 |
| Heating time (min) | G | 4 | 8 |
| Dummy1 | H | −1 | 1 |
| Dummy2 | I | −1 | 1 |
| Dummy3 | J | −1 | 1 |
| Dummy4 | K | −1 | 1 |
| Run Order | Experimental Value | Recovery Rate (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | ||
| 1 | −1 | 1 | 1 | −1 | 1 | 1 | 1 | −1 | −1 | −1 | 1 | 88.3 |
| 2 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | 72.9 |
| 3 | 1 | −1 | 1 | 1 | −1 | 1 | 1 | 1 | −1 | −1 | −1 | 94.5 |
| 4 | −1 | 1 | 1 | 1 | −1 | −1 | −1 | 1 | −1 | 1 | 1 | 72.5 |
| 5 | −1 | 1 | −1 | 1 | 1 | −1 | 1 | 1 | 1 | −1 | −1 | 84.6 |
| 6 | −1 | −1 | −1 | 1 | −1 | 1 | 1 | −1 | 1 | 1 | 1 | 88.1 |
| 7 | 1 | 1 | −1 | −1 | −1 | 1 | −1 | 1 | 1 | −1 | 1 | 94.6 |
| 8 | 1 | −1 | 1 | 1 | 1 | −1 | −1 | −1 | 1 | −1 | 1 | 81.6 |
| 9 | 1 | 1 | −1 | 1 | 1 | 1 | −1 | −1 | −1 | 1 | −1 | 94.8 |
| 10 | 1 | −1 | −1 | −1 | 1 | −1 | 1 | 1 | −1 | 1 | 1 | 88.3 |
| 11 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | 1 | 1 | 1 | −1 | 81.2 |
| 12 | 1 | −1 | 1 | −1 | −1 | −1 | 1 | −1 | 1 | 1 | −1 | 90.5 |
| Coefficient | Sum of Squares | Degrees of Freedom | Mean Square | F-Value | p-Value | ||
|---|---|---|---|---|---|---|---|
| Model | 85.99 | 647.61 | 7 | 92.52 | 38.97 | 0.0016 | ** |
| A | 4.73 | 267.91 | 1 | 267.91 | 112.84 | 0.0004 | *** |
| B | 1.56 | 29.14 | 1 | 29.14 | 12.27 | 0.0248 | * |
| C | −1.23 | 18.01 | 1 | 18.01 | 7.58 | 0.0512 | |
| D | 0.025 | 0.0075 | 1 | 0.0075 | 0.0032 | 0.9579 | |
| E | 0.475 | 2.71 | 1 | 2.71 | 1.14 | 0.3457 | |
| F | 4.26 | 217.60 | 1 | 217.60 | 91.65 | 0.0007 | *** |
| G | 3.06 | 112.24 | 1 | 112.24 | 47.28 | 0.0023 | ** |
| Residual | 9.50 | 4 | 2.37 | ||||
| Correlation Total | 657.11 | 11 |
| Items | X1 | X2 | X3 | X4 | Recovery Rate (%) |
|---|---|---|---|---|---|
| No. 1. Base point (zero level in Plackett-Burman design) | 70 | 1:3 | 33.5 | 6 | |
| No. 2. Origin step unit (range of unity level) | 10 | 1 | 3.5 | 2 | |
| No. 3. Slope (estimated coefficient ratio from Equation (5) | +4.73 | +1.56 | +4.26 | +3.06 | |
| No. 4. Correspondent range = 2 × 3 | 47.3 | 1.56 | 14.91 | 7.12 | |
| No. 5. New step unit = (4) × 0.1 a | 4.73 | 0.156 | 1.49 | 0.712 | |
| No. 6. New step unit with a decimal | 5.0 | 0.2 | 1.5 | 1.0 | |
| Experiment No. 1 | 65 | 1:2.8 | 30 | 4 | 71.8 |
| Experiment No. 2 | 70 | 1:3.0 | 31.5 | 5 | 79.4 |
| Experiment No. 3 | 75 | 1:3.2 | 33 | 6 | 96.1 |
| Experiment No. 4 | 80 | 1:3.4 | 35.5 | 7 | 90.1 |
| Experiment No. 5 | 85 | 1:3.6 | 37 | 8 | 82.2 |
| Variable | Symbol | Coded Level | ||||
