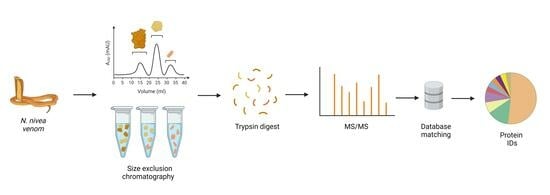
Proteomic Investigation of Cape Cobra (Naja nivea) Venom Reveals First Evidence of Quaternary Protein Structures
Abstract
1. Introduction
2. Results and Discussion
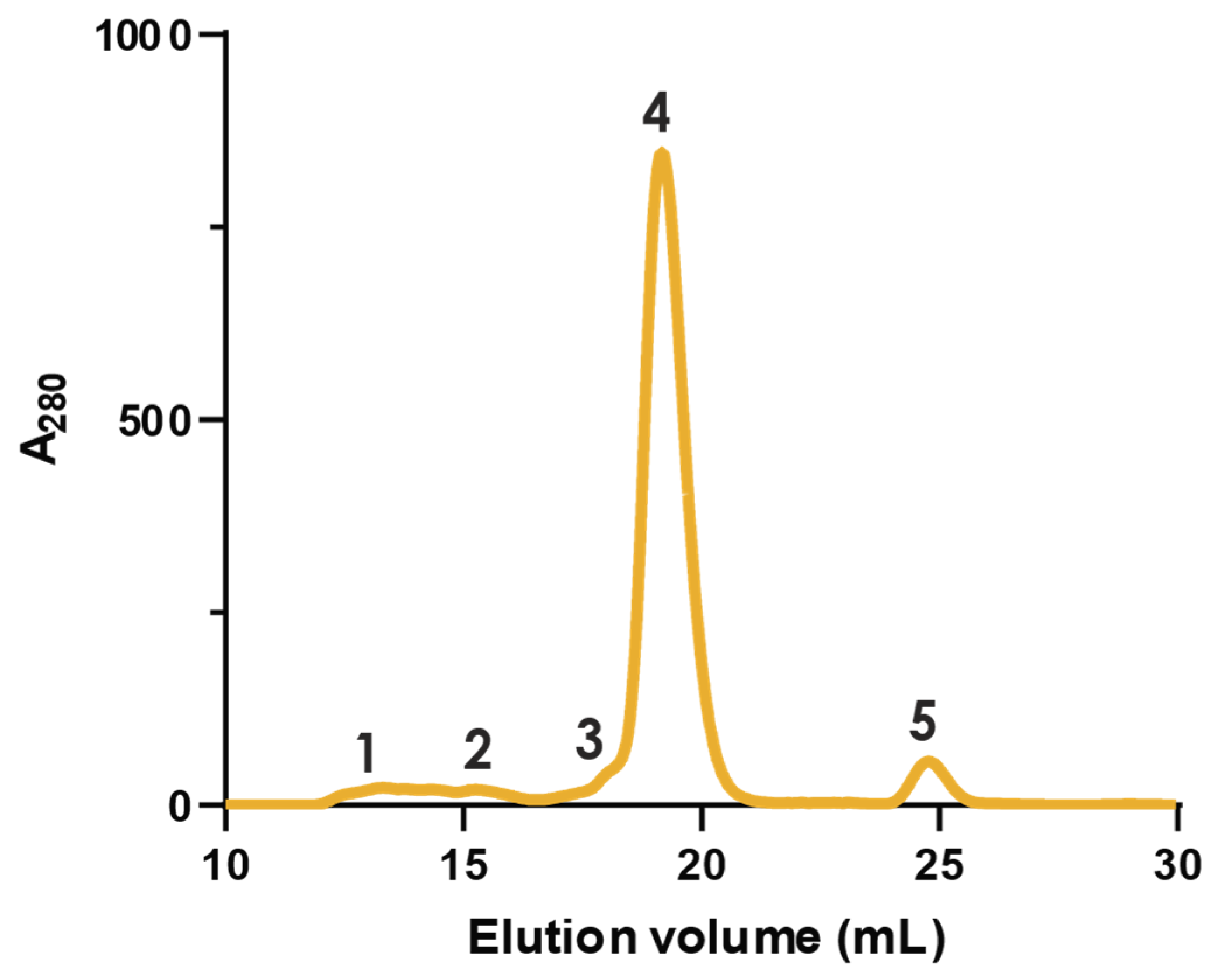
2.1. Fractionation with Size Exclusion Chromatography
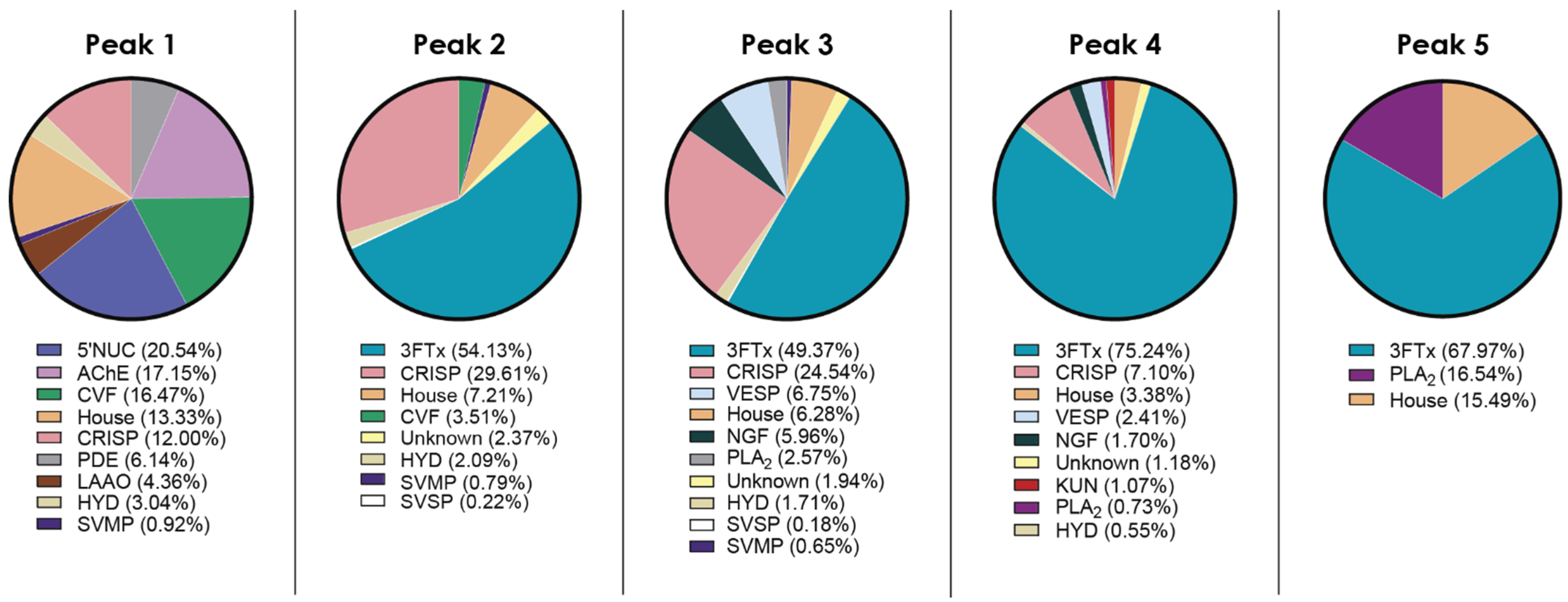
2.2. Venom Proteome of N. nivea
3. Conclusions and Future Directions
4. Materials and Methods
4.1. Materials
4.2. Chromatographic Separation
4.3. One-Dimensional SDS-Polyacrylamide Gel Electrophoresis
4.4. In-Gel Tryptic Digestion
4.5. LC-MS/MS Analysis
4.6. MS/MS Data Analysis
4.7. Estimation of Protein Relative Abundance
4.8. LC-MS Analysis of Venom Fractions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chippaux, J.-P. Snakebite envenomation turns again into a neglected tropical disease! J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 38. [Google Scholar] [CrossRef] [PubMed]
- WHO. Ending the Neglect to Attain the Sustainable Development Goals: A Road Map for Neglected Tropical Diseases 2021–2030; WHO: Geneva, Switzerland, 2021; ISBN 9789240010352. [Google Scholar]
- Gutiérrez, J.M. Global Availability of Antivenoms: The Relevance of Public Manufacturing Laboratories. Toxins 2019, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Halilu, S.; Iliyasu, G.; Hamza, M.; Chippaux, J.P.; Kuznik, A.; Habib, A.G. Snakebite Burden in Sub-Saharan Africa: Estimates from 41 Countries. Toxicon 2019, 159, 1–4. [Google Scholar] [CrossRef]
- Benjamin, J.M.; Abo, B.N.; Brandehoff, N. Review Article: Snake Envenomation in Africa. Curr. Trop. Med. Rep. 2020, 7, 1–10. [Google Scholar] [CrossRef]
- WHO—Regional Office for Africa. Guidelines for the Prevention and Clinical Management of Snakebite in Africa; WHO—Regional Office for Africa: Brazzaville, Republic of Congo, 2010; pp. 1–145. [Google Scholar]
