Unveiling the Protein Components of the Secretory-Venom Gland and Venom of the Scorpion Centruroides possanii (Buthidae) through Omic Technologies
Abstract
1. Introduction
2. Results and Discussion
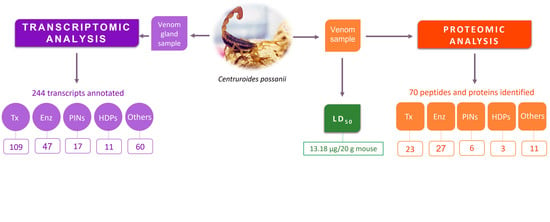
2.1. Determination of LD50 of C. possanii Venom
2.2. RNA Integrity and Sequenciation Quality
2.3. Bioinformatics
2.4. Venom Components Found in the Secretory Venom Gland of C. possanii
2.4.1. Ion Channel-Acting Toxins
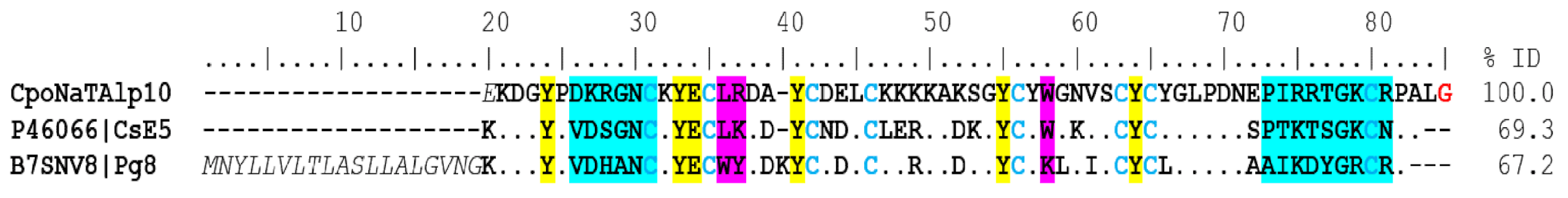
Sodium Channel-Acting Toxins
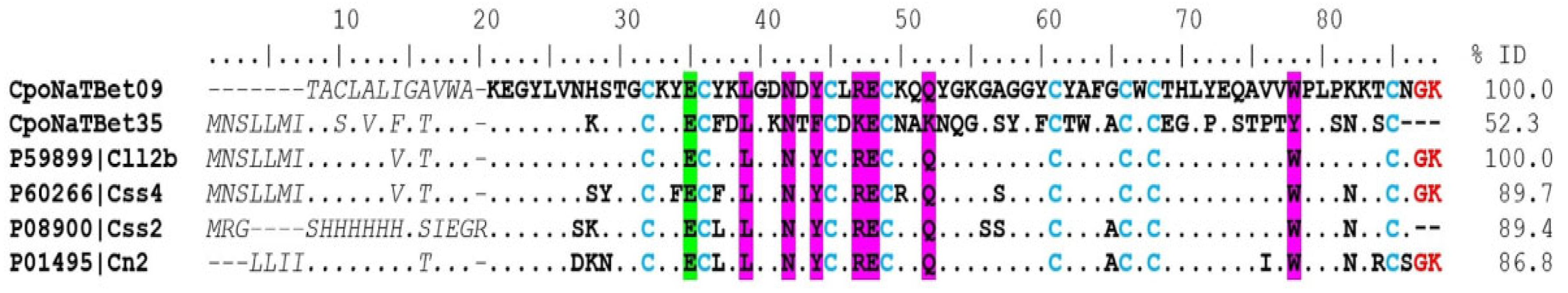
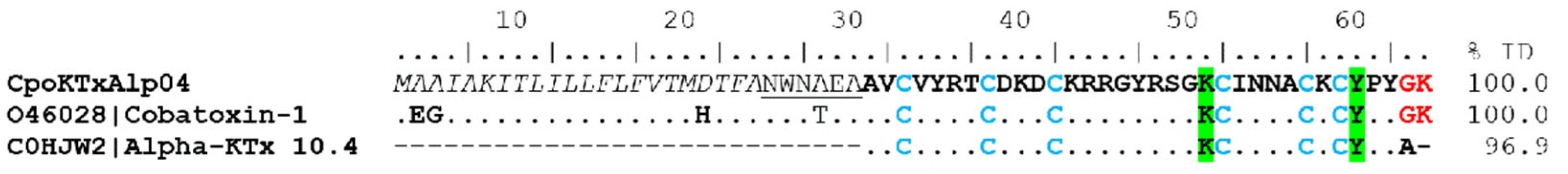
Potassium Channel-Acting Toxins (KTx)
2.4.2. Enzymes
2.4.3. Host Defense Peptides (HDPs)
Defensins
Non-Disulfide-Bridged Peptide (NDBP)
2.4.4. Peptides Inhibitors (PINs)
2.4.5. Other Components
Insulin-like Growth Factor-Binding Proteins (IGFBP)
Cysteine-Rich Secretory Proteins, Antigen 5, and Pathogenesis-Related 1 Proteins (CAP)
Orphan or Undefined Coding Transcripts
La1-like Peptides with Single-Domain von Willebrand Factor
2.5. Venom Proteomic Components of C. possanii Scorpion
2.5.1. Peptide Mass Fingerprinting
2.5.2. Proteomics: Peptides and Proteins Identified in the C. possanii Scorpion Venom
2.6. Correlation between Transcriptomics and Proteomics Results
3. Conclusions
4. Materials and Methods
4.1. Collection of Scorpions
4.2. Determination of the Medium Lethal Dose (LD50) of the Scorpion C. possanii
4.3. RNA Isolation, Sequencing and Reads Quality
4.4. Bioinformatics: Assembly, Annotation and Data Mining
4.5. Soluble Venom Collection and Quantification
4.6. Fingerprinting Mass Analysis of Soluble Venom
