Histone Methyltransferases AcDot1 and AcRmtA Are Involved in Growth Regulation, Secondary Metabolism, and Stress Response in Aspergillus carbonarius
Abstract
:1. Introduction
2. Results
2.1. Phylogenetic Analysis and Generation of A. carbonarius Mutants
2.2. Characterization of Mutants
2.3. Gene Expression Analysis
2.4. Response to Osmotic and Oxidtive Stresses
2.5. In Vivo Evaluations
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Strains and Growing Conditions
5.2. Gene Analysis and Phylogenetic Studies
5.3. Obtaining A. carbonarius Mutants
5.4. Phenotypic Characterization and OTA Extraction
5.5. Gene Expression Study
5.6. Response to Osmotic and Oxidative Stress
5.7. In Vivo Assay
5.8. OTA Quantification
5.9. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Houbraken, J.; Kocsubé, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.-C.; Meijer, M.; Kraak, B.; Hubka, V.; Samson, R.A.; Frisvad, J.C. Classification of Aspergillus, Penicillium, Talaromyces and Related Genera (Eurotiales): An Overview of Families, Genera, Subgenera, Sections, Series and Species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef] [PubMed]
- Zimmerli, B.; Dick, R. Ochratoxin A in Table Wine and Grape-juice: Occurrence and Risk Assesment. Food Addit. Contam. 1996, 13, 655–668. [Google Scholar] [CrossRef]
- Markaki, P.; Delpont-Binet, C.; Grosso, F.; Dragacci, S. Determination of Ochratoxin A in Red Wine and Vinegar by Immunoaffinity High-Pressure Liquid Chromatography. J. Food Prot. 2001, 64, 533–537. [Google Scholar] [CrossRef]
- Abarca, M.L.; Accensi, F.; Bragulat, M.R.; Castellá, G.; Cabañes, F.J. Aspergillus carbonarius as the Main Source of Ochratoxin A Contamination in Dried Vine Fruits from the Spanish Market. J. Food Prot. 2003, 66, 504–506. [Google Scholar] [CrossRef]
- Solfrizzo, M.; Gambacorta, L.; Visconti, A. Assessment of Multi-Mycotoxin Exposure in Southern Italy by Urinary Multi-Biomarker Determination. Toxins 2014, 6, 523–538. [Google Scholar] [CrossRef]
- Bellí, N.; Marín, S.; Duaigües, A.; Ramos, A.J.; Sanchis, V. Ochratoxin A in Wines, Musts and Grape Juices from Spain. J. Sci. Food Agric. 2004, 84, 591–594. [Google Scholar] [CrossRef]
- Cabañes, F.J.; Accensi, F.; Bragulat, M.R.; Abarca, M.L.; Castellá, G.; Minguez, S.; Pons, A. What Is the Source of Ochratoxin A in Wine? Int. J. Food Microbiol. 2002, 79, 213–215. [Google Scholar] [CrossRef]
- Paterson, R.R.M.; Venâncio, A.; Lima, N.; Guilloux-Bénatier, M.; Rousseaux, S. Predominant Mycotoxins, Mycotoxigenic Fungi and Climate Change Related to Wine. Food Res. Int. 2018, 103, 478–491. [Google Scholar] [CrossRef] [PubMed]
- Lasram, S.; Mani, A.; Zaied, C.; Chebil, S.; Abid, S.; Bacha, H.; Mliki, A.; Ghorbel, A. Evolution of Ochratoxin A Content during Red and Rose Vinification. J. Sci. Food Agric. 2008, 88, 1696–1703. [Google Scholar] [CrossRef]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins; IARC monographs on the evaluation of carcinogenic risks to humans; World Health Organization, International Agency for Research on Cancer: Lyon, France, 1993; ISBN 978-92-832-1256-0. [Google Scholar]