- Broadley, D.G.; Wüster, W. A Review of the Southern African ‘Non-Spitting’ Cobras (Serpentes: Elapidae: Naja). Afr. J. Herpetol. 2004, 53, 101–122. [Google Scholar] [CrossRef]
- Menezes, M.C.; Furtado, M.F.; Travaglia-Cardoso, S.R.; Camargo, A.C.M.; Serrano, S.M.T. Sex-Based Individual Variation of Snake Venom Proteome among Eighteen Bothrops jararaca Siblings. Toxicon 2006, 47, 304–312. [Google Scholar] [CrossRef]
- Alape-Girón, A.; Sanz, L.; Escolano, J.; Flores-Díaz, M.; Madrigal, M.; Sasa, M.; Calvete, J.J. Snake Venomics of the Lancehead Pitviper Bothrops asper. Geographic, Individual, and Ontogenetic Variations. J. Proteome Res. 2008, 7, 3556–3571. [Google Scholar] [CrossRef]
- Massey, D.J.; Calvete, J.J.; Sánchez, E.E.; Sanz, L.; Richards, K.; Curtis, R.; Boesen, K. Venom Variability and Envenoming Severity Outcomes of the Crotalus scutulatus scutulatus (Mojave rattlesnake) from Southern Arizona. J. Proteom. 2012, 75, 2576–2587. [Google Scholar] [CrossRef]
- Rey-Suárez, P.; Núñez, V.; Gutiérrez, J.M.; Lomonte, B. Proteomic and Biological Characterization of the Venom of the Redtail Coral Snake, Micrurus mipartitus (Elapidae), from Colombia and Costa Rica. J. Proteom. 2011, 75, 655–667. [Google Scholar] [CrossRef]
- Gutiérrez, J.M.; Sanz, L.; Flores-Díaz, M.; Figueroa, L.; Madrigal, M.; Herrera, M.; Villalta, M.; León, G.; Estrada, R.; Borges, A.; et al. Impact of Regional Variation in Bothrops Asper Snake Venom on the Design of Antivenoms: Integrating Antivenomics and Neutralization Approaches. J. Proteome Res. 2010, 9, 564–577. [Google Scholar] [CrossRef]