|---|---|---|---|---|---|---|
| −2 | −1 | 0 | 1 | 2 | ||
| Methanol-water volume fraction | X1 | 65 | 70 | 75 | 80 | 85 |
| Sample to extraction solvent ratio | X2 | 1:2.8 | 1:3.0 | 1:3.2 | 1:3.4 | 1:3.6 |
| Heating temperature | X3 | 29 | 31 | 33 | 35 | 37 |
| Heating time | X4 | 4 | 5 | 6 | 7 | 8 |
| Run Order | Code Level | Recovery Rate (%) | Run Order | Code Level | Recovery Rate (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| X1 | X2 | X3 | X4 | X1 | X2 | X3 | X4 | ||||
| 1 | 1 | 1 | −1 | −1 | 83.2 | 16 | −1 | 1 | 1 | 1 | 79.5 |
| 2 | 0 | 0 | 0 | 0 | 95.7 | 17 | 0 | 0 | 0 | 0 | 96.7 |
| 3 | 0 | 0 | 0 | 2 | 83.1 | 18 | 0 | 0 | −2 | 0 | 80.2 |
| 4 | 1 | 1 | −1 | 1 | 88.6 | 19 | −1 | 1 | −1 | 1 | 78.9 |
| 5 | −2 | 0 | 0 | 0 | 78.5 | 20 | 1 | −1 | −1 | 1 | 82.3 |
| 6 | 0 | 2 | 0 | 0 | 86.3 | 21 | 1 | 1 | 1 | 1 | 92.1 |
| 7 | 0 | 0 | 0 | −2 | 75.5 | 22 | −1 | 1 | 1 | −1 | 80.2 |
| 8 | −1 | −1 | −1 | 1 | 76.6 | 23 | 2 | 0 | 0 | 0 | 88.7 |
| 9 | 1 | 1 | 1 | −1 | 83.3 | 24 | 0 | 0 | 0 | 0 | 96.4 |
| 10 | −1 | −1 | 1 | 1 | 82.9 | 25 | −1 | −1 | 1 | −1 | 80.8 |
| 11 | 1 | −1 | 1 | 1 | 92.6 | 26 | 0 | 0 | 0 | 0 | 94.9 |
| 12 | 0 | 0 | 2 | 0 | 87.9 | 27 | 0 | −2 | 0 | 0 | 82.6 |
| 13 | 1 | −1 | −1 | −1 | 73.3 | 28 | 0 | 0 | 0 | 0 | 95.1 |
| 14 | −1 | 1 | −1 | −1 | 81.2 | 29 | 0 | 0 | 0 | 0 | 95.2 |
| 15 | −1 | −1 | −1 | −1 | 78.6 | 30 | 1 | −1 | 1 | −1 | 84.2 |
| Source | Sum of Squares | Degrees of Freedom | Mean Square | F-Value | p-Value | |
|---|---|---|---|---|---|---|
| Model | 1371.77 | 14 | 97.98 | 121.96 | <0.0001 | significant |
| X1-X1 | 156.57 | 1 | 156.57 | 194.89 | <0.0001 | |
| X2-X2 | 22.23 | 1 | 22.23 | 27.67 | <0.0001 | |
| X3-X3 | 97.20 | 1 | 97.20 | 120.99 | <0.0001 | |
| X4-X4 | 80.30 | 1 | 80.30 | 99.95 | <0.0001 | |
| X1X2 | 12.08 | 1 | 12.08 | 15.03 | 0.0015 | |
| X1X3 | 17.43 | 1 | 17.43 | 21.70 | 0.0003 | |
| X1X4 | 74.39 | 1 | 74.39 | 92.60 | <0.0001 | |
| X2X3 | 43.89 | 1 | 43.89 | 54.63 | <0.0001 | |
| X2X4 | 2.48 | 1 | 2.48 | 3.09 | 0.0993 | |
| X3X4 | 4.52 | 1 | 4.52 | 5.62 | 0.0316 | |
| X1X1 | 262.35 | 1 | 262.35 | 326.55 | <0.0001 | |
| X2X2 | 227.54 | 1 | 227.54 | 283.22 | <0.0001 | |
| X3X3 | 243.61 | 1 | 243.61 | 303.23 | <0.0001 | |
| X4X4 | 476.43 | 1 | 476.43 | 593.02 | <0.0001 | |
| Residual | 12.05 | 15 | 0.8034 | |||
| Lack of fit | 9.32 | 10 | 0.9318 | 1.70 | 0.2893 | not significant |
| Pure error | 2.73 | 5 | 0.5467 | |||
| Correlation total | 1383.82 | 29 |