- Wang, Y.; Wang, L.; Wu, F.; Liu, F.; Wang, Q.; Zhang, X.; Selvaraj, J.N.; Zhao, Y.; Xing, F.; Yin, W.-B.; et al. A Consensus Ochratoxin A Biosynthetic Pathway: Insights from the Genome Sequence of Aspergillus ochraceus and a Comparative Genomic Analysis. Appl. Environ. Microbiol. 2018, 84, e01009-18. [Google Scholar] [CrossRef]
- Ferrara, M.; Perrone, G.; Gambacorta, L.; Epifani, F.; Solfrizzo, M.; Gallo, A. Identification of a Halogenase Involved in the Biosynthesis of Ochratoxin A in Aspergillus carbonarius. Appl. Environ. Microbiol. 2016, 82, 5631–5641. [Google Scholar] [CrossRef]
- Gallo, A.; Bruno, K.S.; Solfrizzo, M.; Perrone, G.; Mulè, G.; Visconti, A.; Baker, S.E. New Insight into the Ochratoxin A Biosynthetic Pathway through Deletion of a Nonribosomal Peptide Synthetase Gene in Aspergillus carbonarius. Appl. Environ. Microbiol. 2012, 78, 8208–8218. [Google Scholar] [CrossRef]
- Gallo, A.; Knox, B.P.; Bruno, K.S.; Solfrizzo, M.; Baker, S.E.; Perrone, G. Identification and Characterization of the Polyketide Synthase Involved in Ochratoxin A Biosynthesis in Aspergillus Carbonarius. Int. J. Food Microbiol. 2014, 179, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Gerin, D.; Garrapa, F.; Ballester, A.-R.; González-Candelas, L.; De Miccolis Angelini, R.M.; Faretra, F.; Pollastro, S. Functional Role of Aspergillus carbonarius AcOTAbZIP Gene, a bZIP Transcription Factor within the OTA Gene Cluster. Toxins 2021, 13, 111. [Google Scholar] [CrossRef] [PubMed]
- Barda, O.; Maor, U.; Sadhasivam, S.; Bi, Y.; Zakin, V.; Prusky, D.; Sionov, E. The pH-Responsive Transcription Factor PacC Governs Pathogenicity and Ochratoxin A Biosynthesis in Aspergillus carbonarius. Front. Microbiol. 2020, 11, 210. [Google Scholar] [CrossRef]
- Cervini, C.; Gallo, A.; Piemontese, L.; Magistà, D.; Logrieco, A.F.; Ferrara, M.; Solfrizzo, M.; Perrone, G. Effects of Temperature and Water Activity Change on Ecophysiology of Ochratoxigenic Aspergillus carbonarius in Field-Simulating Conditions. Int. J. Food Microbiol. 2020, 315, 108420. [Google Scholar] [CrossRef]
- Cheong, K.K.; Strub, C.; Montet, D.; Durand, N.; Alter, P.; Meile, J.-C.; Schorr Galindo, S.; Fontana, A. Effect of Different Light Wavelengths on the Growth and Ochratoxin A Production in Aspergillus carbonarius and Aspergillus westerdijkiae. Fungal Biol. 2016, 120, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Crespo-Sempere, A.; Marín, S.; Sanchis, V.; Ramos, A.J. VeA and LaeA Transcriptional Factors Regulate Ochratoxin A Biosynthesis in Aspergillus carbonarius. Int. J. Food Microbiol. 2013, 166, 479–486. [Google Scholar] [CrossRef]
- Yang, K.; Tian, J.; Keller, N.P. Post-Translational Modifications Drive Secondary Metabolite Biosynthesis in Aspergillus: A Review. Environ. Microbiol. 2022, 24, 2857–2881. [Google Scholar] [CrossRef]
- Cichewicz, R.H. Epigenome Manipulation as a Pathway to New Natural Product Scaffolds and Their Congeners. Nat. Prod. Rep. 2010, 27, 11–22. [Google Scholar] [CrossRef]
