Bacterial Microbiota in Soil Amended with Deoxynivalenol-Contaminated Wheat
Abstract
1. Introduction
2. Results
2.1. Chemical Analysis of Soil Samples
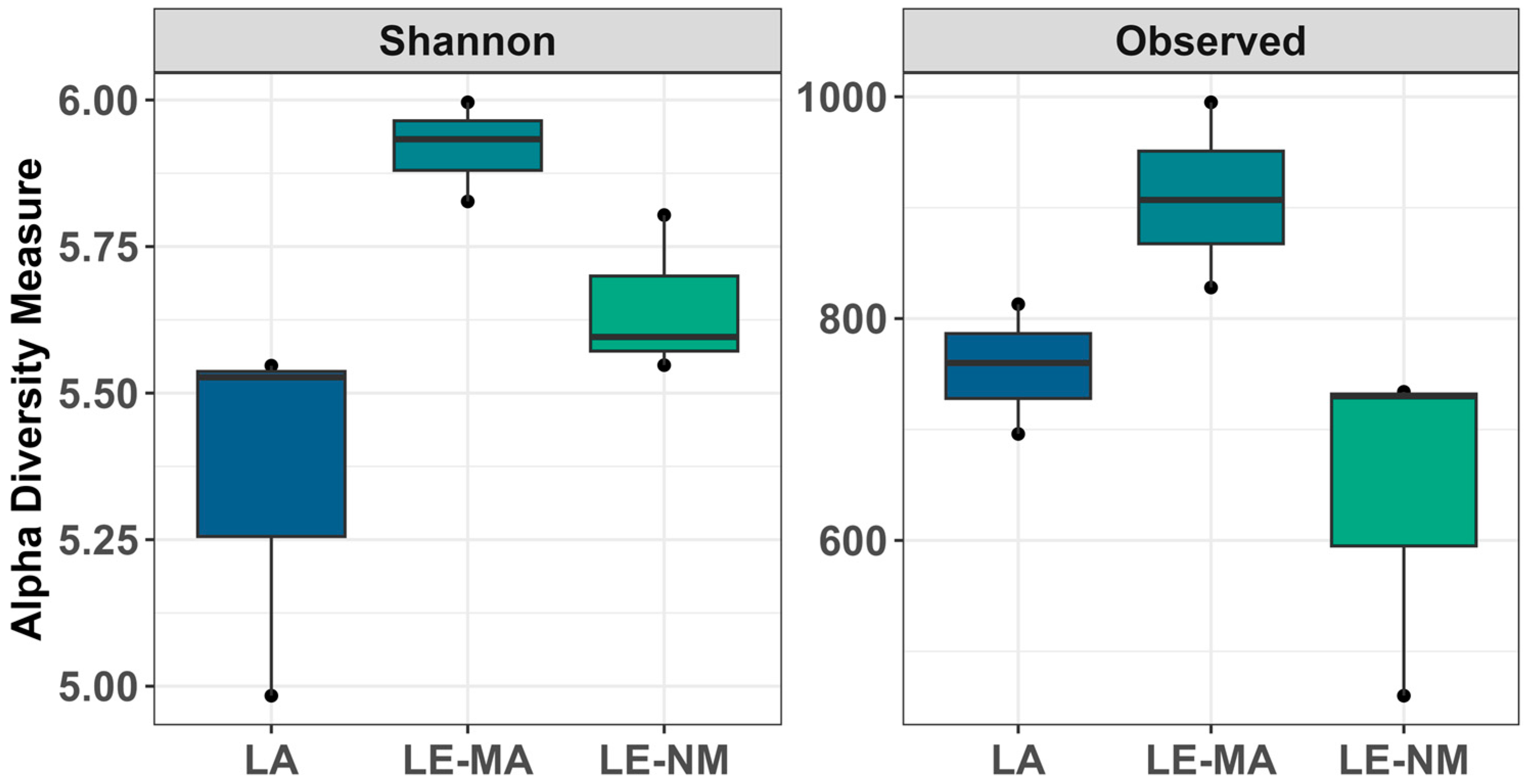
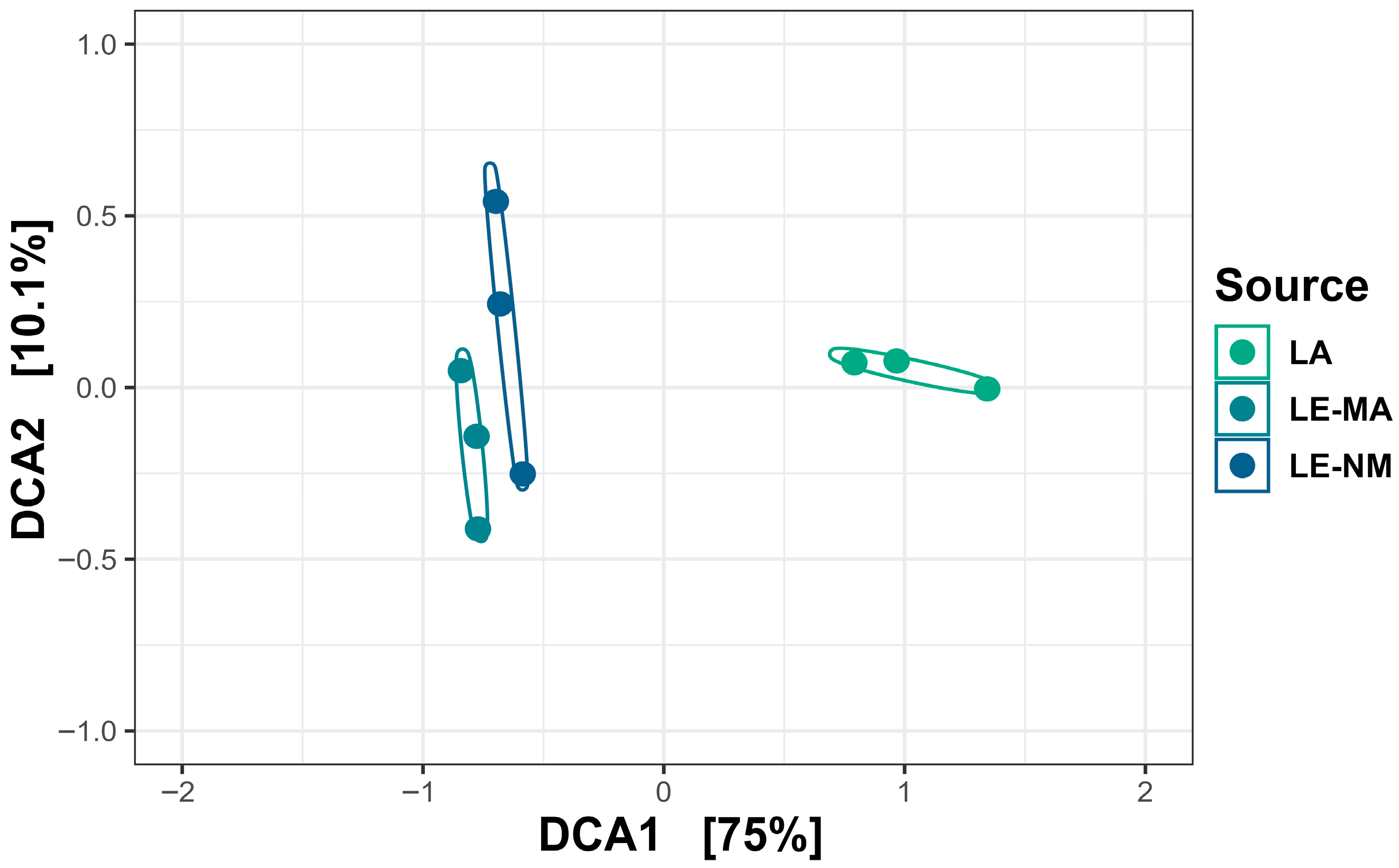
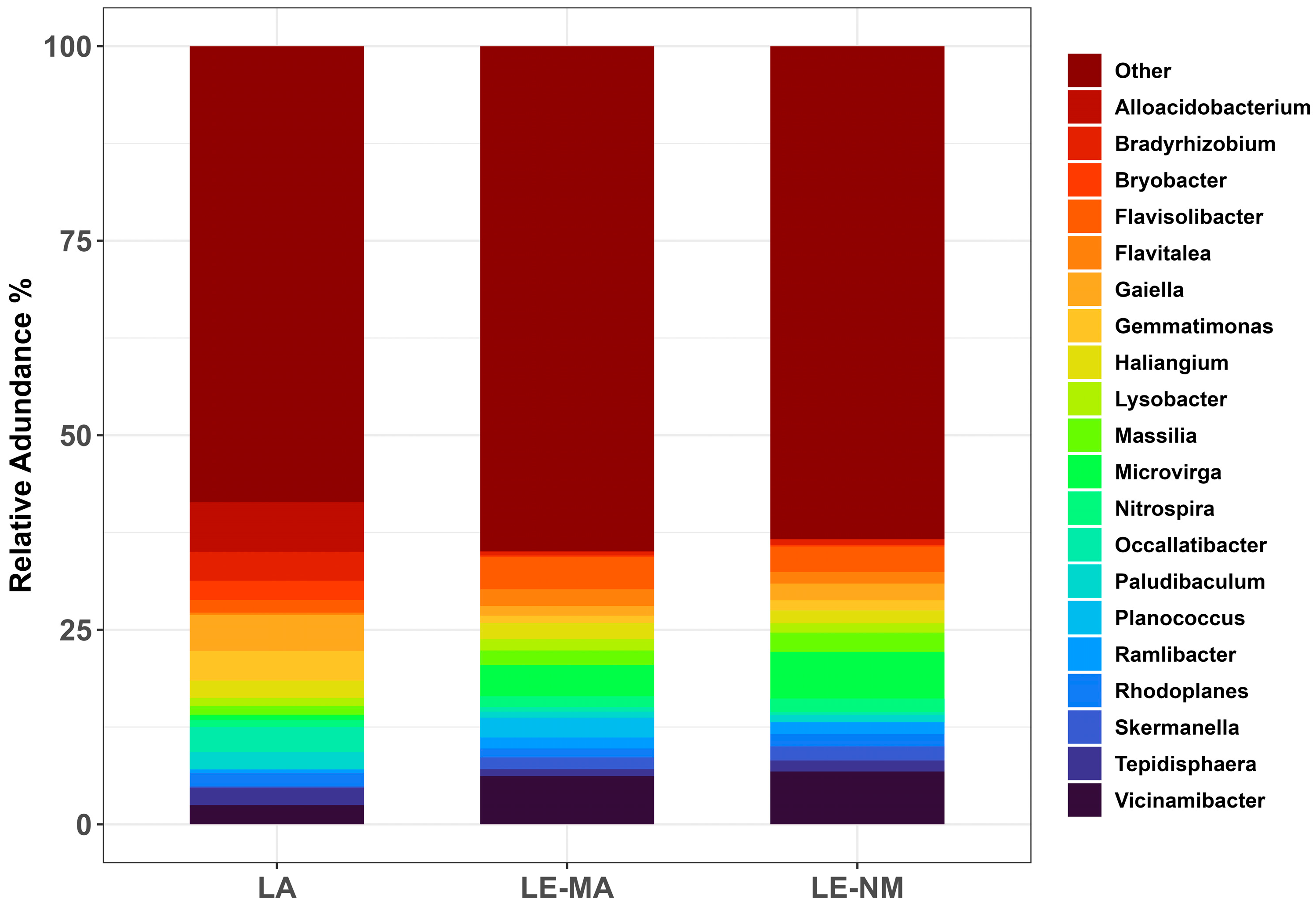
2.2. Community Structure and Composition of Soil Bacteria
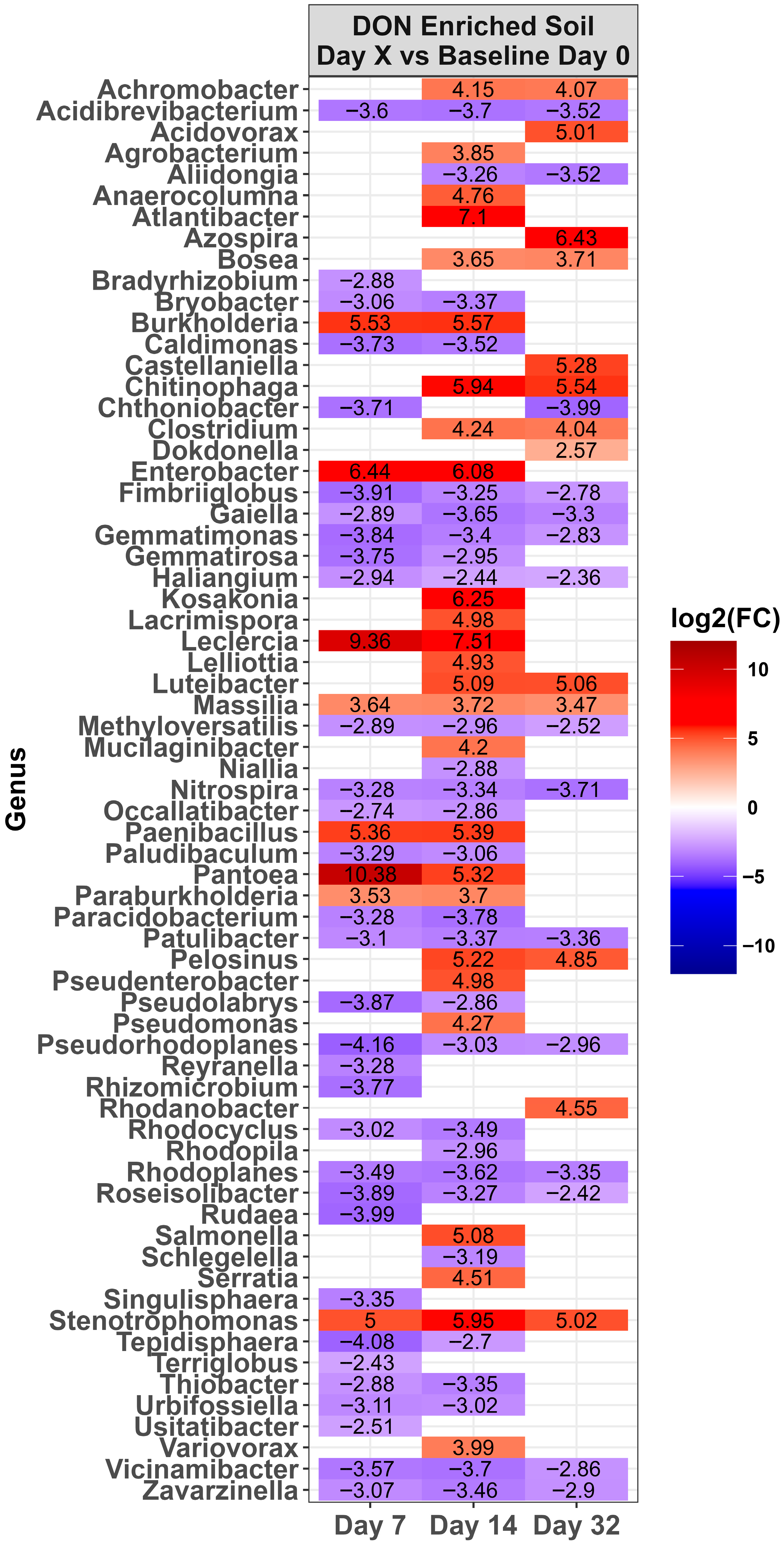
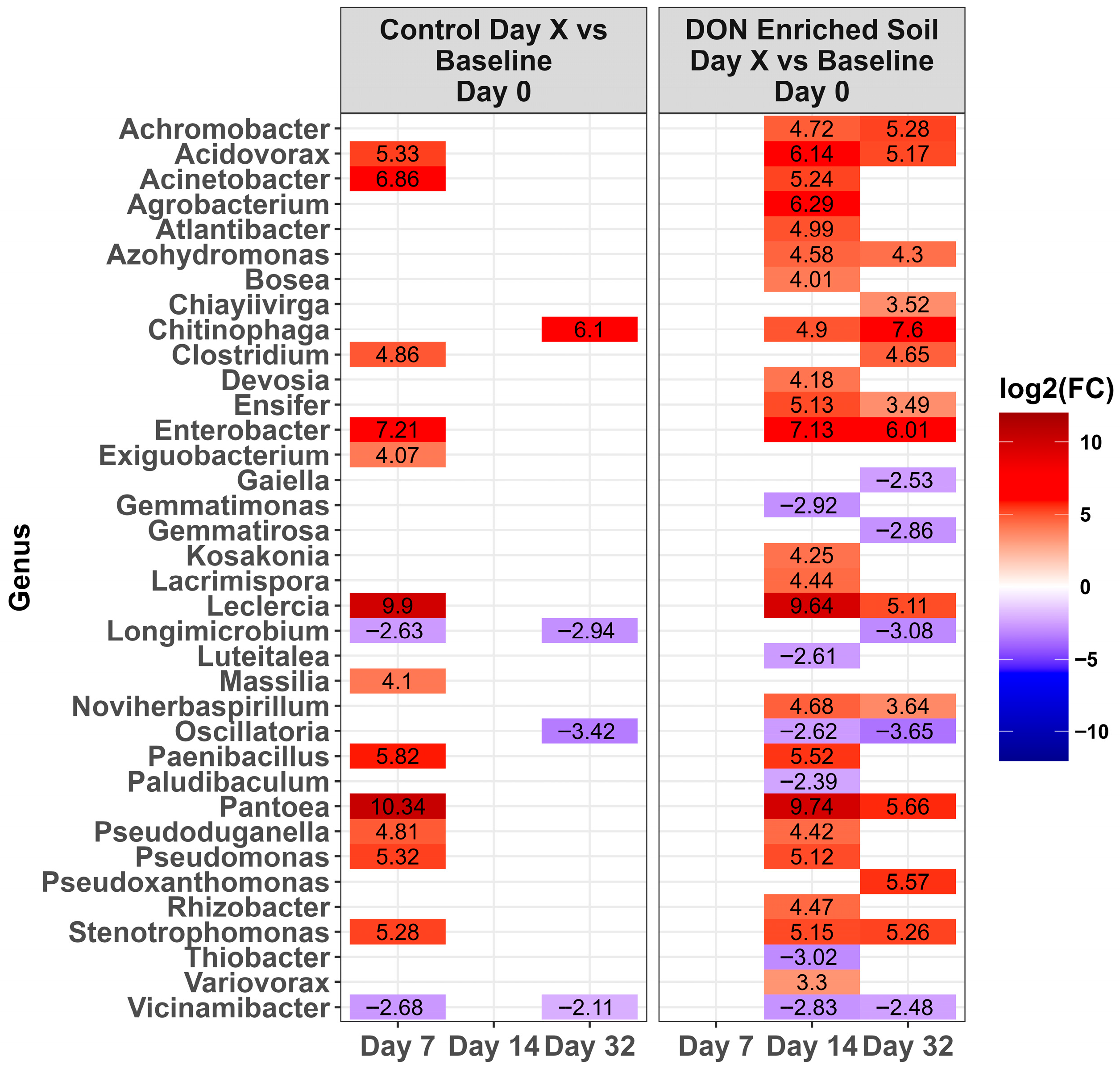
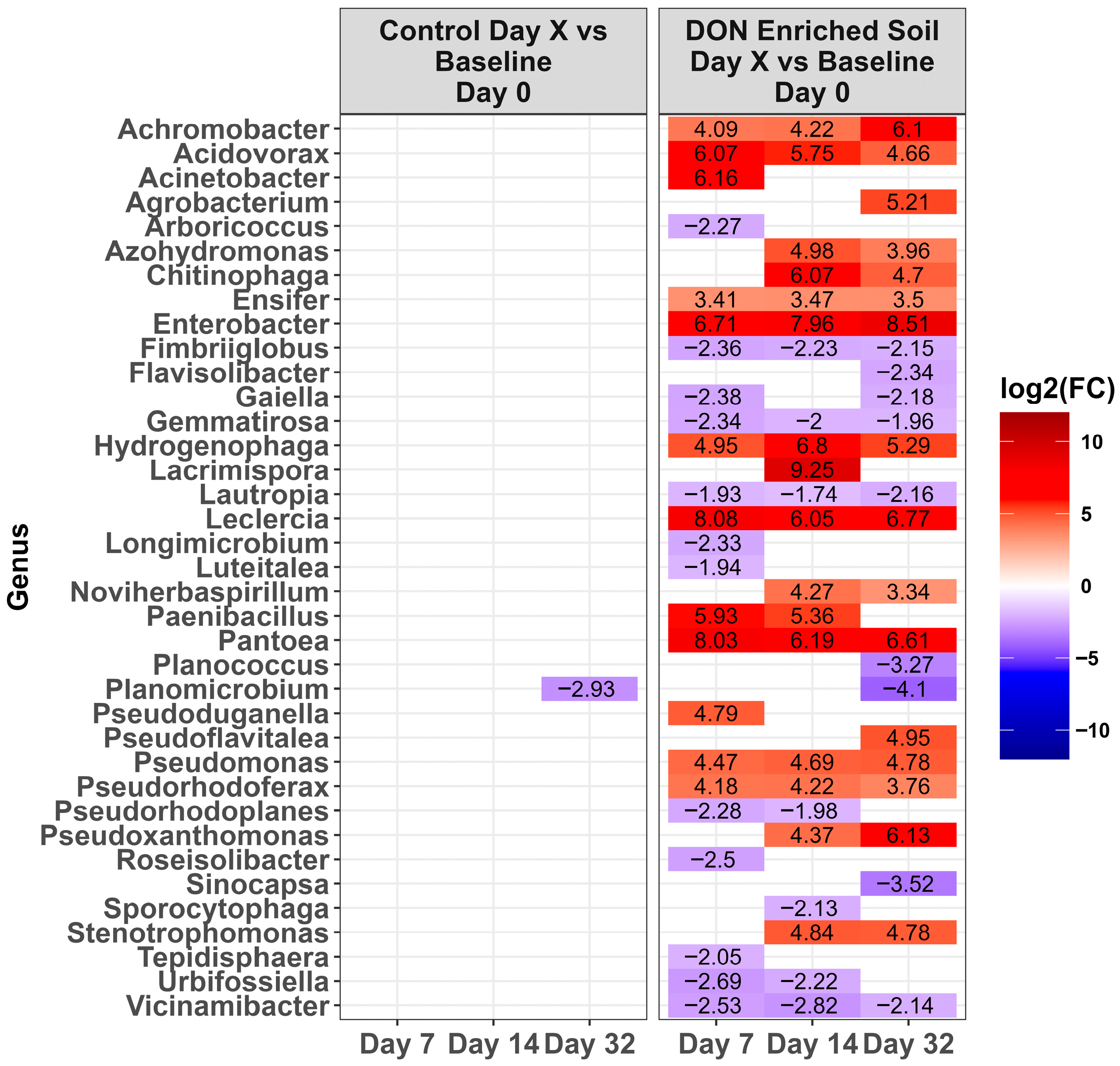
2.3. Changes in the Microbiota Across Time
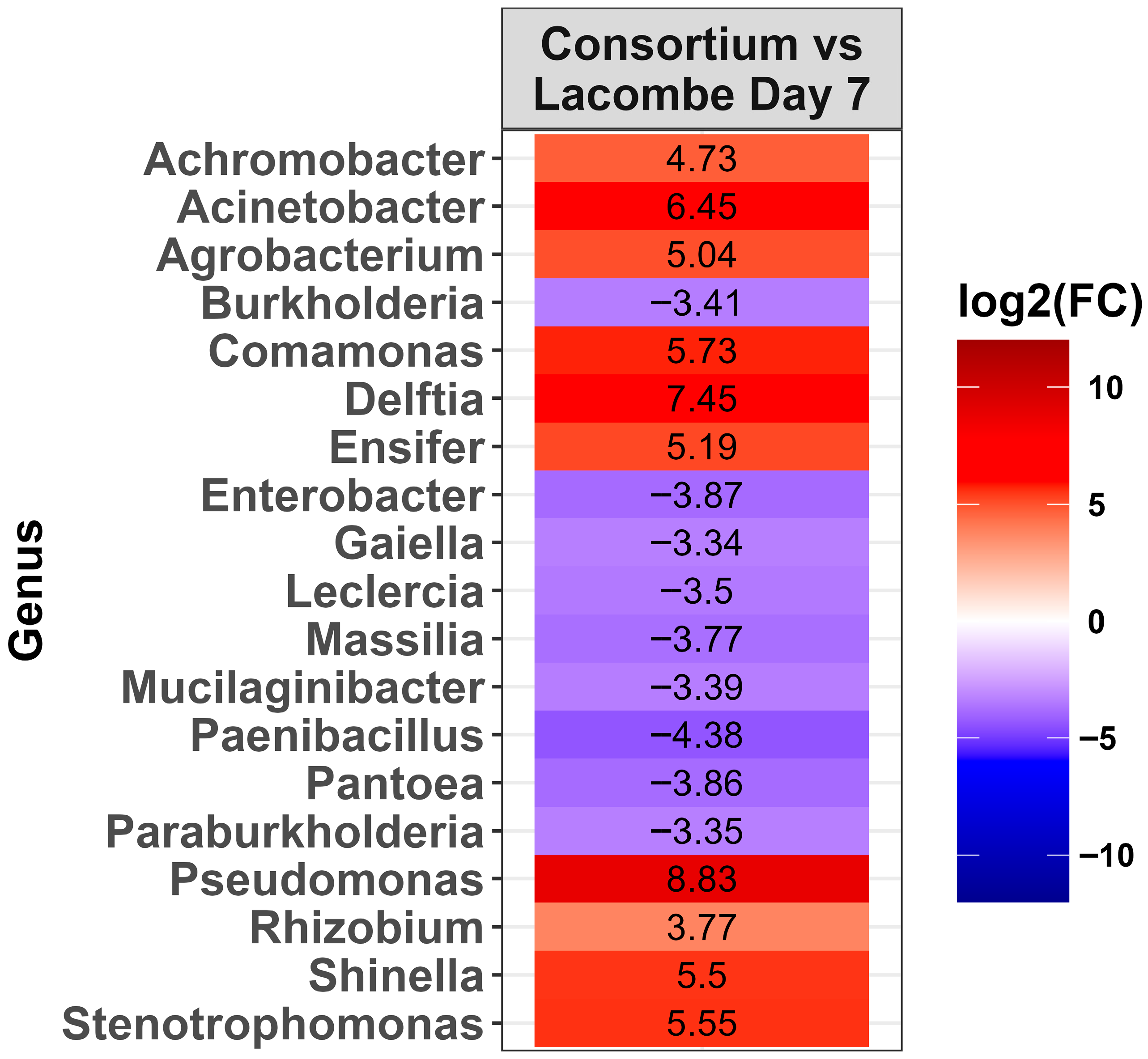
2.4. Changes in the Microbiota Within the LA Consortium
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Soil Samples, Culture Media, and Reagents
5.2. In Vitro Soil Cultures and Isolation of DON-Degrading Consortia
5.3. DNA Isolation and 16S rRNA Sequencing
5.4. Analysis of Bacterial Community Diversity from Different Environmental Sources and Consortia
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chin, T.; Pleskach, K.; Tittlemier, S.A.; Henriquez, M.A.; Bamforth, J.; Withana Gamage, N.; Ashfaq, T.; Lee, S.-J.; Kurera, M.S.; Patel, B. A status update on fusarium head blight on Western Canadian wheat. Can. J. Plant. Pathol. 2023, 45, 277–289. [Google Scholar] [CrossRef]