4.7. Proteomic Analysis of Venom
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lourenço, W.R. Scorpion Diversity and Distribution: Past and Present Patterns. In Scorpion Venoms; Springer: Dordrecht, The Netherlands, 2015; pp. 3–23. [Google Scholar]
- Lourenço, W.R. The Evolution and Distribution of Noxious Species of Scorpions (Arachnida: Scorpiones). J. Venom. Anim. Toxins Incl. Trop. Dis. 2018, 24, 1. [Google Scholar] [CrossRef] [PubMed]
- Rein, J. The Scorpion Files. Norwegian University of Science and Technology: Trondheim, Norway, 2022. Available online: https://www.ntnu.no/ub/scorpion-files/ (accessed on 29 May 2023).
- González, J.A.; Vallejo, J.R. The Scorpion in Spanish Folk Medicine: A Review of Traditional Remedies for Stings and Its Use as a Therapeutic Resource. J. Ethnopharmacol. 2013, 146, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Zhang, R.; Ge, X.; Yang, B.; Zhang, J. Analgesic Peptides in Buthus martensii Karsch: A Traditional Chinese Animal Medicine. Asian J. Tradit. Med. 2007, 2, 45–50. [Google Scholar]
- Bosmans, F.; Tytgat, J. Voltage-Gated Sodium Channel Modulation by Scorpion α-Toxins. Toxicon 2007, 49, 142–158. [Google Scholar] [CrossRef]
- González-Santillán, E.; Galán-Sánchez, M.A.; Valdez-Velázquez, L.L. A New Species of Centruroides (Scorpiones, Buthidae) from Colima, Mexico. C. R. Biol. 2019, 342, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Secretaría de Salud Datos Abiertos—Dirección General de Epidemiología. Available online: https://epidemiologia.salud.gob.mx/anuario/html/morbilidad_estatal.html (accessed on 1 June 2023).
