Molecular Quantification of Total and Toxigenic Microcystis Using Digital-Droplet-Polymerase-Chain-Reaction-Based Multiplex Assay
Abstract
1. Introduction
2. Results
2.1. Verification of secA and mcyA ddPCR Primers and Probes
2.2. Comparison of Sensitivity Between secA and mcyA Singleplex and Multiplex PCR Using ddPCR
2.3. Copy Number per Cell of secA and mcyA Genes in Microcystis Cells
2.4. Comparison Between Microscopic Analysis and Genetic Quantification of Field Samples
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Cyanobacterial Strains and Culture Conditions
5.2. Field Sample Collection and Pretreatment
5.3. DNA Extraction
5.4. Primer Design and Validation
5.5. The ddPCR Analysis
5.6. Calculation of Microcystis Gene Copy Number per Cell Using ddPCR Analysis and Field Application
5.7. Analysis of Microcystins
5.8. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paerl, H.W.; Hall, N.S.; Calandrino, E.S. Controlling harmful cyanobacterial blooms in a world experiencing anthropogenic and climatic-induced change. Sci. Total Environ. 2011, 409, 739–1745. [Google Scholar] [CrossRef] [PubMed]
- Beaver, J.R.; Manis, E.E.; Loftin, K.A.; Graham, J.L.; Pollard, A.I.; Mitchell, R.M. Land use patterns, ecoregion, and microcystin relationships in U.S. lakes and reservoirs: A preliminary evaluation. Harmful Algae 2014, 36, 57–62. [Google Scholar] [CrossRef]
- Huisman, J.M.; Matthijs, H.C.P.; Visser, P.M. Harmful Cyanobacteria; Springer Aquatic Ecology Series 3; Springer: Dordrecht, The Netherlands, 2005; p. 243. [Google Scholar]
- Paerl, H.W.; Otten, T.G. Harmful cyanobacterial blooms: Causes, consequences, and controls. Microbiol. Ecol. 2013, 65, 995–1010. [Google Scholar] [CrossRef] [PubMed]
- Codd, G.A.; Meriluoto, J.; Metcalf, J.S. Introduction: Cyanobacteria, cyanotoxins, their human impact, and risk management. In Handbook of Cyanobacterial Monitoring and Cyanotoxin Analysis; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017. [Google Scholar]
- Mirasbekov, Y.; Abdimanova, A.; Sarkytbayev, K.; Samarkhanov, K.; Abilkas, A.; Potashnikova, D.; Arbuz, G.; Issayev, Z.; Vorobjev, I.A.; Malashenkov, D.V.; et al. Combining Imaging Flow Cytometry and Molecular Biological Methods to Reveal Presence of Potentially Toxic Algae at the Ural River in Kazakhstan. Front. Mar. Sci. 2021, 8, 680482. [Google Scholar] [CrossRef]
- Chorus, I.; Welker, M. (Eds.) Toxic Cyanobacteria in Water, 2nd ed.; CRC Press: Boca Raton, FL, USA; on behalf of the World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Adekolurejo, O.A.; Floyd, M.; Dunn, A.M.; Kay, P.; Dean, A.P.; Hassall, C. Combined effects of increased water temperature and cyanobacterial compounds exert heterogeneous effects on survival and ecological processes in key freshwater species. Oecologia 2022, 200, 515–528. [Google Scholar] [CrossRef]
- Erratt, K.J.; Creed, I.F.; Lobb, D.A.; Smol, J.P.; Trick, C.G. Climate change amplifies the risk of potentially toxigenic cyanobacteria. Glob. Change Biol. 2023, 29, 5240–5249. [Google Scholar] [CrossRef]