- Bamforth, J.; Chin, T.; Ashfaq, T.; Gamage, N.W.; Pleskach, K.; Tittlemier, S.A.; Henriquez, M.A.; Kurera, S.; Lee, S.-J.; Patel, B. A survey of Fusarium species and ADON genotype on Canadian wheat grain. Front. Fungal Biol. 2022, 3, 1062444. [Google Scholar] [CrossRef]
- Islam, R.; Zhou, T.; Christopher Young, J.; Goodwin, P.H.; Peter Pauls, K. Aerobic and anaerobic de-epoxydation of mycotoxin deoxynivalenol by bacteria originating from agricultural soil. World J. Microbiol. Biotechnol. 2012, 28, 7–13. [Google Scholar] [CrossRef]
- He, W.-J.; Yuan, Q.-S.; Zhang, Y.-B.; Guo, M.-W.; Gong, A.-D.; Zhang, J.-B.; Wu, A.-B.; Huang, T.; Qu, B.; Li, H.-P. Aerobic de-epoxydation of trichothecene mycotoxins by a soil bacterial consortium isolated using in situ soil enrichment. Toxins 2016, 8, 277. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, G.; Dai, Y.; Wang, Y.; Lee, Y.-W.; Shi, J.; Xu, J. Biodegradation of deoxynivalenol by a novel microbial consortium. Front. Microbiol. 2020, 10, 2964. [Google Scholar] [CrossRef]