- Avella, I.; Calvete, J.J.; Sanz, L.; Wüster, W.; Licata, F.; Quesada-Bernat, S.; Rodríguez, Y.; Martínez-Freiría, F. Interpopulational Variation and Ontogenetic Shift in the Venom Composition of Lataste’s Viper (Vipera latastei, Boscá 1878) from Northern Portugal. J. Proteom. 2022, 263, 104613. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Wong, K.Y.; Huang, L.K.; Tan, K.Y.; Tan, N.H.; Wu, W.G. Snake Venomics and Antivenomics of Cape Cobra (Naja nivea) from South Africa: Insights into Venom Toxicity and Cross-Neutralization Activity. Toxins 2022, 14, 860. [Google Scholar] [CrossRef] [PubMed]
- Blaylock, R.S.; Lichtman, A.R.; Potgieter, P.D. Clinical Manifestations of Cape Cobra (Naja nivea) Bites. A Report of 2 Cases. S. Afr. Med. J. 1985, 68, 342–344. [Google Scholar]
- Botes, D.P.; Strydom, D.J.; Anderson, C.G.; Christensen, P.A. Snake Venom Toxins. Purification and Properties of Three Toxins from Naja nivea (Linnaeus) (Cape cobra) Venom and the Amino Acid Sequence of Toxin Delta. J. Biol. Chem. 1971, 246, 3132–3139. [Google Scholar] [CrossRef] [PubMed]
- Botes, D.P. Snake Venom Toxins. The Amino Acid Sequences of Toxins and from Naja nivea Venom and the Disulfide Bonds of Toxin. J. Biol. Chem. 1971, 246, 7383–7391. [Google Scholar] [CrossRef]
- Dawie, P.; Botes, C.C.V. The Amino Acid Sequence of Three Non-Curarimimetic Toxins from Naja nivea Venom. Biochim. Biophys. Acta 1976, 13, 258–283. [Google Scholar]
- Hokama, Y.; Iwanaga, S.; Tatsuki, T.; Suzuki, T. Isolation of Five Polypeptide Inhibitors from the Venoms of Htmachatus haemachatus (Ringhal’s Cobra) and Naja nivea (Cape cobra) and the Complete Amino Acid Sequences of Two of Them. J. Biol. Chem. 1976, 79, 559–578. [Google Scholar]
- Botes, D.P. Snake Venom Toxins, the Reactivity of the Disulphide Bonds of Naja nivea Toxin α. Biochim. Biophys. Acta 1974, 359, 242–247. [Google Scholar] [CrossRef]