- Smucker, N.J.; Beaulieu, J.J.; Nietch, C.T.; Young, J.L. Increasingly severe cyanobacterial blooms and deep water hypoxia coincide with warming water temperatures in reservoirs. Glob. Change Biol. 2021, 27, 2507–2519. [Google Scholar] [CrossRef]
- Merder, J.; Harris, R.T.; Zhao, G.; Stasinopoulos, D.M.; Rigby, R.A.; Michalak, A.M. Geographic redistribution of microcystin hotspots in response to climate warming. Nat. Water 2023, 1, 844–859. [Google Scholar] [CrossRef]
- Kleinteich, J.; Frassl, M.A.; Schulz, M.; Fischer, H. Climate change triggered planktonic cyanobacterial blooms in a regulated temperate river. Sci. Rep. 2024, 14, 16298. [Google Scholar] [CrossRef]
- Saleem, F.; Jiang, J.L.; Atrache, R.; Paschos, A.; Edge, T.A.; Schellhorn, H.E. Cyanobacterial Algal Bloom Monitoring: Molecular Methods and Technologies for Freshwater Ecosystems. Microorganisms 2023, 11, 851. [Google Scholar] [CrossRef]
- Park, H.-D.; Watanabe, M.F.; Harada, K.I.; Nagai, H.; Suzuki, M.; Watanabe, M.; Hayashi, H. Hepatotoxin (microcystin) and neurotoxin (anatoxin-a) contained in natural blooms and strains of cyanobacteria from Japanese freshwaters. Nat. Toxins 1993, 1, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Otten, T.G.; Xu, H.; Qin, B.; Zhu, G.; Paerl, H.W. Spatiotemporal patterns and ecophysiology of toxigenic Microcystis blooms in Lake Taihu, China: Implications for water quality management. Environ. Sci. Technol. 2012, 46, 3480–3488. [Google Scholar] [CrossRef] [PubMed]
- Guedes, I.A.; da Costa Leite, D.M.; Manhães, L.A.; Bisch, P.M.; Azevedo, S.M.; Pacheco, A.B.F. Fluctuations in microcystin concentrations, potentially toxic Microcystis and genotype diversity in a cyanobacterial community from a tropical reservoir. Harmful Algae 2014, 39, 303–309. [Google Scholar] [CrossRef]
- Wu, Y.; Li, L.; Zheng, L.; Dai, G.; Ma, H.; Shan, K.; Wu, H.; Zhou, Q.; Song, L. Patterns of succession between bloom-forming cyanobacteria Aphanizomenon flos-aquae and Microcystis and related environmental factors in large, shallow Dianchi Lake. China Hydrobiol. 2016, 765, 1–13. [Google Scholar] [CrossRef]
- Harke, M.J.; Steffen, M.M.; Gobler, C.J.; Otten, T.G.; Wilhelm, S.W.; Wood, S.A.; Paerl, H.W. A review of the global ecology, genomics, and biogeography of the toxic cyanobacterium, Microcystis spp. Harmful Algae 2016, 54, 4–20. [Google Scholar] [CrossRef]
- Jablonska, M.; Cerasino, L.; Boscaini, A.; Capelli, C.; Greco, C.; Klemenčič, A.K.; Mischke, U.; Salmaso, N.; Kurmayer, R. Distribution of toxigenic cyanobacteria in Alpine lakes and rivers as revealed by molecular screening. Water Res. 2024, 258, 121783. [Google Scholar] [CrossRef]
- Joung, S.-H.; Oh, H.-M.; Ko, S.-R.; Ahn, C.-Y. Correlations between environmental factors and toxic and non–toxic Microcystis dynamics during bloom in Daechung Reservoir, Korea. Harmful Algae 2011, 10, 188–193. [Google Scholar] [CrossRef]
- Park, H.K.; Lee, H.J.; Heo, J.; Yun, J.H.; Kim, Y.J.; Kim, H.M.; Hong, D.G.; Lee, I.J. Deciphering the key factors determining spatio-temporal heterogeneity of cyanobacterial bloom dynamics in the Nakdong River with consecutive large weirs. Sci. Total Environ. 2020, 755, 143079. [Google Scholar] [CrossRef]