- Pinto, A.C.S.M.; De Pierri, C.R.; Evangelista, A.G.; Gomes, A.S.d.L.P.B.; Luciano, F.B. Deoxynivalenol: Toxicology, degradation by bacteria, and phylogenetic analysis. Toxins 2022, 14, 90. [Google Scholar] [CrossRef]
- Cai, C.; Zhao, M.; Yao, F.; Zhu, R.; Cai, H.; Shao, S.; Li, X.-Z.; Zhou, T. Deoxynivalenol degradation by various microbial communities and its impacts on different bacterial flora. Toxins 2022, 14, 537. [Google Scholar] [CrossRef] [PubMed]
- Sato, I.; Ito, M.; Ishizaka, M.; Ikunaga, Y.; Sato, Y.; Yoshida, S.; Koitabashi, M.; Tsushima, S. Thirteen novel deoxynivalenol-degrading bacteria are classified within two genera with distinct degradation mechanisms. FEMS Microbiol. Lett. 2012, 327, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Zhong, L.; Gao, H.; Lu, Z.; Bie, X.; Zhao, H.; Zhang, C.; Lu, F. Detoxification of deoxynivalenol by a mixed culture of soil bacteria with 3-epi-deoxynivalenol as the main intermediate. Front. Microbiol. 2019, 10, 2172. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hao, X.; Thomas, B.W.; McAllister, T.A.; Workentine, M.; Jin, L.; Shi, X.; Alexander, T.W. Soil antibiotic resistance genes accumulate at different rates over four decades of manure application. J. Hazard. Mater. 2023, 443, 130136. [Google Scholar] [CrossRef]
- Lagomarsino, A.; Grego, S.; Kandeler, E. Soil organic carbon distribution drives microbial activity and functional diversity in particle and aggregate-size fractions. Pedobiologia 2012, 55, 101–110. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J. Soil moisture as a factor affecting the microbiological and biochemical activity of soil. Open Access CAAS Agric. J. 2016, 62, 250–255. [Google Scholar] [CrossRef]
- Kim, B.-R.; Shin, J.; Guevarra, R.B.; Lee, J.H.; Kim, D.W.; Seol, K.-H.; Lee, J.-H.; Kim, H.B.; Isaacson, R.E. Deciphering diversity indices for a better understanding of microbial communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef]
- Sahu, K.P.; Kumar, A.; Patel, A.; Kumar, M.; Gopalakrishnan, S.; Prakash, G.; Rathour, R.; Gogoi, R. Rice blast lesions: An unexplored phyllosphere microhabitat for novel antagonistic bacterial species against Magnaporthe oryzae. Microb. Ecol. 2021, 81, 731–745. [Google Scholar] [CrossRef]