- González-Santillán, E.; Possani, L.D. North American Scorpion Species of Public Health Importance with a Reappraisal of Historical Epidemiology. Acta Trop. 2018, 187, 264–274. [Google Scholar] [CrossRef]
- Cid-Uribe, J.I.; Meneses, E.P.; Batista, C.V.F.; Ortiz, E.; Possani, L.D. Dissecting Toxicity: The Venom Gland Transcriptome and the Venom Proteome of the Highly Venomous Scorpion Centruroides Limpidus (Karsch, 1879). Toxins 2019, 11, 247. [Google Scholar] [CrossRef]
- Riaño-Umbarila, L.; Rodríguez-Rodríguez, E.R.; Santibañez-López, C.E.; Güereca, L.; Uribe-Romero, S.J.; Gómez-Ramírez, I.V.; Cárcamo-Noriega, E.N.; Possani, L.D.; Becerril, B. Updating Knowledge on New Medically Important Scorpion Species in Mexico. Toxicon 2017, 138, 130–137. [Google Scholar] [CrossRef]
- Natsidis, P.; Schiffer, P.H.; Salvador-Martínez, I.; Telford, M.J. Computational Discovery of Hidden Breaks in 28S Ribosomal RNAs across Eukaryotes and Consequences for RNA Integrity Numbers. Sci. Rep. 2019, 9, 19477. [Google Scholar] [CrossRef]
- Jablonsky, M.J.; Watt, D.D.; Rama Krishna, N. Solution Structure of an Old World-like Neurotoxin from the Venom of the New World Scorpion Centruroides sculpturatus Ewing. J. Mol. Biol. 1995, 248, 449–458. [Google Scholar] [CrossRef]
- Rojas-Azofeifa, D.; Sasa, M.; Lomonte, B.; Diego-García, E.; Ortiz, N.; Bonilla, F.; Murillo, R.; Tytgat, J.; Díaz, C. Biochemical Characterization of the Venom of Central American Scorpion Didymocentrus krausi Francke, 1978 (Diplocentridae) and Its Toxic Effects in vivo and in vitro. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2019, 217, 54–67. [Google Scholar] [CrossRef] [PubMed]
- Stevens, M.; Peigneur, S.; Tytgat, J. Neurotoxins and Their Binding Areas on Voltage-Gated Sodium Channels. Front. Pharmacol. 2011, 2, 71. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Tobar, L.L.; Meza-Cabrera, I.A.; Sepúlveda-Arias, J.C.; Guerrero-Vargas, J.A. Comparison of the Scorpionism Caused by Centruroides margaritatus, Tityus pachyurus and Tityus n. sp. aff. metuendus Scorpion Venoms in Colombia. Toxins 2021, 13, 757. [Google Scholar] [CrossRef]
- Valdez-Velázquez, L.L.; Cid-Uribe, J.; Romero-Gutierrez, M.T.; Olamendi-Portugal, T.; Jimenez-Vargas, J.M.; Possani, L.D. Transcriptomic and Proteomic Analyses of the Venom and Venom Glands of Centruroides hirsutipalpus, a Dangerous Scorpion from Mexico. Toxicon 2020, 179, 21–32. [Google Scholar] [CrossRef]
- de Oliveira, U.C.; Nishiyama, M.Y.; dos Santos, M.B.V.; Santos-da-Silva, A.d.P.; Chalkidis, H.d.M.; Souza-Imberg, A.; Candido, D.M.; Yamanouye, N.; Dorce, V.A.C.; Junqueira-de-Azevedo, I.d.L.M. Proteomic Endorsed Transcriptomic Profiles of Venom Glands from Tityus obscurus and T. serrulatus Scorpions. PLoS ONE 2018, 13, e0193739. [Google Scholar] [CrossRef] [PubMed]
- Ward, M.J.; Ellsworth, S.A.; Rokyta, D.R. Venom-Gland Transcriptomics and Venom Proteomics of the Hentz Striped Scorpion (Centruroides hentzi; Buthidae) Reveal High Toxin Diversity in a Harmless Member of a Lethal Family. Toxicon 2018, 142, 14–29. [Google Scholar] [CrossRef]