- Komáreck, J.; Anagnostidis, K. Freshwater Flora of Central Europe Vol. 19/1 Cyanoprokaryota, 1st Part: Chroococcales; Gustav Fisher: Jena, Germany, 1999. [Google Scholar]
- Carmichael, W.W. Toxic Microcystis and the environment. In Toxic Microcystis; Watanabe, M., Harada, K., Carmichael, W., Fujiki, H., Eds.; CRC Press: Boca Raton, FL, USA, 1995; pp. 1–12. [Google Scholar]
- Preece, E.P.; Hardy, F.J.; Moore, B.C.; Bryan, M. A review of microcystin detections in Estuarine and Marine waters: Environmental implications and human health risk. Harmful Algae 2017, 61, 31–45. [Google Scholar] [CrossRef]
- Humpage, A.; Fastner, J. Cyanobacterial toxins: Hepatotoxic cyclic peptides—Microcystins and nodularins. In Toxic Cyanobacteria in Water, 2nd ed.; Chorus, I., Welker, M., Eds.; CRC Press: Boca Raton, FL, USA; on behalf of the World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Mallia, V.; Uhlig, S.; Rafuse, C.; Meija, J.; Miles, C.O. Novel Microcystins from Planktothrix prolifica NIVA–CYA 544 Identified by LC–MS/MS, Functional Group Derivatization and 15N–labeling. Mar. Drugs 2019, 17, 643. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Sinha, R.P.; Incharoensakdi, A. The cyanotoxin–microcystins: Current overview. Rev. Environ. Sci. Biotechnol. 2014, 13, 215–249. [Google Scholar] [CrossRef]
- World Health Organization. Guidelines for Drinking–Water Quality: Fourth Edition Incorporating the First and Second Addenda; WHO: Geneva, Switzerland, 2022.
- Kouzminov, A.; Ruck, J.; Wood, S.A. New Zealand risk management approach for toxic cyanobacteria in drinking water. Aust. N. Z. J. Public Health 2007, 31, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Izydorczyk, K.; Carpentier, C.; Mrówczyński, J.; Wagenvoort, A.; Jurczak, T.; Tarczyńska, M. Establishment of an Alert Level Framework for cyanobacteria in drinking water resources by using the Algae Online Analyser for monitoring cyanobacterial chlorophyll a. Water Res. 2009, 43, 989–996. [Google Scholar] [CrossRef]
- Ibelings, B.W.; Backer, L.C.; Kardinaal, W.E.A.; Chorus, I. Current approaches to cyanotoxin risk assessment and risk management around the globe. Harmful Algae 2014, 40, 63–74. [Google Scholar] [CrossRef]
- Park, H.-K.; Kim, H.; Lee, J.J.; Lee, J.A.; Lee, H.; Park, J.-H.; Seo, J.; Youn, S.-J.; Moon, J. Investigation of criterion on harmful algae alert system using correlation between cell numbers and cellular microcystins content of Korean toxic cyanobacteria. J. Kor. Soc. Water Env. 2011, 27, 491–498. [Google Scholar]
- Schürmann, Q.J.F.; Visser, P.M.; Sollie, S.; Kardinaal, W.E.A.; Faassen, E.J.; Lokmani, R.; van der Oost, R.; van de Waal, D.B. Risk assessment of toxic cyanobacterial blooms in recreational waters: A comparative study of monitoring methods. Harmful Algae 2024, 138, 102683. [Google Scholar] [CrossRef]