- Siani, R.; Stabl, G.; Gutjahr, C.; Schloter, M.; Radl, V. Acidovorax pan-genome reveals specific functional traits for plant beneficial and pathogenic plant-associations. Microb. Genom. 2021, 7, 000666. [Google Scholar] [CrossRef] [PubMed]
- Davin-Regli, A.; Lavigne, J.-P.; Pagès, J.-M. Enterobacter spp.: Update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin. Microbiol. Rev. 2019, 32, e00002-19. [Google Scholar] [CrossRef]
- Kenngott, K.G.; Muñoz, K. The potential of soil microbial communities to transform deoxynivalenol in agricultural soils—A soil microcosm study. Mycotoxin Res. 2024, 40, 295–307. [Google Scholar] [CrossRef]
- Ahad, R.; Zhou, T.; Lepp, D.; Pauls, K.P. Microbial detoxification of eleven food and feed contaminating trichothecene mycotoxins. BMC Biotech. 2017, 17, 30. [Google Scholar] [CrossRef] [PubMed]
- Wilson, N.M.; McMaster, N.; Gantulga, D.; Soyars, C.; McCormick, S.P.; Knott, K.; Senger, R.S.; Schmale, D.G. Modification of the mycotoxin deoxynivalenol using microorganisms isolated from environmental samples. Toxins 2017, 9, 141. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Niu, J.-f.; Yang, H.; Lu, Z.-x.; Chen, M.-r.; Lü, F.-x. High-throughput sequencing analysis of vomitoxin-degrading bacteria in bacterial consortium. Food Sci. 2021, 42, 123–130. [Google Scholar]
- Wang, Y.; Zhao, D.; Zhang, W.; Wang, S.; Huang, K.; Guo, B. Biotransformation of deoxynivalenol by a dual-member bacterial consortium isolated from Tenebrio molitor larval feces. Toxins 2023, 15, 492. [Google Scholar] [CrossRef] [PubMed]
- Shima, J.; Takase, S.; Takahashi, Y.; Iwai, Y.; Fujimoto, H.; Yamazaki, M.; Ochi, K. Novel detoxification of the trichothecene mycotoxin deoxynivalenol by a soil bacterium isolated by enrichment culture. Appl. Environ. Microbiol. 1997, 63, 3825–3830. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, H.; Zhao, C.; Han, Y.; Liu, Y.; Zhang, X. Isolation and characterization of a novel deoxynivalenol-transforming strain Paradevosia shaoguanensis DDB001 from wheat field soil. Lett. Appl. Microbiol. 2017, 65, 414–422. [Google Scholar] [CrossRef]