- Martin, M.F.; Garcia y Perez, L.G.; el Ayeb, M.; Kopeyan, C.; Bechis, G.; Jover, E.; Rochat, H. Purification and Chemical and Biological Characterizations of Seven Toxins from the Mexican Scorpion, Centruroides suffusus Suffusus. J. Biol. Chem. 1987, 262, 4452–4459. [Google Scholar] [CrossRef]
- Zamudio, F.; Saavedra, R.; Martin, B.M.; Gurrola-Briones, G.; Herion, P.; Possani, L.D. Amino Acid Sequence and Immunological Characterization with Monoclonal Antibodies of Two Toxins from the Venom of the Scorpion Centruroides noxius Hoffmann. Eur. J. Biochem. 1992, 204, 281–292. [Google Scholar] [CrossRef]
- Riaño-Umbarila, L.; Ledezma-Candanoza, L.M.; Serrano-Posada, H.; Fernández-Taboada, G.; Olamendi-Portugal, T.; Rojas-Trejo, S.; Gómez-Ramírez, I.V.; Rudiño-Piñera, E.; Possani, L.D.; Becerril, B. Optimal Neutralization of Centruroides noxius Venom Is Understood through a Structural Complex between Two Antibody Fragments and the Cn2 Toxin. J. Biol. Chem. 2016, 291, 1619–1630. [Google Scholar] [CrossRef]
- Selisko, B.; Licea, A.F.; Becerril, B.; Zamudio, F.; Possani, L.D.; Horjales, E. Antibody BCF2 against Scorpion Toxin Cn2 from Centuroides noxius Hoffmann: Primary Structure and Three-Dimensional Model as Free Fv Fragment and Complexed with Its Antigen. Proteins 1999, 37, 130–143. [Google Scholar] [CrossRef]
- Schiavon, E.; Pedraza-Escalona, M.; Gurrola, G.B.; Olamendi-Portugal, T.; Corzo, G.; Wanke, E.; Possani, L.D. Negative-Shift Activation, Current Reduction and Resurgent Currents Induced by β-Toxins from Centruroides Scorpions in Sodium Channels. Toxicon 2012, 59, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Jouirou, B.; Mosbah, A.; Visan, V.; Grissmer, S.; M’Barek, S.; Fajloun, Z.; Van Rietschoten, J.; Devaux, C.; Rochat, H.; Lippens, G.; et al. Cobatoxin 1 from Centruroides noxius Scorpion Venom: Chemical Synthesis, Three-Dimensional Structure in Solution, Pharmacology and Docking on K+ Channels. Biochem. J. 2004, 377, 37–49. [Google Scholar] [CrossRef]
- Torres, A.M.; Bansal, P.; Alewood, P.F.; Bursill, J.A.; Kuchel, P.W.; Vandenberg, J.I. Solution Structure of CnErg1 (Ergtoxin), a HERG Specific Scorpion Toxin. FEBS Lett. 2003, 539, 138–142. [Google Scholar] [CrossRef]
- Arcangeli, A.; Becchetti, A. New Trends in Cancer Therapy: Targeting Ion Channels and Transporters. Pharmaceuticals 2010, 3, 1202–1224. [Google Scholar] [CrossRef]
- Mendes, L.C.; Viana, G.M.M.; Nencioni, A.L.A.; Pimenta, D.C.; Beraldo-Neto, E. Scorpion Peptides and Ion Channels: An Insightful Review of Mechanisms and Drug Development. Toxins 2023, 15, 238. [Google Scholar] [CrossRef]
- Chen, Z.; Luo, F.; Feng, J.; Yang, W.; Zeng, D.; Zhao, R.; Cao, Z.; Liu, M.; Li, W.; Jiang, L.; et al. Genomic and Structural Characterization of Kunitz-Type Peptide LmKTT-1a Highlights Diversity and Evolution of Scorpion Potassium Channel Toxins. PLoS ONE 2013, 8, e60201. [Google Scholar] [CrossRef]