- Preece, E.P.; Otten, T.G.; Cooke, J. Use of multiple sampling techniques for cyanobacteria and cyanotoxin monitoring in the Sacramento–San Joaquin Delta under different hydrologic regimes. Mar. Pollut. Bull. 2024, 205, 116585. [Google Scholar] [CrossRef]
- Fickas, K.C.; O’Shea, R.E.; Pahlevan, N.; Smith, B.; Bartlett, S.L.; Wolny, J.L. Leveraging multimission satellite data for spatiotemporally coherent cyanoHAB monitoring. Front. Remote Sens. 2023, 4, 1157609. [Google Scholar] [CrossRef]
- Mishra, S.; Stumpf, R.P.; Meredith, A. Constructing a Consistent and Continuous Cyanobacteria Bloom Monitoring Product from Multi–Mission Ocean Color Instruments. Remote Sens. 2023, 15, 5291. [Google Scholar] [CrossRef]
- Gaget, V.; Lau, M.; Scendall, B.; Froscio, S.; Humpage, A.R. Cyanotoxins: Which detection technique for an optimum risk assessment? Water Res. 2017, 118, 227–238. [Google Scholar] [CrossRef]
- Huisman, J.; Codd, G.A.; Paerl, H.W.; Ibelings, B.W.; Verspagen, J.M.H.; Visser, P.M. Cyanobacterial blooms. Nat. Rev. Micrbiol 2018, 16, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Park, R.; Yu, M.; Byeon, M.; Kang, T. qPCR–Based Monitoring of 2–Methylisoborneol/Geosmin–Producing Cyanobacteria in Drinking Water Reservoirs in South Korea. Microorganisms 2023, 11, 2332. [Google Scholar] [CrossRef] [PubMed]
- Te, S.H.; Chen, E.Y.; Gin, K.Y. Comparison of Quantitative PCR and Droplet Digital PCR Multiplex Assays for Two Genera of Bloom–Forming Cyanobacteria, Cylindrospermopsis and Microcystis. Appl. Environ. Microbiol. 2015, 81, 5203–5211. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Park, H.-K.; Kim, I.-S. Assessment of the Appearance and Toxin Production Potential of Invasive Nostocalean Cyanobacteria Using Quantitative Gene Analysis in Nakdong River, Korea. Toxins 2022, 14, 294. [Google Scholar] [CrossRef] [PubMed]
- Panksep, K.; Tamm, M.; Mantzouki, E.; Rantala–Ylinen, A.; Laugaste, R.; Sivonen, K.; Tammeorg, O.; Kisand, V. Using Microcystin Gene Copies to Determine Potentially–Toxic Blooms, Example from Shallow Eutrophic Lake Peipsi. Toxins 2020, 12, 211. [Google Scholar] [CrossRef]
- Padovan, A.; Kennedy, K.; Gibb, K. A Microcystin synthesis mcyE/ndaF gene assay enables early detection of microcystin production in a tropical wastewater pond. Harmful Algae 2023, 127, 102476. [Google Scholar] [CrossRef]
- Wang, Z.; Song, G.; Shao, J.; Tan, W.; Li, Y.; Li, L. Establishment and field applications of real-time PCR methods for the quantification of potential MIB-producing cyanobacteria in aquatic systems. J. Appl. Phycol. 2016, 28, 325–333. [Google Scholar] [CrossRef]
- Kurmayer, R.; Kutzenberger, T. Application of Real-Time PCR for Quantification of Microcystin Genotypes in a Population of the Toxic Cyanobacterium Microcystis sp. Appl. Env. Mocrobiol 2003, 69, 6723–6730. [Google Scholar] [CrossRef]
- Te, S.H.; Gin, K.Y. The dynamics of cyanobacteria and microcystin production in a tropical reservoir of Singapore. Harmful Algae 2011, 10, 319–329. [Google Scholar] [CrossRef]
- Kataoka, T.; Homma, T.; Nakano, S.; Hodoki, Y.; Ohbayashi, K.; Kondo, R. PCR primers for selective detection of intra–Species variations in the bloom–forming cyanobacterium, Microcystis. Harmful Algae 2013, 23, 46–54. [Google Scholar] [CrossRef]