- Jaenisch, R.; Bird, A. Epigenetic Regulation of Gene Expression: How the Genome Integrates Intrinsic and Environmental Signals. Nat. Genet. 2003, 33, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Tsankova, N.; Renthal, W.; Kumar, A.; Nestler, E.J. Epigenetic Regulation in Psychiatric Disorders. Focus 2010, 8, 435–448. [Google Scholar] [CrossRef]
- Aldholmi, M.; Wilkinson, B.; Ganesan, A. Epigenetic Modulation of Secondary Metabolite Profiles in Aspergillus calidoustus and Aspergillus westerdijkiae through Histone Deacetylase (HDAC) Inhibition by Vorinostat. J. Antibiot. 2020, 73, 410–413. [Google Scholar] [CrossRef] [PubMed]
- Bok, J.W.; Soukup, A.A.; Chadwick, E.; Chiang, Y.; Wang, C.C.C.; Keller, N.P. VeA and MvlA Repression of the Cryptic Orsellinic Acid Gene Cluster in Aspergillus nidulans Involves Histone 3 Acetylation. Mol. Microbiol. 2013, 89, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Ciesielska, A.; Kowalczyk, A.; Paneth, A.; Stączek, P. Evaluation of the Antidermatophytic Activity of Potassium Salts of N-Acylhydrazinecarbodithioates and Their Aminotriazole-Thione Derivatives. Sci. Rep. 2024, 14, 3521. [Google Scholar] [CrossRef]
- Nützmann, H.-W.; Fischer, J.; Scherlach, K.; Hertweck, C.; Brakhage, A.A. Distinct Amino Acids of Histone H3 Control Secondary Metabolism in Aspergillus nidulans. Appl. Environ. Microbiol. 2013, 79, 6102–6109. [Google Scholar] [CrossRef]
- Yang, K.; Liang, L.; Ran, F.; Liu, Y.; Li, Z.; Lan, H.; Gao, P.; Zhuang, Z.; Zhang, F.; Nie, X.; et al. The DmtA Methyltransferase Contributes to Aspergillus flavus Conidiation, Sclerotial Production, Aflatoxin Biosynthesis and Virulence. Sci. Rep. 2016, 6, 23259. [Google Scholar] [CrossRef]
- Zhuang, Z.; Pan, X.; Zhang, M.; Liu, Y.; Huang, C.; Li, Y.; Hao, L.; Wang, S. Set2 Family Regulates Mycotoxin Metabolism and Virulence via H3K36 Methylation in Pathogenic Fungus Aspergillus flavus. Virulence 2022, 13, 1358–1378. [Google Scholar] [CrossRef]
- Zhang, X.; Noberini, R.; Bonaldi, T.; Collemare, J.; Seidl, M.F. The Histone Code of the Fungal Genus Aspergillus Uncovered by Evolutionary and Proteomic Analyses. Microb. Genom. 2022, 8, 000856. [Google Scholar] [CrossRef]
- Bauer, I.; Graessle, S.; Loidl, P.; Hohenstein, K.; Brosch, G. Novel Insights into the Functional Role of Three Protein Arginine Methyltransferases in Aspergillus nidulans. Fungal Genet. Biol. 2010, 47, 551–561. [Google Scholar] [CrossRef]
- Li, Y.; He, Y.; Li, X.; Fasoyin, O.E.; Hu, Y.; Liu, Y.; Yuan, J.; Zhuang, Z.; Wang, S. Histone Methyltransferase aflrmtA Gene Is Involved in the Morphogenesis, Mycotoxin Biosynthesis, and Pathogenicity of Aspergillus flavus. Toxicon 2017, 127, 112–121. [Google Scholar] [CrossRef]
- Satterlee, T.; Cary, J.W.; Calvo, A.M. RmtA, a Putative Arginine Methyltransferase, Regulates Secondary Metabolism and Development in Aspergillus flavus. PLoS ONE 2016, 11, e0155575. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.T.; Zhang, Y. The Diverse Functions of Dot1 and H3K79 Methylation. Genes Dev. 2011, 25, 1345–1358. [Google Scholar] [CrossRef]