- Ding, L.; Wang, X.; Liu, H.; San, M.; Xu, Y.; Li, J.; Li, S.; Cao, Z.; Li, W.; Wu, Y.; et al. A New Kunitz-Type Plasmin Inhibitor from Scorpion Venom. Toxicon 2015, 106, 7–13. [Google Scholar] [CrossRef]
- NaderiSoorki, M.; Galehdari, H.; Baradaran, M.; Jalali, A. First Venom Gland Transcriptomic Analysis of Iranian Yellow Scorpion “Odonthubuthus doriae” with Some New Findings. Toxicon 2016, 120, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Ruiming, Z.; Yibao, M.; Yawen, H.; Zhiyong, D.; Yingliang, W.; Zhijian, C.; Wenxin, L. Comparative Venom Gland Transcriptome Analysis of the Scorpion Lychas Mucronatus Reveals Intraspecific Toxic Gene Diversity and New Venomous Components. BMC Genom. 2010, 11, 452. [Google Scholar] [CrossRef] [PubMed]
- Rokyta, D.R.; Ward, M.J. Venom-Gland Transcriptomics and Venom Proteomics of the Black-Back Scorpion (Hadrurus spadix) Reveal Detectability Challenges and an Unexplored Realm of Animal Toxin Diversity. Toxicon 2017, 128, 23–37. [Google Scholar] [CrossRef]
- Oliveira, A.L.; Viegas, M.F.; da Silva, S.L.; Soares, A.M.; Ramos, M.J.; Fernandes, P.A. The Chemistry of Snake Venom and Its Medicinal Potential. Nat. Rev. Chem. 2022, 6, 451–469. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional Classification of Proteins via Subfamily Domain Architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Masood, R. The Sequence and Three-Dimensional Structure Characterization of Snake Venom Phospholipases B. Front. Mol. Biosci. 2020, 7, 175. [Google Scholar] [CrossRef] [PubMed]
- Kemparaju, K.; Girish, K.S. Snake Venom Hyaluronidase: A Therapeutic Target. Cell Biochem. Funct. 2006, 24, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Hubert, C.; Houot, A.M.; Corvol, P.; Soubrier, F. Structure of the Angiotensin I-Converting Enzyme Gene. Two Alternate Promoters Correspond to Evolutionary Steps of a Duplicated Gene. J. Biol. Chem. 1991, 266, 15377–15383. [Google Scholar] [CrossRef]
- Morgenstern, D.; Rohde, B.H.; King, G.F.; Tal, T.; Sher, D.; Zlotkin, E. The Tale of a Resting Gland: Transcriptome of a Replete Venom Gland from the Scorpion Hottentotta judaicus. Toxicon 2011, 57, 695–703. [Google Scholar] [CrossRef]
- de Oliveira, U.C.; Candido, D.M.; Coronado Dorce, V.A.; Junqueira-de-Azevedo, I.d.L.M. The Transcriptome Recipe for the Venom Cocktail of Tityus bahiensis Scorpion. Toxicon 2015, 95, 52–61. [Google Scholar] [CrossRef]
- Almeida, D.D.; Scortecci, K.C.; Kobashi, L.S.; Agnez-Lima, L.F.; Medeiros, S.R.B.; Silva-Junior, A.A.; Junqueira-de-Azevedo, I.d.L.M.; Fernandes-Pedrosa, M.d.F. Profiling the Resting Venom Gland of the Scorpion Tityus stigmurus through a Transcriptomic Survey. BMC Genom. 2012, 13, 362. [Google Scholar] [CrossRef]
- Alvarenga, E.R.; Mendes, T.M.; Magalhaes, B.F.; Siqueira, F.F.; Dantas, A.E.; Barroca, T.M.; Horta, C.C.; Kalapothakis, E. Transcriptome Analysis of the Tityus serrulatus Scorpion Venom Gland. Open J. Genet. 2012, 2, 210–220. [Google Scholar] [CrossRef]