- Oh, K.-H.; Jeong, D.-H.; Shin, S.-H.; Cho, Y.-C. Simultaneous Quantification of Cyanobacteria and Microcystis spp. Using Real–Time PCR. J. Microbiol. Biotechnol. 2012, 22, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Strain, M.C.; Lada, S.M.; Luong, T.; Rought, S.E.; Gianella, S.; Terry, V.H.; Spina, C.A.; Woelk, C.H.; Richman, D.D. Highly precise measurement of HIV DNA by Droplet Digital PCR. PLoS ONE 2013, 8, e55943. [Google Scholar] [CrossRef] [PubMed]
- Vynck, M.; Vandesompeleb, J.; Nijs, N.; Mentenb, B.; Ganck, A.D.; Thasa, O. Flexible analysis of digital PCR experiments using generalized linear mixed models. Biomol. Detect. Quantif. 2016, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Fan, D.; Wang, N.; Wang, F.; Wang, B.; Zhu, L.; Guo, Y. Applications of digital PCR in COVID-19 pandemic. View 2021, 2, 20200082. [Google Scholar] [CrossRef]
- Dominguez, S.L.; Suárez, C.A.; Ledesma, S.M.; Bellido, M.J.L. Present and future applications of digital PCR in infectious diseases diagnosis. Diagnostics 2024, 14, 931. [Google Scholar] [CrossRef]
- Murthy, S.; Suresh, A.; Dandasena, D.; Singh, S.; Subudhi, M.; Bhandari, V.; Bhanot, V.; Arora, J.S.; Sharma, P. Multiplex ddPCR: A promising diagnostic assay for early detection and drug monitoring in bovine theileriosis. Pathogens 2023, 12, 296. [Google Scholar] [CrossRef]
- Hayden, R.T.; Ingersoll, J.; Abdul–Ali, D.; Shi, L.; Pounds, S.; Caliendo, A.M. Comparison of droplet digital PCR to real–time PCR for quantitative detection of cytomegalovirus. J. Clin. Microbiol. 2013, 51, 540–546. [Google Scholar] [CrossRef]
- Dong, L.; Yang, S.; Chen, J.; Wu, X.; Sun, D.; Song, C.; Li, L. Structural basis of secA–mediated protein translocation. Proc. Natl. Acad. Sci. USA 2023, 120, e2208070120. [Google Scholar] [CrossRef]
- Rinta–kanto, J.M.; Ouellette, A.J.A.; Boyer, G.L.; Twiss, M.R.; Bridgeman, T.B.; Wilhelm, S.W. Quantification of toxic Microcystis spp. during the 2003 and 2004 Blooms in Western Lake Erie using quantitative real–time PCR. Environ. Sci. Technol. 2005, 39, 4198–4205. [Google Scholar]
- Hisbergues, M.; Christiansen, G.; Rouhiainen, L.; Sivonen, K.; Börner, T. PCR–Based Identification of microcystin–producing genotypes of cifferent cyanobacterial genera. Arch. Microbiol. 2003, 180, 402–410. [Google Scholar] [CrossRef]
- Saker, M.L.; Welker, M.; Vasconcelos, V.M. Multiplex PCR for the detection of toxigenic cyanobacteria in dietary supplements produced for human consumption. Appl. Microbiol. Biotechnol. 2007, 73, 1136–1142. [Google Scholar] [CrossRef] [PubMed]
- Sneideris, D.; Ivanauskas, A.; Zizyte, M.; Valiunas, D. secA Gene suitability for fast and easy identification of phytoplasmas by RFLP analysis. Eur. J. Plant Pathol. 2021, 160, 737–743. [Google Scholar] [CrossRef]
- Kumar, S.; Jadon, V.S.; Rao, G.P. Use of secA gene for characterization of Phytoplasmas associated with sugarcane grassy shoot disease in India. Sugar Tech 2017, 19, 632–637. [Google Scholar] [CrossRef]