- Williams, H.F.; Layfield, H.J.; Vallance, T.; Patel, K.; Bicknell, A.B.; Trim, S.A.; Vaiyapuri, S. The Urgent Need to Develop Novel Strategies for the Diagnosis and Treatment of Snakebites. Toxins 2019, 11, 363. [Google Scholar] [CrossRef]
- Wachtel, L.W.; Cole, L.J. Guidelines for the Production, Control and Regulation of Snake Antivenom Immunoglobulins; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Vaiyapuri, S.; Vaiyapuri, R.; Ashokan, R.; Ramasamy, K.; Nattamaisundar, K.; Jeyaraj, A.; Chandran, V.; Gajjeraman, P.; Baksh, M.F.; Gibbins, J.M.; et al. Snakebite and Its Socio-Economic Impact on the Rural Population of Tamil Nadu, India. PLoS ONE 2013, 8, e80090. [Google Scholar] [CrossRef]
- Potet, J.; Smith, J.; McIver, L. Reviewing Evidence of the Clinical Effectiveness of Commercially Available Antivenoms in Sub-Saharan Africa Identifies the Need for a Multi-Centre, Multi-Antivenom Clinical Trial. PLoS Negl. Trop. Dis. 2019, 13, e0007551. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.S.; Georgieva, D.; Genov, N.; Murakami, M.T.; Sinha, M.; Kumar, R.P.; Kaur, P.; Kumar, S.; Dey, S.; Sharma, S.; et al. Enzymatic Toxins from Snake Venom: Structural Characterization and Mechanism of Catalysis. FEBS J. 2011, 278, 4544–4576. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y.; Yu, T.F.; Lian, E.C.Y. Purification and Characterization of L-Amino Acid Oxidase from King Cobra (Ophiophagus hannah) Venom and Its Effects on Human Platelet Aggregation. Toxicon 1994, 32, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- McCleary, R.J.R.; Kini, R.M. Non-Enzymatic Proteins from Snake Venoms: A Gold Mine of Pharmacological Tools and Drug Leads. Toxicon 2013, 62, 56–74. [Google Scholar] [CrossRef] [PubMed]
- Tasoulis, T.; Isbister, G.K. A Review and Database of Snake Venom Proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef] [PubMed]