- Goudarzi, M.H.; Eagles, D.A.; Lim, J.; Biggs, K.A.; Kotze, A.C.; Ruffell, A.P.; Fairlie, D.P.; King, G.F.; Walker, A.A. Venom Composition and Bioactive RF-Amide Peptide Toxins of the Saddleback caterpillar, Acharia stimulea (Lepidoptera: Limacodidae). Biochem. Pharmacol. 2023, 213, 115598. [Google Scholar] [CrossRef]
- Vaiyapuri, S.; Wagstaff, S.C.; Harrison, R.A.; Gibbins, J.M.; Hutchinson, E.G. Evolutionary Analysis of Novel Serine Proteases in the Venom Gland Transcriptome of Bitis Gabonica Rhinoceros. PLoS ONE 2011, 6, e21532. [Google Scholar] [CrossRef]
- Baradaran, M.; Salabi, F. Identification, Classification and Characterization of the Dermonecrotic Toxins in Venom Glands of Hottentotta saulcyi, Androctonus crassicauda and Hemiscorpius lepturus Using Transcriptome Analysis. Res. Sq. 01 February 2023, preprint, 1–32. [Google Scholar] [CrossRef]
- Fox, J.W. A Brief Review of the Scientific History of Several Lesser-Known Snake Venom Proteins: L-Amino Acid Oxidases, Hyaluronidases and Phosphodiesterases. Toxicon 2013, 62, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Pedroso, A.; Matioli, S.R.; Murakami, M.T.; Pidde-Queiroz, G.; Tambourgi, D.V. Adaptive Evolution in the Toxicity of a Spider’s Venom Enzymes. BMC Evol. Biol. 2015, 15, 290. [Google Scholar] [CrossRef]
- Boto, A.; Pérez de la Lastra, J.; González, C. The Road from Host-Defense Peptides to a New Generation of Antimicrobial Drugs. Molecules 2018, 23, 311. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Zhao, Y.; Qu, D.; Xie, Z.; Guo, X.; Zhu, Z.; Chen, Z.; Zhang, L.; Li, W.; Cao, Z.; et al. Ion Channel Modulation by Scorpion Hemolymph and Its Defensin Ingredients Highlights Origin of Neurotoxins in Telson Formed in Paleozoic Scorpions. Int. J. Biol. Macromol. 2020, 148, 351–363. [Google Scholar] [CrossRef]
- Cociancich, S.; Goyffon, M.; Bontems, F.; Bulet, P.; Bouet, F.; Menez, A.; Hoffmann, J. Purification and Characterization of a Scorpion Defensin, a 4 kDa Antibacterial Peptide Presenting Structural Similarities with Insect Defensins and Scorpion Toxins. Biochem. Biophys. Res. Commun. 1993, 194, 17–22. [Google Scholar] [CrossRef]
- Ferreira, L.A.F.; Alves, E.W.; Henriques, O.B. Peptide T, a Novel Bradykinin Potentiator Isolated from Tityus serrulatus Scorpion Venom. Toxicon 1993, 31, 941–947. [Google Scholar] [CrossRef]
- Fan, Z.; Cao, L.; He, Y.; Hu, J.; Di, Z.; Wu, Y.; Li, W.; Cao, Z. Ctriporin, a New Anti-Methicillin-Resistant Staphylococcus aureus Peptide from the Venom of the Scorpion Chaerilus tricostatus. Antimicrob. Agents Chemother. 2011, 55, 5220–5229. [Google Scholar] [CrossRef]
- Hoskin, D.W.; Ramamoorthy, A. Studies on Anticancer Activities of Antimicrobial Peptides. Biochim. Biophys. Acta-Biomembr. 2008, 1778, 357–375. [Google Scholar] [CrossRef]