- Bloch, M.; Helmick, E.E.; Bahder, B.W. Differentiation of palm–infecting Phytoplasmas in the caribbean basin using high resolution melt curve analysis of the secA gene. Plant Dis. 2022, 106, 2480–2489. [Google Scholar] [CrossRef]
- Zhang, W.; Lou, I.; Ung, W.K.; Kong, Y.; Mok, K.M. Application of PCR and real–time PCR for monitoring cyanobacteria, Microcystis spp. and Cylindrospermopsis raciborskii in Macau freshwater reservoir. Front. Earth Sci. 2014, 8, 291–301. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Y.; Guo, L.; Wu, Y.; Wang, Y.; Xu, L.; Xu, M.; Huang, S.; Chen, P.; Wang, T.; et al. Multiplex asymmetric PCR by combining the amplification refractory mutation system with the homo–tag–assisted nondimer system. Anal. Chem. 2024, 96, 9200–9208. [Google Scholar] [CrossRef]
- Wang, D.; Wang, S.; Du, X.; He, Q.; Liu, Y.; Wang, Z.; Feng, K.; Li, Y.; Deng, Y. ddPCR surpasses classical qPCR technology in quantitating bacteria and fungi in the environment. Mol. Ecol. Resour. 2022, 22, 2587–2598. [Google Scholar] [CrossRef]
- Cox, C.R.; Weghorn, K.N.; Ruger, K.; Powers–Fletcher, M.V.; Powell, E.A.; Mortensen, J.E. Clinical utility of multiplex PCR in the detection of pathogens from sterile body fluids. J. Clin. Microbiol. 2024, 62, e0161123. [Google Scholar] [CrossRef]
- Mori, C.; Matsumura, S. Development and validation of simultaneous identification of 26 mammalian and poultry species by a multiplex assay. Int. J. Leg. Med. 2022, 136, 1–12. [Google Scholar] [CrossRef]
- Yuan, J.; Yi, J.; Zhan, M.; Xie, Q.; Zhen, T.T.; Zhou, J.; Li, Z.; Li, Z. The web–based multiplex PCR primer design software ultiplex and the associated experimental workflow: Up to 100–Plex multiplicity. BMC Genom. 2021, 22, 835. [Google Scholar] [CrossRef]
- Kim, H.-R.; Kim, C.-K.; Ahn, T.-S.; Yoo, S.-A.; Lee, D.-H. Effects of temperature and light on microcystin synthetase gene transcription in Microcystis aeruginosa. Key Eng. Mater. 2005, 277–279, 606–611. [Google Scholar] [CrossRef]
- Riaz, S.; Xiao, M.; Chen, P.; Li, M.; Cui, Y.; Daroch, M. The genome copy number of the thermophilic cyanobacterium Thermosynechococcus elongatus E542 is controlled by growth phase and nutrient availability. Appl. Environ. Microbiol. 2021, 87, e02993-20. [Google Scholar] [CrossRef] [PubMed]
- Riaz, S.; Jiang, Y.; Xiao, M.; You, D.; Klepacz–Smółka, A.; Rasul, F.; Daroch, M. Generation of miniploid cells and improved natural transformation procedure for a model cyanobacterium Synechococcus elongatus PCC7942. Front. Microbiol. 2022, 13, 959043. [Google Scholar] [CrossRef] [PubMed]
- Schirrmeister, B.E.; Dalquen, D.A.; Anisimova, M.; Bagheri, H.C. Gene copy number variation and its significance in cyanobacterial phylogeny. BMC Microbiol. 2012, 12, 177. [Google Scholar] [CrossRef]
- Miller, S.R.; Wood, A.M.; Blankenship, R.E.; Kim, M.; Ferriera, S. Dynamics of gene duplication in the genomes of chlorophyll d–producing cyanobacteria: Implications for the ecological niche. Genome Biol. Evol. 2011, 3, 601–613. [Google Scholar] [CrossRef]