- Doley, R.; Kini, R.M. Protein Complexes in Snake Venom. Cell. Mol. Life Sci. 2009, 66, 2851–2871. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Huang, C. Synergistic Strategies of Predominant Toxins in Snake Venoms. Toxicol. Lett. 2018, 287, 142–154. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.K. Non-Covalent Interaction of Phospholipase A2 (PLA2) and Kaouthiotoxin (KTX) from Venom of Naja kaouthia Exhibits Marked Synergism to Potentiate Their Cytotoxicity on Target Cells. J. Venom Res. 2010, 1, 37–42. [Google Scholar]
- Laustsen, A.H. Toxin Synergism in Snake Venoms. Toxin Rev. 2016, 35, 165–170. [Google Scholar] [CrossRef]
- Wang, C.R.; Bubner, E.R.; Jovcevski, B.; Mittal, P.; Pukala, T.L. Interrogating the Higher Order Structures of Snake Venom Proteins Using an Integrated Mass Spectrometric Approach. J. Proteom. 2020, 216, 103680. [Google Scholar] [CrossRef]
- Hilton, G.R.; Benesch, J.L.P. Two Decades of Studying Non-Covalent Biomolecular Assemblies by Means of Electrospray Ionization Mass Spectrometry. J. R. Soc. Interface 2012, 9, 801–816. [Google Scholar] [CrossRef] [PubMed]
- Kini, R.M.; Doley, R. Structure, Function and Evolution of Three-Finger Toxins: Mini Proteins with Multiple Targets. Toxicon 2010, 56, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Deng, N.; Li, Z.; Pan, C.; Duan, H. FreeQuant: A Mass Spectrometry Label-Free Quantification Software Tool for Complex Proteome Analysis. Sci. World J. 2015, 2015, 137076. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.Y.; Tan, K.Y.; Tan, N.H.; Tan, C.H. A Neurotoxic Snake Venom without Phospholipase A2: Proteomics and Cross-Neutralization of the Venom from Senegalese Cobra, Naja senegalensis (Subgenus: Uraeus). Toxins 2021, 13, 60. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.Y.; Wong, K.Y.; Tan, N.H.; Tan, C.H. Quantitative Proteomics of Naja annulifera (Sub-Saharan Snouted cobra) Venom and Neutralization Activities of Two Antivenoms in Africa. Int. J. Biol. Macromol. 2020, 158, 605–616. [Google Scholar] [CrossRef]
- Tan, C.H.; Wong, K.Y.; Tan, N.H.; Ng, T.S.; Tan, K.Y. Distinctive Distribution of Secretory Phospholipases A2 in the Venoms of Afro-Asian Cobras (Subgenus: Naja, Afronaja, Boulengerina and Uraeus). Toxins 2019, 11, 116. [Google Scholar] [CrossRef]
- Vogel, C.W.; Fritzinger, D.C. Cobra Venom Factor: Structure, Function, and Humanization for Therapeutic Complement Depletion. Toxicon 2010, 56, 1198–1222. [Google Scholar] [CrossRef]
- Winkler, R. ESIprot: A Universal Tool for Charge State Determination and Molecular Weight Calculation of Proteins from Electrospray Ionization Mass Spectrometry Data. Rapid Commun. Mass Spectrom. 2010, 24, 285–294. [Google Scholar] [CrossRef]
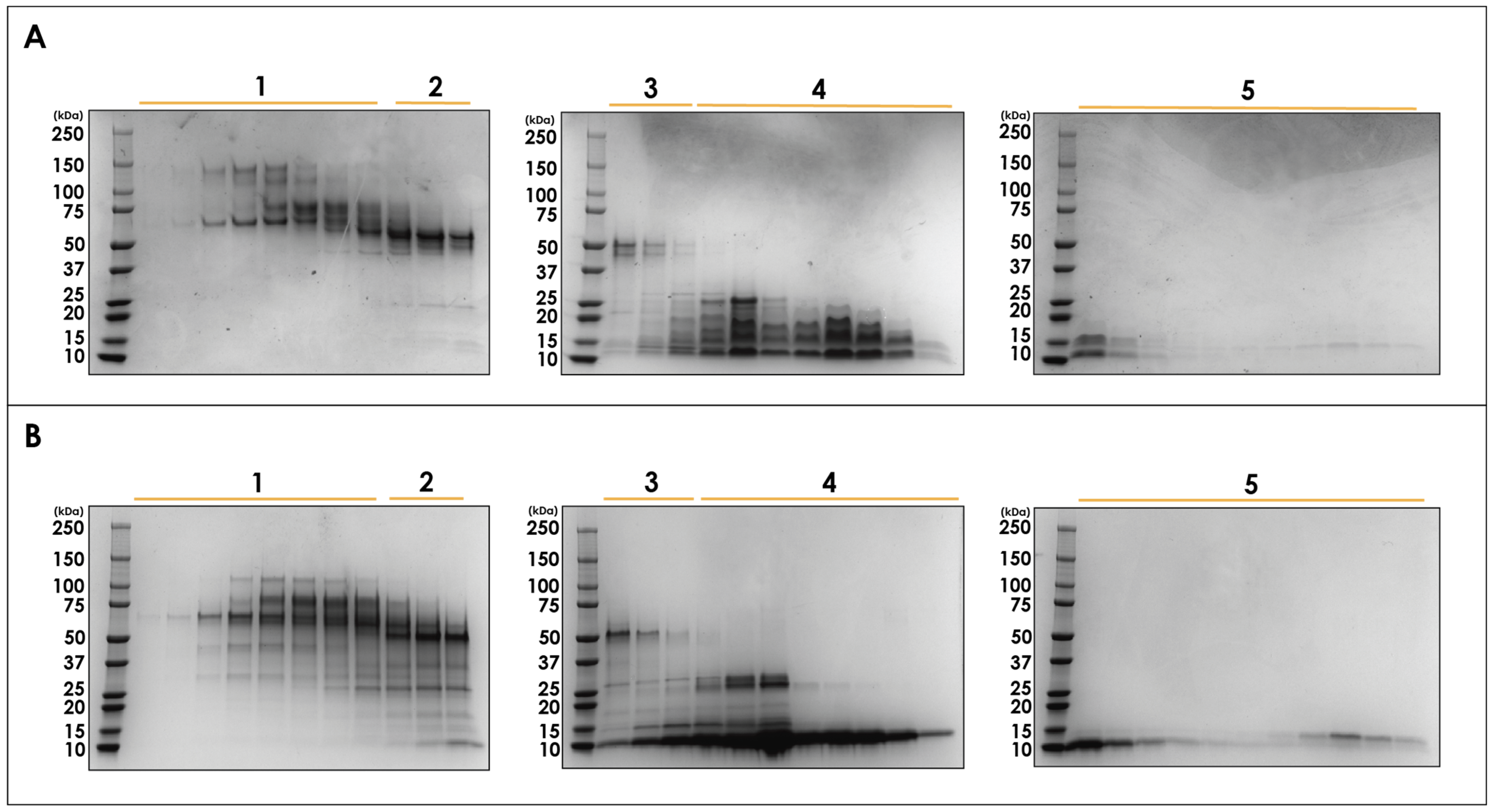


| Protein Family | Number of Proteins | R.A (%) |
|---|---|---|
| Band 1 | ||
| PDE | 3 | 0.41 |
| Band 2 | ||
| AChE | 5 | 0.26 |
| PDE | 2 | 0.08 |
| Band 3 | ||
| CVF | 7 | 0.22 |
| 5′NUC | 3 | 0.66 |
| LAAO | 4 | 0.35 |
| AChE | 14 | 0.90 |
| SVMP | 2 | 0.07 |
| Band 4 | ||
| CVF | 7 | 0.49 |
| 5′NUC | 3 | 1.02 |
| LAAO | 1 | 0.13 |
| AChE | 2 | 0.24 |
| Band 5 | ||
| CVF | 3 | 0.24 |
| House | 4 | 0.12 |
| Band 6 | ||
| CVF | 1 | 0.06 |
| Band 7 | ||
| CVF | 6 | 0.70 |
| Band 8 | ||
| No hits > 1 unique pep | - | - |
| Band 9 | ||
| SVMP | 1 | 0.12 |
| Unknown | 3 | 0.32 |
| Band 10 | ||
| SVMP | 2 | 0.03 |
| 3FTx | 6 | 2.97 |
| SVSP | 2 | 0.04 |
| Housekeeping | 6 | 0.16 |
| Unknown | 4 | 0.14 |
| Band 11 | ||
| CRISP | 7 | 0.99 |
| HYD | 1 | 0.25 |