- Liang, L.; Liu, Y.; Yang, K.; Lin, G.; Xu, Z.; Lan, H.; Wang, X.; Wang, S. The Putative Histone Methyltransferase DOT1 Regulates Aflatoxin and Pathogenicity Attributes in Aspergillus flavus. Toxins 2017, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F.; et al. MycoCosm Portal: Gearing up for 1000 Fungal Genomes. Nucleic Acids Res. 2014, 42, D699–D704. [Google Scholar] [CrossRef] [PubMed]
- Frandsen, R.J.; Andersson, J.A.; Kristensen, M.B.; Giese, H. Efficient Four Fragment Cloning for the Construction of Vectors for Targeted Gene Replacement in Filamentous Fungi. BMC Mol. Biol. 2008, 9, 70. [Google Scholar] [CrossRef]
- Sawan, C.; Herceg, Z. Histone Modifications and Cancer. In Advances in Genetics; Elsevier: Amsterdam, The Netherlands, 2010; Volume 70, pp. 57–85. ISBN 978-0-12-380866-0. [Google Scholar]
- Tessarz, P.; Kouzarides, T. Histone Core Modifications Regulating Nucleosome Structure and Dynamics. Nat. Rev. Mol. Cell Biol. 2014, 15, 703–708. [Google Scholar] [CrossRef]
- Dubey, A.; Jeon, J. Epigenetic Regulation of Development and Pathogenesis in Fungal Plant Pathogens. Mol. Plant Pathol. 2017, 18, 887–898. [Google Scholar] [CrossRef]
- Banerjee, T.; Chakravarti, D. A Peek into the Complex Realm of Histone Phosphorylation. Mol. Cell. Biol. 2011, 31, 4858–4873. [Google Scholar] [CrossRef]
- Kaimori, J.-Y.; Maehara, K.; Hayashi-Takanaka, Y.; Harada, A.; Fukuda, M.; Yamamoto, S.; Ichimaru, N.; Umehara, T.; Yokoyama, S.; Matsuda, R.; et al. Histone H4 Lysine 20 Acetylation Is Associated with Gene Repression in Human Cells. Sci. Rep. 2016, 6, 24318. [Google Scholar] [CrossRef]
- Khan, S.A. Global Histone Post-Translational Modifications and Cancer: Biomarkers for Diagnosis, Prognosis and Treatment? World J. Biol. Chem. 2015, 6, 333. [Google Scholar] [CrossRef]
- Arnaudo, A.M.; Garcia, B.A. Proteomic Characterization of Novel Histone Post-Translational Modifications. Epigenetics Chromatin 2013, 6, 24. [Google Scholar] [CrossRef]
- Duan, G.; Walther, D. The Roles of Post-Translational Modifications in the Context of Protein Interaction Networks. PLOS Comput. Biol. 2015, 11, e1004049. [Google Scholar] [CrossRef] [PubMed]
- Romanoski, C.E.; Glass, C.K.; Stunnenberg, H.G.; Wilson, L.; Almouzni, G. Roadmap for Regulation. Nature 2015, 518, 314–316. [Google Scholar] [CrossRef]
- Flanagan, J.F.; Mi, L.-Z.; Chruszcz, M.; Cymborowski, M.; Clines, K.L.; Kim, Y.; Minor, W.; Rastinejad, F.; Khorasanizadeh, S. Double Chromodomains Cooperate to Recognize the Methylated Histone H3 Tail. Nature 2005, 438, 1181–1185. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Krautkramer, K.A.; Feldman, J.L.; Denu, J.M. Metabolic Regulation of Histone Post-Translational Modifications. ACS Chem. Biol. 2015, 10, 95–108. [Google Scholar] [CrossRef]