- Ohbayashi, R.; Nakamachi, A.; Hatakeyama, T.S.; Watanabe, S.; Kanesaki, Y.; Chibazakura, T.; Yoshikawa, H.; Miyagishima, S.-Y. Coordination of polyploid chromosome replication with cell size and growth in a cyanobacterium. mBio 2019, 10, e00510-19. [Google Scholar] [CrossRef]
- Griese, M.; Lange, C.; Soppa, J. Ploidy in cyanobacteria. FEMS Microbiol. Lett. 2011, 323, 124–131. [Google Scholar] [CrossRef]
- Davis, T.W.; Berry, D.L.; Boyer, G.L.; Gobler, C.J. The effects of temperature and nutrients on the growth and dynamics of toxic and non–toxic strains of Microcystis during cyanobacterial blooms. Harmful Algae 2009, 8, 715–725. [Google Scholar] [CrossRef]
- Gaget, V.; Humpage, A.R.; Huang, Q.; Monis, P.; Burch, M.D. Water treatment of cyanobacterial blooms: Understanding cyanobacteria cell behaviour and its impact on drinking water treatment processes. Water Res. 2017, 113, 61–77. [Google Scholar]
- Tanabe, Y.; Kaya, K.; Watanabe, M.M. Evidence for reclassification of toxic and non–toxic Microcystis strains on the basis of genetic analysis. FEMS Microbiol. Lett. 2007, 264, 15–19. [Google Scholar]
- Puddick, J.; Prinsep, M.R.; Wood, S.A.; Kaufononga, S.A.; Cary, S.C.; Hamilton, D.P. Further characterization of glycosylated microcystins from Microcystis spp. using LC–MS/MS and NMR analysis. Toxins 2014, 6, 4435–4451. [Google Scholar]
- Shirai, M.; Matumaru, K.; Ohotake, A.; Takamura, Y.; Aida, T.; Nakano, M. Development of a solid medium for growth and isolation of axenic Microcystis strains (Cyanobacteria). Appl. Environ. Microbiol. 1989, 55, 2569–2571. [Google Scholar] [CrossRef] [PubMed]
- John, D.M.; Whitton, B.A.; Brook, A.J. The Freshwater Algal Flora of the British Isles. In An Identification Guide to Freshwater and Terrestrial Algae, 2nd ed.; Cambridge University Press: Cambridge, UK, 2011. [Google Scholar]
- Lee, H.-J.; Park, H.-K.; Heo, J.; Lee, H.-J.; Hong, D.-G. Colonial cyanobacteria, Microcystis cell density variations using ultrasonic treatment. J. Korean Soc. Water Environ. 2018, 34, 210–215. [Google Scholar]
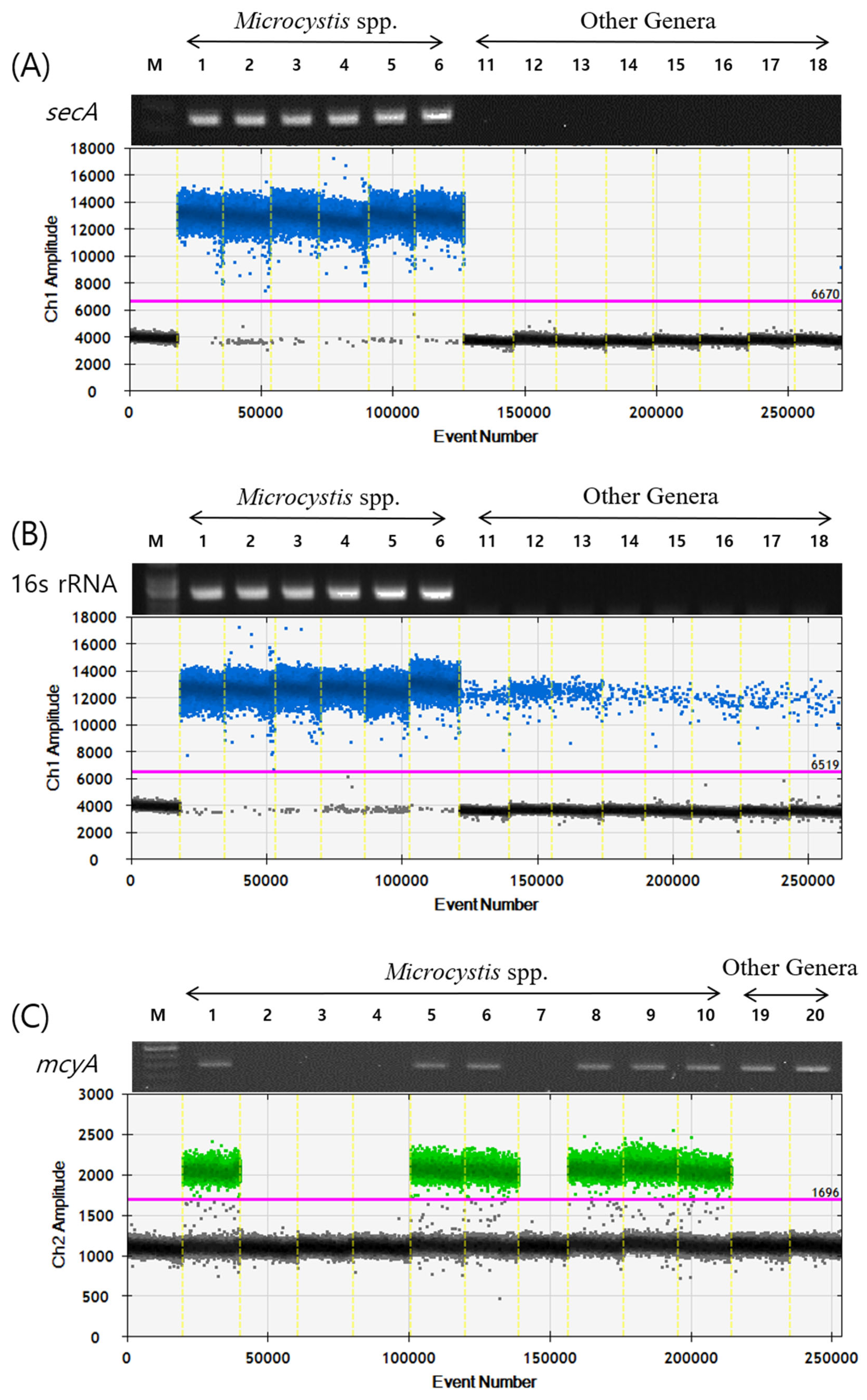
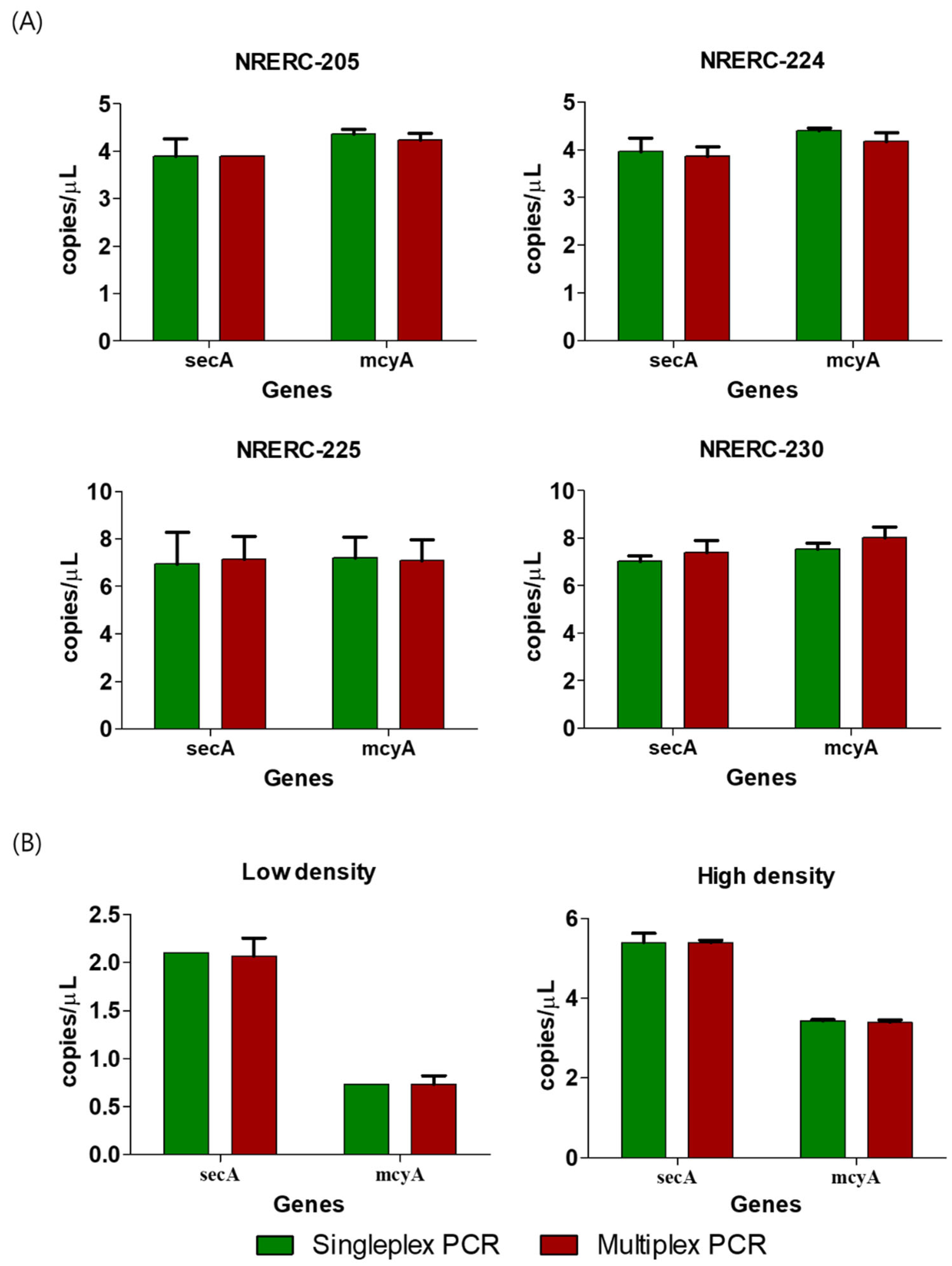
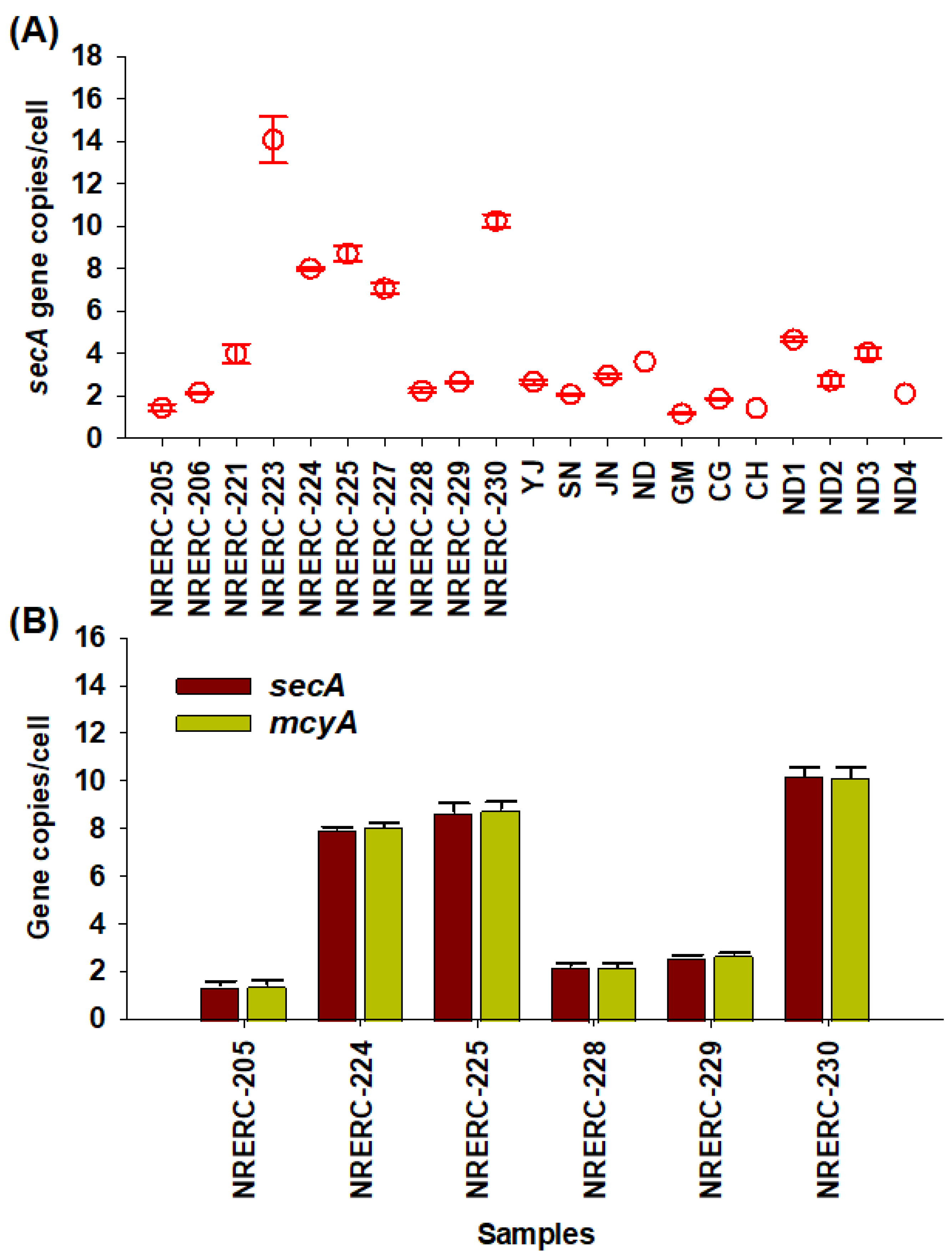
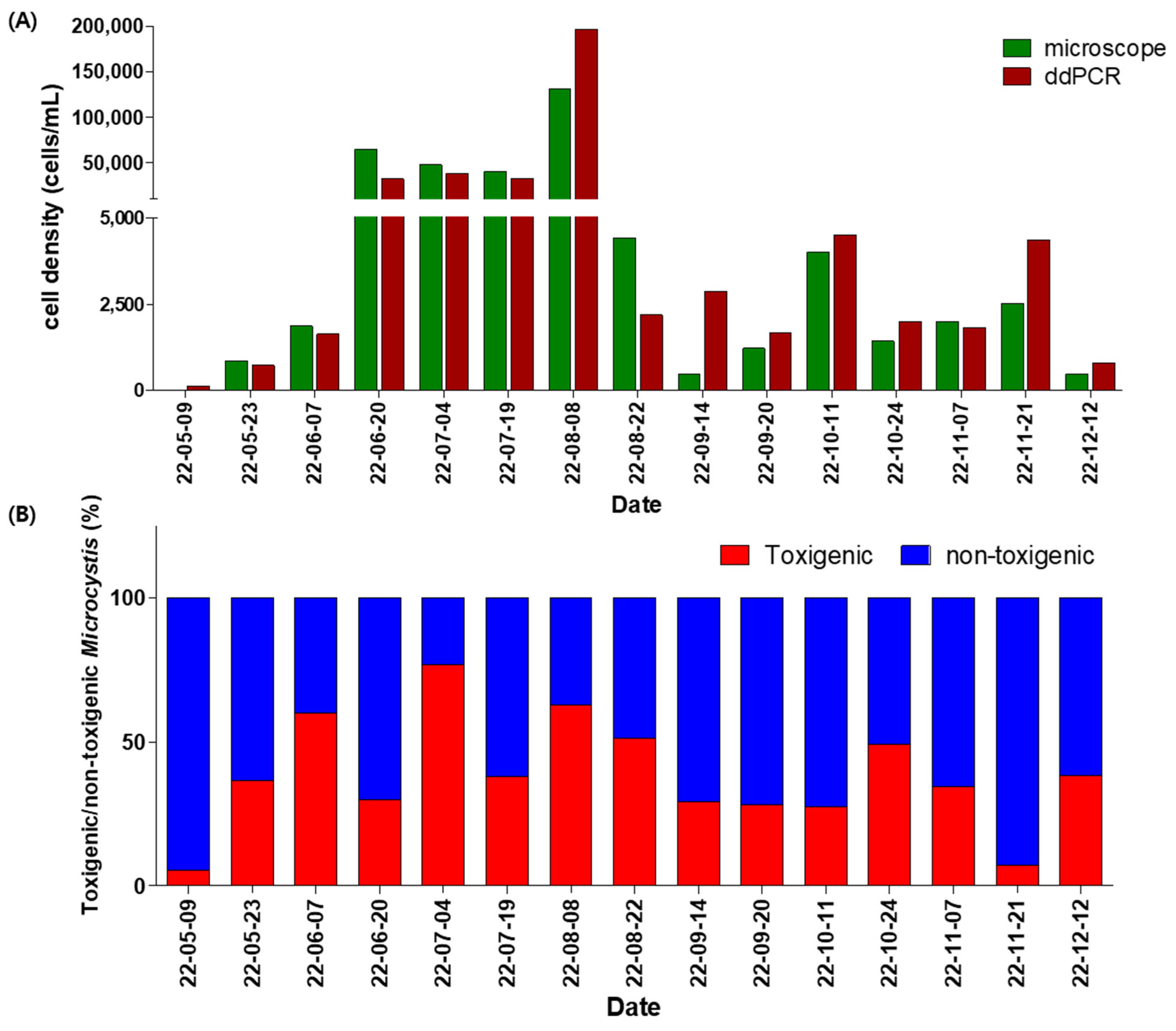

| Platform | Target Gene | Name | Sequence(5′ -> 3′) | Reference |
|---|---|---|---|---|
| ddPCR | secA | secA-F | GTGTGGGACTAATTCAAGCC | This study |
| secA-R | GGCCTCATCGATAAGAATGG | |||
| secA-probe | TACACCACTAACAGCGAACTCGGCT | |||
| mcyA | mcyA-F | GGTTAGAAGCAGCCGATGG | This study | |
| mcyA-R | GCTCCAAGAACCTCCAGATAAC | |||
| mcyA-probe | TTTTGAATACTTTGCCCCTACGTTTAGA | |||
| 16S rRNA | MICR 184F | GCCGCRAGGTGAAAMCTAA | [56] | |
| MICR 431R | AATCCAAARACCTTCCTCCC | |||
| MICR 228F | AAGAGCTTGCGTCTGATTAGCTAGT | |||
| Conventional PCR | mcyA | mcyA-Cd1F | AAAATTAAAAGCCGTATCAAA | [57] |
| mcyA-Cd1R | AAAAGTGTTTTATTAGCGGCTCAT |
| Survey Date | Dominant Species (Relative Abundance %) | Sub-Dominant Species (Relative Abundance %) |
|---|---|---|
| 2022.5.23 | Microcystis sp. (100) | |
| 2022.6.7 | M. aeruginosa (46) | M. viridis (38) |
| 2022.6.20 | M. aeruginosa (41) | M. novacekii (26) |