- Almaaytah, A.; Albalas, Q. Scorpion Venom Peptides with No Disulfide Bridges: A Review. Peptides 2014, 51, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.-C.; Corzo, G.; Hahin, R. Scorpion Venom Peptides without Disulfide Bridges. IUBMB Life Int. Union Biochem. Mol. Biol. Life 2005, 57, 13–21. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.-C.; Nie, Y.; Luo, X.; Wu, S.; Shi, W.; Zhang, L.; Liu, Y.; Cao, H.; Yang, Y.; Zhou, J. Molecular and Bioinformatical Characterization of a Novel Superfamily of Cysteine-Rich Peptides from Arthropods. Peptides 2013, 41, 45–58. [Google Scholar] [CrossRef]
- Law, R.H.P.; Zhang, Q.; McGowan, S.; Buckle, A.M.; Silverman, G.A.; Wong, W.; Rosado, C.J.; Langendorf, C.G.; Pike, R.N.; Bird, P.I.; et al. An Overview of the Serpin Superfamily. Genome Biol. 2006, 7, 216. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, L.; Shi, W.; Zeng, X.-C.; Ge, F.; Yang, M.; Nie, Y.; Bao, A.; Wu, S.; E, G. Unique Diversity of the Venom Peptides from the Scorpion Androctonus bicolor Revealed by Transcriptomic and Proteomic Analysis. J. Proteom. 2015, 128, 231–250. [Google Scholar] [CrossRef]
- Reddy, T.; Gibbs, G.M.; Merriner, D.J.; Kerr, J.B.; O’Bryan, M.K. Cysteine-Rich Secretory Proteins Are Not Exclusively Expressed in the Male Reproductive Tract. Dev. Dyn. 2008, 237, 3313–3323. [Google Scholar] [CrossRef]
- Fry, B.G.; Vidal, N.; Norman, J.A.; Vonk, F.J.; Scheib, H.; Ramjan, S.F.R.; Kuruppu, S.; Fung, K.; Blair Hedges, S.; Richardson, M.K.; et al. Early Evolution of the Venom System in Lizards and Snakes. Nature 2006, 439, 584–588. [Google Scholar] [CrossRef]
- Milne, T.J.; Abbenante, G.; Tyndall, J.D.A.; Halliday, J.; Lewis, R.J. Isolation and Characterization of a Cone Snail Protease with Homology to CRISP Proteins of the Pathogenesis-Related Protein Superfamily. J. Biol. Chem. 2003, 278, 31105–31110. [Google Scholar] [CrossRef]
- Zhang, Q.; Xu, J.; Zhou, X.; Liu, Z. CAP Superfamily Proteins from Venomous Animals: Who We Are and What to Do? Int. J. Biol. Macromol. 2022, 221, 691–702. [Google Scholar] [CrossRef]
- Qin, N.; Sun, H.; Lu, M.; Wang, J.; Tang, T.; Liu, F. A Single von Willebrand Factor C-Domain Protein Acts as an Extracellular Pattern-Recognition Receptor in the River Prawn Macrobrachium Nipponense. J. Biol. Chem. 2020, 295, 10468–10477. [Google Scholar] [CrossRef] [PubMed]
- Dixon, W.J. The Up-and-Down Method for Small Samples. J. Am. Stat. Assoc. 1965, 60, 967–978. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De Novo Transcript Sequence Reconstruction from RNA-Seq Using the Trinity Platform for Reference Generation and Analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Bryant, D.M.; Johnson, K.; DiTommaso, T.; Tickle, T.; Couger, M.B.; Payzin-Dogru, D.; Lee, T.J.; Leigh, N.D.; Kuo, T.-H.; Davis, F.G.; et al. A Tissue-Mapped Axolotl De Novo Transcriptome Enables Identification of Limb Regeneration Factors. Cell Rep. 2017, 18, 762–776. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef] [PubMed]






| Entry | Quantity |
|---|---|
| Sequence length | 76 base pairs (bp) |
| Reads | 19,158,736 |
| Total transcripts | 835,204 |
| Annotated transcripts | 720,463 |
| Putative transcripts | 598,252 |
| Venom transcripts | 434,492 |