- Jeon, J.; Choi, J.; Lee, G.-W.; Park, S.-Y.; Huh, A.; Dean, R.A.; Lee, Y.-H. Genome-Wide Profiling of DNA Methylation Provides Insights into Epigenetic Regulation of Fungal Development in a Plant Pathogenic Fungus, Magnaporthe oryzae. Sci. Rep. 2015, 5, 8567. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Nie, X.; Wang, H.; Gao, N.; Liu, H.; Chen, J. SUMOylation of Wor1 by a Novel SUMO E3 Ligase Controls Cell Fate in Andida albicans. Mol. Microbiol. 2015, 98, 69–89. [Google Scholar] [CrossRef]
- Zemach, A.; McDaniel, I.E.; Silva, P.; Zilberman, D. Genome-Wide Evolutionary Analysis of Eukaryotic DNA Methylation. Science 2010, 328, 916–919. [Google Scholar] [CrossRef]
- Hao, L.; Zhang, M.; Yang, C.; Pan, X.; Wu, D.; Lin, H.; Ma, D.; Yao, Y.; Fu, W.; Chang, J.; et al. The Epigenetic Regulator Set9 Harmonizes Fungal Development, Secondary Metabolism, and Colonization Capacity of Aspergillus flavus. Int. J. Food Microbiol. 2023, 403, 110298. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, M.; Xie, R.; Zhang, F.; Wang, S.; Pan, X.; Wang, S.; Zhuang, Z. The Methyltransferase AflSet1 Is Involved in Fungal Morphogenesis, AFB1 Biosynthesis, and Virulence of Aspergillus flavus. Front. Microbiol. 2020, 11, 234. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Dominguez, Y.; Bok, J.W.; Berger, H.; Shwab, E.K.; Basheer, A.; Gallmetzer, A.; Scazzocchio, C.; Keller, N.; Strauss, J. Heterochromatic Marks Are Associated with the Repression of Secondary Metabolism Clusters in Aspergillus Nidulans: Heterochromatin Regulation of Secondary Metabolism. Mol. Microbiol. 2010, 76, 1376–1386. [Google Scholar] [CrossRef]
- Bok, J.W.; Keller, N.P. LaeA, a Regulator of Secondary Metabolism in Aspergillus spp. Eukaryot. Cell 2004, 3, 527–535. [Google Scholar] [CrossRef]
- Gacek-Matthews, A.; Berger, H.; Sasaki, T.; Wittstein, K.; Gruber, C.; Lewis, Z.A.; Strauss, J. KdmB, a Jumonji Histone H3 Demethylase, Regulates Genome-Wide H3K4 Trimethylation and Is Required for Normal Induction of Secondary Metabolism in Aspergillus Nidulans. PLOS Genet. 2016, 12, e1006222. [Google Scholar] [CrossRef]
- Nie, X.; Li, B.; Wang, S. Chapter Five—Epigenetic and Posttranslational Modifications in Regulating the Biology of Aspergillus Species. In Advances in Applied Microbiology; Gadd, G.M., Sariaslani, S., Eds.; Academic Press: Cambridge, MA, USA, 2018; Volume 105, pp. 191–226. ISBN 0065-2164. [Google Scholar]
- Lee, S.; Oh, S.; Jeong, K.; Jo, H.; Choi, Y.; Seo, H.D.; Kim, M.; Choe, J.; Kwon, C.S.; Lee, D. Dot1 Regulates Nucleosome Dynamics by Its Inherent Histone Chaperone Activity in Yeast. Nat. Commun. 2018, 9, 240. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Huang, S.; Liu, J.; Gai, Y.; Li, M.; Duan, S.; Zhang, S.; Sun, X.; Yang, Q.; Wang, Y.; et al. Histone Methylation Is Required for Virulence, Conidiation, and Multi-Stress Resistance of Alternaria alternata. Front. Microbiol. 2022, 13, 924476. [Google Scholar] [CrossRef] [PubMed]