| 2022.7.4 | M. aeruginosa (46) | M. viridis (33) |
| 2022.7.19 | M. wesenbergii (65) | M. aeruginosa (23) |
| 2022.8.8 | M. aeruginosa (61) | M. novacekii (12) |
| 2022.8.22 | M. aeruginosa (70) | Microcystis sp. (20) |
| 2022.9.14 | Microcystis sp. (100) | |
| 2022.9.20 | M. aeruginosa (56) | M. ichthyoblabe (44) |
| 2022.10.11 | M. aeruginosa (46) | M. wesenbergii (31) |
| 2022.10.24 | M. aeruginosa (83) | Microcystis sp. (17) |
| 2022.11.7 | M. wesenbergii (50) | Microcystis sp. (50) |
| 2022.11.21 | Microcystis sp. (100) | |
| 2022.12.12 | Microcystis sp. (100) |
| No. | Strain No. | Scientific Name | Isolation Site | secA Gene | mcyA Gene | MCY |
|---|---|---|---|---|---|---|
| 1 | NRERC-205 | Microcystis flos-aquae | Nakdong River | + | + | + |
| 2 | NRERC-206 | Microcystis ichthyoblabe | Nakdong River | + | − | − |
| 3 | NRERC-221 | Microcystis aeruginosa | Youngju Lake | + | − | − |
| 4 | NRERC-223 | Microcystis wesenbergii | Youngju Lake | + | − | − |
| 5 | NRERC-224 | Microcystis viridis | Nakdong River | + | + | + |
| 6 | NRERC-225 | Microcystis novacekii | Banbyeon River | + | + | + |
| 7 | NRERC-227 | Microcystis viridis | Youngsan River | + | − | − |
| 8 | NRERC-228 | Microcystis flos-aquae | Youngsan River | + | + | + |
| 9 | NRERC-229 | Microcystis flos-aquae | Youngsan River | + | + | + |
| 10 | NRERC-230 | Microcystis aeruginosa | Youngsan River | + | + | + |
| 11 | NRERC-008 | Aphanizomenon flos-aquae | Nakdong River | − | − | − |
| 12 | NRERC-101 | Dolichospermum planctonicum | Nakdong River | − | − | − |
| 13 | NRERC-450 | Geitlerinema amphibium | Nakdong River | − | − | − |
| 14 | NRERC-501 | Cylindrospermopsis raciborskii | Nakdong River | − | − | − |
| 15 | NRERC-601 | Sphaerospermopsis aphanizomenoides | Nakdong River | − | − | − |
| 16 | NRERC-650 | Cuspidothrix issatchenkoi | Nakdong River | − | − | − |
| 17 | AG10016 | Aphanocapsa sp. | − | − | − | |
| 18 | AG20470 | Synechococcus sp. | − | − | − | |
| 19 | NIVA-CYA 656 | Dolichospermum flos-aquae | − | + | + | |
| 20 | NIVA-CYA 855 | Planktothrix agardhii | − | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, I.-S.; Park, H.-K. Molecular Quantification of Total and Toxigenic Microcystis Using Digital-Droplet-Polymerase-Chain-Reaction-Based Multiplex Assay. Toxins 2025, 17, 242. https://doi.org/10.3390/toxins17050242
Kim I-S, Park H-K. Molecular Quantification of Total and Toxigenic Microcystis Using Digital-Droplet-Polymerase-Chain-Reaction-Based Multiplex Assay. Toxins. 2025; 17(5):242. https://doi.org/10.3390/toxins17050242
Chicago/Turabian StyleKim, In-Su, and Hae-Kyung Park. 2025. "Molecular Quantification of Total and Toxigenic Microcystis Using Digital-Droplet-Polymerase-Chain-Reaction-Based Multiplex Assay" Toxins 17, no. 5: 242. https://doi.org/10.3390/toxins17050242
APA StyleKim, I.-S., & Park, H.-K. (2025). Molecular Quantification of Total and Toxigenic Microcystis Using Digital-Droplet-Polymerase-Chain-Reaction-Based Multiplex Assay. Toxins, 17(5), 242. https://doi.org/10.3390/toxins17050242