| Spiked Level (μg·kg−1) | Average Finding (μg·kg−1) | Recovery Rate (%) | Standard Deviation (μg·kg−1) | RSD (%) |
|---|---|---|---|---|
| 1 | 0.879 | 87.9 | 0.0550 | 6.26 |
| 5 | 4.885 | 97.7 | 0.2345 | 4.80 |
| 10 | 10.569 | 105.7 | 0.4734 | 4.48 |
| Spiked Level (μg·kg−1) | Average Finding (μg·kg−1) | Recovery Rate (%) | Standard Deviation (μg·kg−1) | RSD (%) |
|---|---|---|---|---|
| 1 | 0.859 | 85.9 | 0.0761 | 8.86 |
| 5 | 4.760 | 95.2 | 0.3380 | 7.10 |
| 10 | 9.980 | 99.8 | 0.6088 | 6.10 |
| Sample Number | Finding by TRFIA (μg·kg−1) | Finding by HPLC (μg·kg−1) | Relative Error (%) |
|---|---|---|---|
| Sample 1 | 1.646 | 1.543 | 6.68 |
| Sample 2 | N.D. | N.D. | / |
| Sample 3 | N.D. | 0.157 | / |
| Sample 4 | 2.105 | 2.185 | 3.66 |
| Sample 5 | 6.710 | 6.910 | 2.89 |
| Sample 6 | N.D. | N.D. | / |
| Sample 7 | 1.200 | 1.330 | 7.69 |
| Sample 8 | N.D. | N.D. | / |
| Sample 9 | N.D. | 0.09 | / |
| Sample 10 | 0.612 | 0.684 | 10.53 |
| Parameters | TRFIA | ELISA [12,24] | HPLC [24] | LC-MS/MS [24,26,27] |
|---|---|---|---|---|
| Detection limit | 0.3 μg/L | 0.05–1 μg/L | 0.02–0.03 μg/L | 0.03–0.5 μg/L |
| Limit of quantitation | 0.8 μg/L | 0.1–3 μg/L | 0.05–0.1 μg/L | 0.1–0.25 μg/L |
| Analysis time | 30–60 min | 45–90 min | 2–3 h | 2–3 h |
| Specificity | high | low | extremely high | extremely high |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, M.; Wang, D.; Wang, D.; Dong, J.; Wang, X.; Zhang, Q.; Xiao, M. Response Surface Methodology Optimization of Time-Resolved Fluorescence Immunoassay for Rapid Detection of AflatoxinB1 in Yellow Rice Wine. Toxins 2025, 17, 248. https://doi.org/10.3390/toxins17050248
Zhu M, Wang D, Wang D, Dong J, Wang X, Zhang Q, Xiao M. Response Surface Methodology Optimization of Time-Resolved Fluorescence Immunoassay for Rapid Detection of AflatoxinB1 in Yellow Rice Wine. Toxins. 2025; 17(5):248. https://doi.org/10.3390/toxins17050248
Chicago/Turabian StyleZhu, Mengjie, Dun Wang, Du Wang, Jing Dong, Xue Wang, Qi Zhang, and Man Xiao. 2025. "Response Surface Methodology Optimization of Time-Resolved Fluorescence Immunoassay for Rapid Detection of AflatoxinB1 in Yellow Rice Wine" Toxins 17, no. 5: 248. https://doi.org/10.3390/toxins17050248
APA StyleZhu, M., Wang, D., Wang, D., Dong, J., Wang, X., Zhang, Q., & Xiao, M. (2025). Response Surface Methodology Optimization of Time-Resolved Fluorescence Immunoassay for Rapid Detection of AflatoxinB1 in Yellow Rice Wine. Toxins, 17(5), 248. https://doi.org/10.3390/toxins17050248