- Giannattasio, M.; Lazzaro, F.; Plevani, P.; Muzi-Falconi, M. The DNA Damage Checkpoint Response Requires Histone H2B Ubiquitination by Rad6-Bre1 and H3 Methylation by Dot1. J. Biol. Chem. 2005, 280, 9879–9886. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, N.; Yin, Y.; Chen, Y.; Jiang, J.; Ma, Z. Histone H3K4 Methylation Regulates Hyphal Growth, Secondary Metabolism and Multiple Stress Responses in Fusarium graminearum. Environ. Microbiol. 2015, 17, 4615–4630. [Google Scholar] [CrossRef]
- Pham, K.T.M.; Inoue, Y.; Vu, B.V.; Nguyen, H.H.; Nakayashiki, T.; Ikeda, K.; Nakayashiki, H. MoSET1 (Histone H3K4 Methyltransferase in Magnaporthe Oryzae) Regulates Global Gene Expression during Infection-Related Morphogenesis. PLOS Genet. 2015, 11, e1005385. [Google Scholar] [CrossRef]
- Bauer, I.; Lechner, L.; Pidroni, A.; Petrone, A.-M.; Merschak, P.; Lindner, H.; Kremser, L.; Graessle, S.; Golderer, G.; Allipour, S.; et al. Type I and II PRMTs Regulate Catabolic as Well as Detoxifying Processes in Aspergillus nidulans. Fungal Genet. Biol. 2019, 129, 86–100. [Google Scholar] [CrossRef]
- Xu, X.; Chen, Y.; Li, B.; Tian, S. Arginine Methyltransferase Permtc Regulates Development and Pathogenicity of Penicillium expansum via Mediating Key Genes in Conidiation and Secondary Metabolism. J. Fungi 2021, 7, 807. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Mo, L.-X.; Li, W.-T.; Jiang, L.-L.; Meng, Y.-Y.; Ou, J.-F.; Liao, L.-S.; Yan, Y.-S.; Luo, X.-M.; Feng, J.-X. Arginine Methyltransferases PRMT2 and PRMT3 Are Essential for Biosynthesis of Plant-Polysaccharide-Degrading Enzymes in Penicillium oxalicum. PLoS Genet. 2023, 19, e1010867. [Google Scholar] [CrossRef] [PubMed]
- Feldman, D.; Ziv, C.; Gorovits, R.; Efrat, M.; Yarden, O. Neurospora crassa Protein Arginine Methyl Transferases Are Involved in Growth and Development and Interact with the NDR Kinase COT1. PLoS ONE 2013, 8, e80756. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Gerin, D.; González-Candelas, L.; Ballester, A.-R.; Pollastro, S.; De Miccolis Angelini, R.; Faretra, F. Functional Characterization of the Alb1 Orthologue Gene in the Ochratoxigenic Fungus Aspergillus carbonarius (AC49 Strain). Toxins 2018, 10, 120. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Abarca, M.L.; Bragulat, M.R.; Cabañes, F.J. A New in Vitro Method to Detect Growth and Ochratoxin A-Producing Ability of Multiple Fungal Species Commonly Found in Food Commodities. Food Microbiol. 2014, 44, 243–248. [Google Scholar] [CrossRef]
- Llobregat, B.; González-Candelas, L.; Ballester, A.-R. Ochratoxin a Defective Aspergillus Carbonarius Mutants as Potential Biocontrol Agents. Toxins 2022, 14, 745. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-DDCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Abbott, W.S. A Method of Computing the Effectiveness of an Insecticide. J. Econ. Entomol. 1925, 18, 265–267. [Google Scholar] [CrossRef]

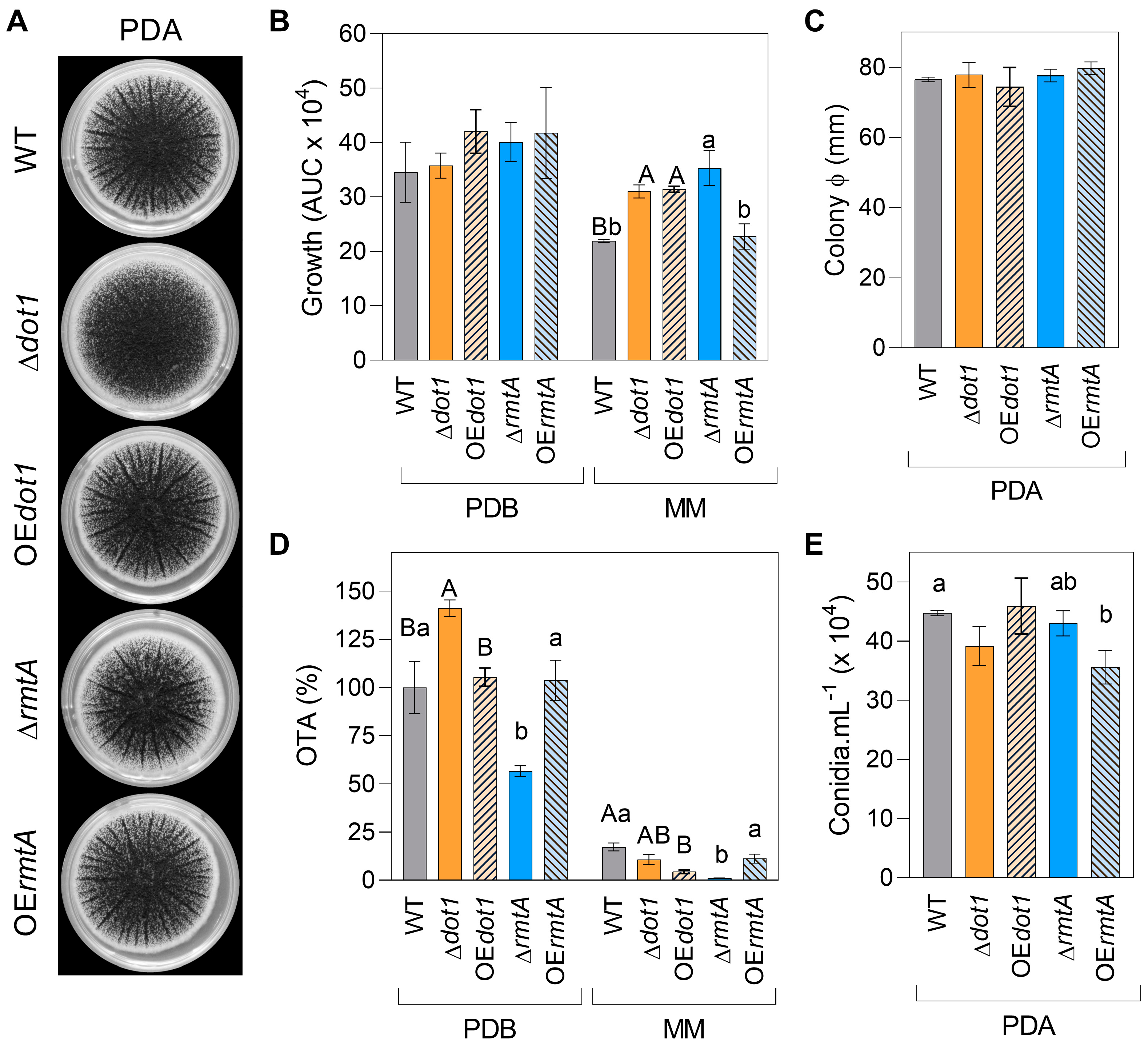

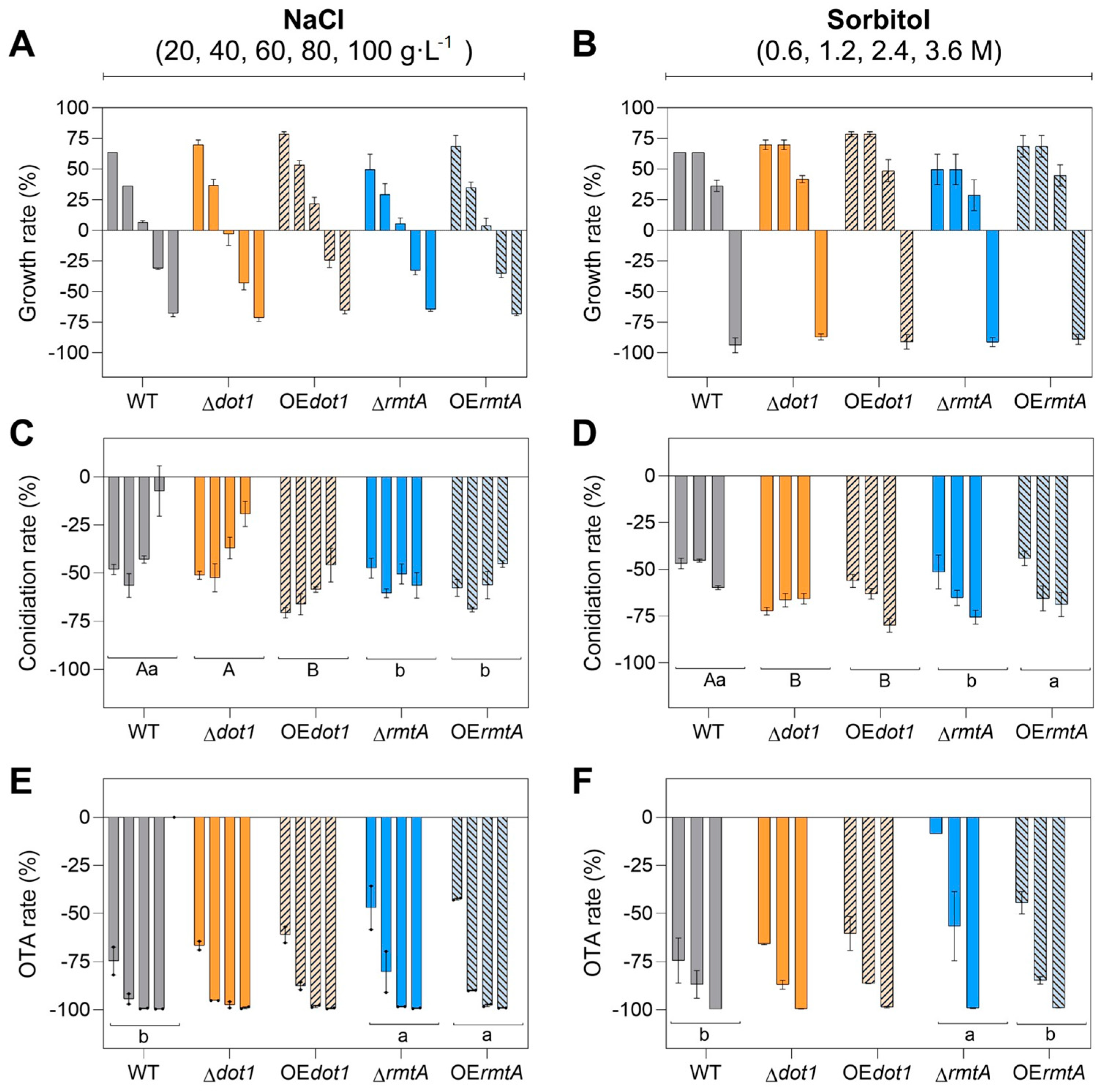
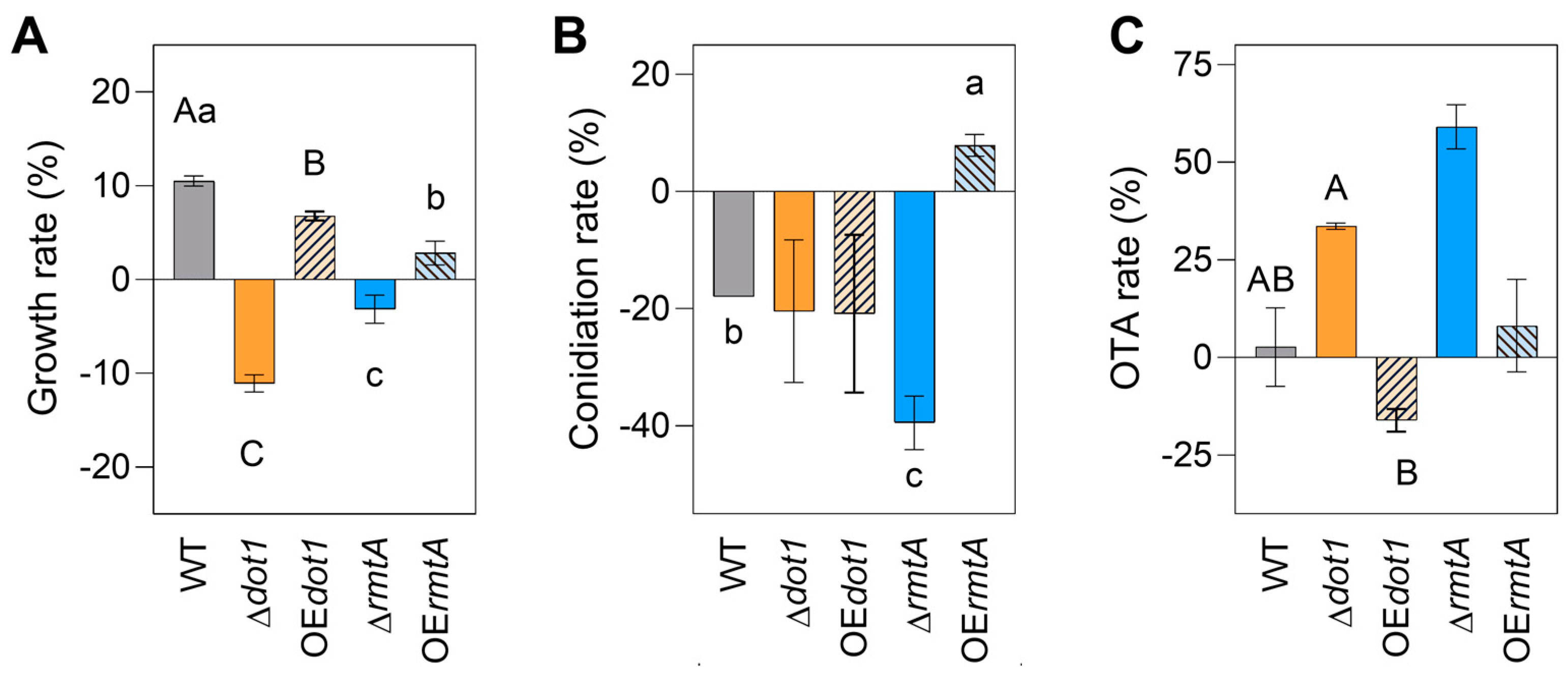
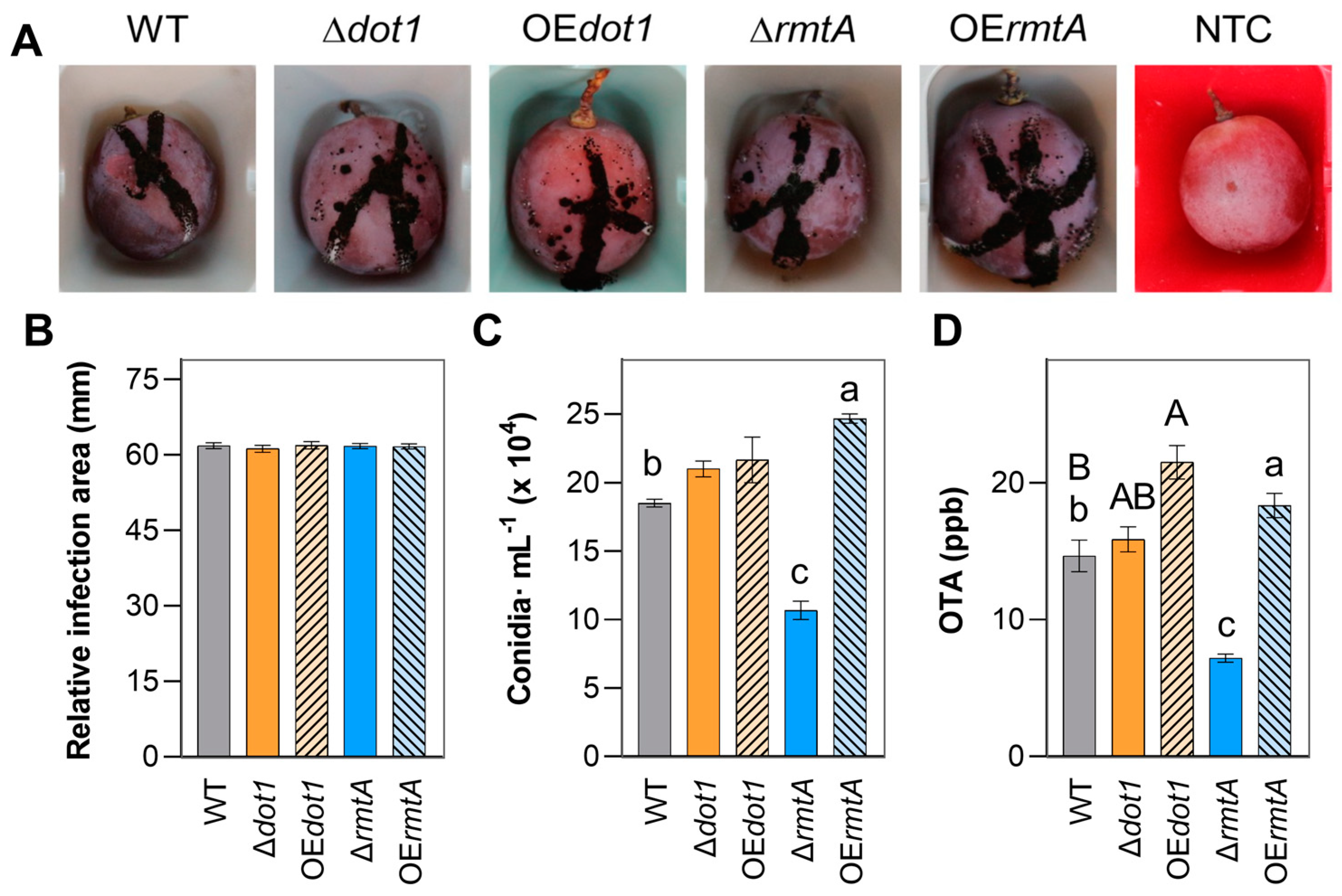
| Strain | Genotype | Cq GOI | Cq NRPS | T-DNA Copies Estimated |
|---|---|---|---|---|