- Nathanail, A.V.; Gibson, B.; Han, L.; Peltonen, K.; Ollilainen, V.; Jestoi, M.; Laitila, A. The lager yeast Saccharomyces pastorianus removes and transforms Fusarium trichothecene mycotoxins during fermentation of brewer’s wort. Food Chem. 2016, 203, 448–455. [Google Scholar] [CrossRef]
- Chlebicz, A.; Śliżewska, K. In vitro detoxification of aflatoxin B 1, deoxynivalenol, fumonisins, T-2 toxin and zearalenone by probiotic bacteria from genus Lactobacillus and Saccharomyces cerevisiae yeast. Probiotics Antimicrob. Proteins 2020, 12, 289–301. [Google Scholar] [CrossRef]
- Ellert, B.; Rock, L. Stable isotopes in soil and environmental research. Soil Sampling and Methods of Analysis, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2008; pp. 693–711. [Google Scholar]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- The R Core Team. R: A Language and Environment for Statistical Computing; Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.; O’hara, R.; Simpson, G.; Solymos, P. Vegan: Community Ecology Package (Version 2.5-6), The Comprehensive R Archive Network. 2019. Available online: https://cran.r-project.org (accessed on 19 May 2023).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bumunang, E.W.; Stanford, K.; Wang, Y.; Ellert, B.; Waldner, M.; Alexander, T.W. Bacterial Microbiota in Soil Amended with Deoxynivalenol-Contaminated Wheat. Toxins 2025, 17, 565. https://doi.org/10.3390/toxins17120565
Bumunang EW, Stanford K, Wang Y, Ellert B, Waldner M, Alexander TW. Bacterial Microbiota in Soil Amended with Deoxynivalenol-Contaminated Wheat. Toxins. 2025; 17(12):565. https://doi.org/10.3390/toxins17120565
Chicago/Turabian StyleBumunang, Emmanuel W., Kim Stanford, Yuxi Wang, Benjamin Ellert, Matthew Waldner, and Trevor W. Alexander. 2025. "Bacterial Microbiota in Soil Amended with Deoxynivalenol-Contaminated Wheat" Toxins 17, no. 12: 565. https://doi.org/10.3390/toxins17120565
APA StyleBumunang, E. W., Stanford, K., Wang, Y., Ellert, B., Waldner, M., & Alexander, T. W. (2025). Bacterial Microbiota in Soil Amended with Deoxynivalenol-Contaminated Wheat. Toxins, 17(12), 565. https://doi.org/10.3390/toxins17120565