| Housekeeping | 9 | 0.97 |
| Band 12 | ||
| CRISP | 11 | 4.76 |
| HYD | 1 | 0.16 |
| Housekeeping | 1 | 0.14 |
| Band 13 | ||
| CRISP | 1 | 0.15 |
| Band 14 | ||
| VESP | 6 | 0.48 |
| NGF | 3 | 0.85 |
| CRISP | 1 | 0.08 |
| Band 15 | ||
| VESP | 2 | 1.16 |
| NGF | 2 | 0.60 |
| PLA2 | 2 | 0.63 |
| 3FTx | 2 | 1.24 |
| Housekeeping | 3 | 0.09 |
| Band 16 | ||
| 3FTx | 5 | 7.82 |
| Housekeeping | 4 | 0.12 |
| Band 17 | ||
| 3FTx | 13 | 31.15 |
| KUN | 1 | 0.39 |
| Housekeeping | 4 | 0.18 |
| Band 18 | ||
| VESP | 5 | 0.42 |
| CRISP | 2 | 0.15 |
| HYD | 1 | 0.6 |
| Housekeeping | 5 | 0.27 |
| Band 19 | ||
| 3FTx | 13 | 29.60 |
| KUN | 1 | 0.52 |
| Housekeeping | 14 | 1.10 |
| Unknown | 2 | 1.01 |
| Band 20 | ||
| No hits > 1 unique pep | - | - |
| Band 21 | ||
| PLA2 | 4 | 0.79 |
| Housekeeping | 13 | 0.55 |
| Band 22 | ||
| 3FTx | 4 | 3.23 |
| Housekeeping | 6 | 0.19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McFarlane, L.O.; Pukala, T.L. Proteomic Investigation of Cape Cobra (Naja nivea) Venom Reveals First Evidence of Quaternary Protein Structures. Toxins 2024, 16, 63. https://doi.org/10.3390/toxins16020063
McFarlane LO, Pukala TL. Proteomic Investigation of Cape Cobra (Naja nivea) Venom Reveals First Evidence of Quaternary Protein Structures. Toxins. 2024; 16(2):63. https://doi.org/10.3390/toxins16020063
Chicago/Turabian StyleMcFarlane, Lewis O., and Tara L. Pukala. 2024. "Proteomic Investigation of Cape Cobra (Naja nivea) Venom Reveals First Evidence of Quaternary Protein Structures" Toxins 16, no. 2: 63. https://doi.org/10.3390/toxins16020063
APA StyleMcFarlane, L. O., & Pukala, T. L. (2024). Proteomic Investigation of Cape Cobra (Naja nivea) Venom Reveals First Evidence of Quaternary Protein Structures. Toxins, 16(2), 63. https://doi.org/10.3390/toxins16020063