| AC49 | Wild type | 19.47 ± 0.15 | 19.62 ± 0.17 | - |
| AcΔdot1-A5 | Knockout | 21.61 ± 0.05 | 21.71 ± 0.05 | 1 |
| AcΔdot1-A6 | Knockout | 20.73 ± 0.11 | 21.09 ± 0.05 | 1 |
| AcΔdot1-A10 | Knockout | 21.33 ± 0.12 | 21.68 ± 0.04 | 1 |
| AC49 | Wild type | 15.35 ± 0.12 | 15.94 ± 0.05 | - |
| AcOEdot1-B3 | Overexpression | 19.82 ± 0.10 | 21.82 ± 0.06 | 3 |
| AcOEdot1-B6 | Overexpression | 19.25 ± 0.12 | 21.82 ± 0.00 | 4 |
| AcOEdot1-B8 | Overexpression | 19.31 ± 0.01 | 22.28 ± 0.07 | 5 |
| AC49 | Wild type | 19.43 ± 0.06 | 19.47 ± 0.10 | - |
| AcΔrmtA-C3 | Knockout | 21.71 ± 0.03 | 21.40 ± 0.08 | 1 |
| AcΔrmtA-C5 | Knockout | 21.54 ± 0.04 | 21.14 ± 0.05 | 1 |
| AcΔrmtA-C8 | Knockout | 21.15 ± 0.11 | 21.14 ± 0.08 | 1 |
| AC49 | Wild type | 16.23 ± 0.13 | 15.86 ± 0.05 | - |
| AcOErmtA-D3 | Overexpression | 21.44 ± 0.08 | 21.70 ± 0.07 | 2 |
| AcOErmtA-D7 | Overexpression | 20.58 ± 0.10 | 21.41 ± 0.13 | 2 |
| AcOErmtA-D9 | Overexpression | 19.91 ± 0.17 | 20.97 ± 0.04 | 3 |
| Name | Target | Sequence (5′-3′) | bp |
|---|---|---|---|
| Amplification of upstream region | |||
| dot1-O1 | Acdot1 upstream | GGTCTTAAUCGAAACTACGGGCGAAGAAG | 29 |
| dot1-O2 | GGCATTAAUGCGACGGTGAGAGGGAAA | 27 | |
| rmtA-O1 | AcrmtA upstream | GGTCTTAAUGTCTATTGGTTGGTTGGTTGCT | 31 |
| rmtA-O2 | GGCATTAAUAGGTTAGCTGATTTGAACTTCCG | 32 | |
| Amplification of downstream region | |||
| dot1-A3 | Acdot1 downstream | GGACTTAAUCCCTAGCACTCTTCACATTCAG | 31 |
| dot1-A4 | GGGTTTAAUTGGACGGTGAACTGAGCC | 27 | |
| rmtA-A3 | AcrmtA downstream | GGACTTAAUTCCGTTCGTGTATTAGCCGA | 29 |
| rmtA-A4 | GGGTTTAAUACAGGGCCAATCAAAGCTTG | 29 | |
| CDS + 3′UTR amplification for overexpression | |||
| dot1-OE-A3 | Acdot1 | GGACTTAAUATGGGATTTTTCGACCACCT | 29 |
| dot1-OE-A4 | GGGTTTAAUAGCACGCGCTGATATGTATG | 29 | |
| rmtA-OE-A3 | AcrmtA | GGACTTAAUATGTCCGGGCAATCCGC | 26 |
| rmtA-OE-A4 | GGGTTTAAUAGCGTCGACTCGGGGTATCTA | 30 | |
| E. coli (DH5α) screening | |||
| RF-2 | p507-H2 | TCTCCTTGCATGCACCATTCCTTG | 24 |
| p507-HUE-Fw11 | CAAGAAAACGCCAGGAAAAG | 20 | |
| RF-1 | p507-H2 | AAATTTTGTGCTCACCGCCTGGAC | 24 |
| RF-6 | ACGCCAGGGTTTTCCCAGTC | 20 | |
| RF-6 | p507-HE | ACGCCAGGGTTTTCCCAGTC | 20 |
| p507-HUE-Fw3 | TCAGTTCGAGCTTTCCCACT | 20 | |
| Sequencing | |||
| RF-3 | p507-H2 | TTGCGTCAGTCCAACATTTGTTGCCA | 26 |
| dot1-3F | Acdot1 | GAGCACTTGCCCATTCCTCT | 20 |
| dot1-4R | CCACCAACATCGGTCCAACT | 20 | |
| dot1-8F | GCTTTCACCCCTCAACTGAA | 20 | |
| dot1-9R | GCCTCCTCATCGGGTATGTA | 20 | |
| rmtA-3F | AcrmtA | AAGGCAGGTGCTAAGCATGT | 20 |
| rmtA-4R | ATCGCGCAGATAGAAGACGG | 20 | |
| rmtA-8F | TGCATGCCATAAGCCTATCA | 20 | |
| rmtA-9R | GACGACCTCTTCCATCTTGC | 20 | |
| A. carbonarius mutants screening | |||
| dot1-1F | Upstream Acdot1/HygB | GGTCAGGGACGTCATTGCA | 19 |
| HPH-TER2 | GCTCCGTAACACCCAATAC | 19 | |
| rmtA-1F | Upstream AcrmtA/HygB | GGCTGACTCCTCCGTACCTA | 20 |
| HPH-TER2 | GCTCCGTAACACCCAATAC | 19 | |
| dot1-2R | Downstream Acdot1/HygB | GGGAGGATGGGAAGTTGAGC | 20 |
| HPHPRO4 | GCACCAAGCAGCAGATGATA | 20 | |