| No venom transcripts | 163,760 |
| Pfam annotation | 28,399 |
| Pfam scorpion domain | 56 |
| Pfam scorpion venom domain | 50 |
| GO terms | 18.4% cell components 30.8% biological process 50.7% molecular function |
| Family | Subfamily | Number of Coding Transcripts |
|---|---|---|
| Toxins | α-NaTx | 21 |
| β-NaTx | 50 | |
| α-KTx | 20 | |
| β-KTx | 1 | |
| γ-KTx | 5 | |
| δ-KTx | 12 | |
| Enzymes | PA1 | 4 |
| PA2 | 4 | |
| PLB | 1 | |
| MtP | 16 | |
| SeP | 14 | |
| Hya | 1 | |
| CiP | 2 | |
| PDE | 3 | |
| Mon | 2 | |
| Inhibitory peptides | SERPIN | 5 |
| TIL | 11 | |
| Kunitz type | 1 | |
| Defense peptides | Defensin | 10 |
| NDBP-3 | 1 | |
| Other components | CAP | 7 |
| IGFBP | 27 | |
| La1-like | 7 | |
| Und | 19 | |
| Total | 244 |
| Family | Subfamily | Number of Peptides and Proteins |
|---|---|---|
| Toxins | α-NaTx | 4 |
| β-NaTx | 14 | |
| α-KTx | 3 | |
| β-KTx | 1 | |
| γ-KTx | 1 | |
| Enzymes | PA1 | 3 |
| PA2 | 1 | |
| PLB | 1 | |
| MtP | 11 | |
| SeP | 4 | |
| Hya | 1 | |
| CiP | 1 | |
| PDE | 3 | |
| Mon | 2 | |
| Inhibitory peptides | SERPIN | 4 |
| TIL | 2 | |
| Defense peptides | Defensin | 2 |
| NDBP-3 | 1 | |
| Other components | CAP | 4 |
| IGFBP | 1 | |
| La1-like | 1 | |
| Und | 5 | |
| Total | 70 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Villalvazo, P.E.; Jiménez-Vargas, J.M.; Lino-López, G.J.; Meneses, E.P.; Bermúdez-Guzmán, M.d.J.; Barajas-Saucedo, C.E.; Delgado Enciso, I.; Possani, L.D.; Valdez-Velazquez, L.L. Unveiling the Protein Components of the Secretory-Venom Gland and Venom of the Scorpion Centruroides possanii (Buthidae) through Omic Technologies. Toxins 2023, 15, 498. https://doi.org/10.3390/toxins15080498
García-Villalvazo PE, Jiménez-Vargas JM, Lino-López GJ, Meneses EP, Bermúdez-Guzmán MdJ, Barajas-Saucedo CE, Delgado Enciso I, Possani LD, Valdez-Velazquez LL. Unveiling the Protein Components of the Secretory-Venom Gland and Venom of the Scorpion Centruroides possanii (Buthidae) through Omic Technologies. Toxins. 2023; 15(8):498. https://doi.org/10.3390/toxins15080498
Chicago/Turabian StyleGarcía-Villalvazo, Patricia Elizabeth, Juana María Jiménez-Vargas, Gisela Jareth Lino-López, Erika Patricia Meneses, Manuel de Jesús Bermúdez-Guzmán, Carlos Eduardo Barajas-Saucedo, Iván Delgado Enciso, Lourival Domingos Possani, and Laura Leticia Valdez-Velazquez. 2023. "Unveiling the Protein Components of the Secretory-Venom Gland and Venom of the Scorpion Centruroides possanii (Buthidae) through Omic Technologies" Toxins 15, no. 8: 498. https://doi.org/10.3390/toxins15080498
APA StyleGarcía-Villalvazo, P. E., Jiménez-Vargas, J. M., Lino-López, G. J., Meneses, E. P., Bermúdez-Guzmán, M. d. J., Barajas-Saucedo, C. E., Delgado Enciso, I., Possani, L. D., & Valdez-Velazquez, L. L. (2023). Unveiling the Protein Components of the Secretory-Venom Gland and Venom of the Scorpion Centruroides possanii (Buthidae) through Omic Technologies. Toxins, 15(8), 498. https://doi.org/10.3390/toxins15080498