| rmtA-2R | Downstream AcrmtA/HygB | GCCGCAATTCAGAGCTTGAC | 20 |
| HPHPRO4 | GCACCAAGCAGCAGATGATA | 20 | |
| hyg-1R | HygB | ATTTGTGTACGCCCGACAGT | 20 |
| hyg-2F | GATGTAGGAGGGCGTGGATA | 20 | |
| dot1-3F | Acdot1 | GAGCACTTGCCCATTCCTCT | 20 |
| dot1-4R | CCACCAACATCGGTCCAACT | 20 | |
| rmtA-3F | AcrmtA | AAGGCAGGTGCTAAGCATGT | 20 |
| rmtA-4R | ATCGCGCAGATAGAAGACGG | 20 | |
| T-DNA copy number | |||
| dot1-5F | Acdot1 | AAGGATTGACCCTCCAAGCG | 20 |
| dot1-6R | ATGAGCTGCCCTTGCCTTAG | 20 | |
| rmtA-5F | AcrmtA | AGGCGGTTGACTTTCCAATG | 20 |
| rmtA-6R | TCAGCAAGGAGGAAGACGTAC | 21 | |
| AcNRP-F | nrps | CTCCACCCATCCTCCCGTTC | 20 |
| AcNRP-R | AATCCATGTCCTCACCATCGC | 21 | |
| Gene expression analysis | |||
| AcbZIP-For | otaR1 (AcOTAbZIP) | TTTCCCTAGGATCTCTCCTA | 20 |
| AcbZIP-Rev | TATTGGGGTCGGACAGGAAT | 20 | |
| Acpks4-For | otaA (AcOTApks) | TCTGTATGAGCGCATCGCC | 20 |
| Acpks4-Rev | GCAGAAGGCCACTTTCCAG | 20 | |
| AcotaYfor | otaY (AcOTAcyc) | ACCATCCTCACCACCCTTGT | |
| AcotaYrev | GGGACTCTGGGCTAACACCT | ||
| Acnrps6-For | otaB (AcOTAnrps) | GATTCCGATGGAACTGCAAT | 20 |
| Acnrps6-Rev | CTGCCCCAGCATATCAATCT | 20 | |
| AcP450-For | otaC (AcOTAP450) | GCCATACCTGACCGGGATCA | 20 |
| AcP450-Rev | GGGAAAATGGTCTCGTCGTG | 20 | |
| Achal-For | otaD (AcOTAhal) | AAAGAAGCCTACACCGACTT | 20 |
| Achal-Rev | GAATTCGATGGATCCCGTGC | 20 | |
| Acub-For | Ubiquitin | CCGAAGGTCAACTTCACCAC | 20 |
| Acub-Rev | GGCATATTTGCGAGTCCATT | 20 | |
| Acdot1-For | Acdot1 | TATTGGTCGAACAGCGTCAG | 20 |
| Acdot1-rev | AGGTTCATTGCATGCTTTCC | 20 | |
| AcrmtA-For | AcrmtA | GGAAGTTCGCTTGTGTGGAT | 20 |
| AcrmtA-rev | GTACCGCGCCAACTGATAAT | 20 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agnusdei, A.; González-García, A.; Gerin, D.; Pollastro, S.; Faretra, F.; González-Candelas, L.; Ballester, A.-R. Histone Methyltransferases AcDot1 and AcRmtA Are Involved in Growth Regulation, Secondary Metabolism, and Stress Response in Aspergillus carbonarius. Toxins 2025, 17, 196. https://doi.org/10.3390/toxins17040196
Agnusdei A, González-García A, Gerin D, Pollastro S, Faretra F, González-Candelas L, Ballester A-R. Histone Methyltransferases AcDot1 and AcRmtA Are Involved in Growth Regulation, Secondary Metabolism, and Stress Response in Aspergillus carbonarius. Toxins. 2025; 17(4):196. https://doi.org/10.3390/toxins17040196
Chicago/Turabian StyleAgnusdei, Angelo, Adrián González-García, Donato Gerin, Stefania Pollastro, Francesco Faretra, Luis González-Candelas, and Ana-Rosa Ballester. 2025. "Histone Methyltransferases AcDot1 and AcRmtA Are Involved in Growth Regulation, Secondary Metabolism, and Stress Response in Aspergillus carbonarius" Toxins 17, no. 4: 196. https://doi.org/10.3390/toxins17040196
APA StyleAgnusdei, A., González-García, A., Gerin, D., Pollastro, S., Faretra, F., González-Candelas, L., & Ballester, A.-R. (2025). Histone Methyltransferases AcDot1 and AcRmtA Are Involved in Growth Regulation, Secondary Metabolism, and Stress Response in Aspergillus carbonarius. Toxins, 17(4), 196. https://doi.org/10.3390/toxins17040196










