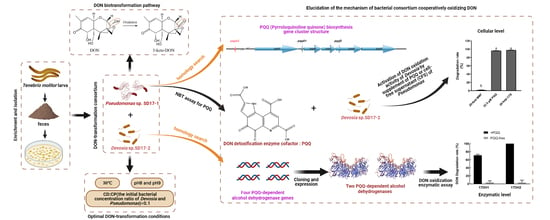
Biotransformation of Deoxynivalenol by a Dual-Member Bacterial Consortium Isolated from Tenebrio molitor Larval Feces
Abstract
1. Introduction
2. Results
2.1. Isolation and Identification of the DON-Transforming Bacterial Community
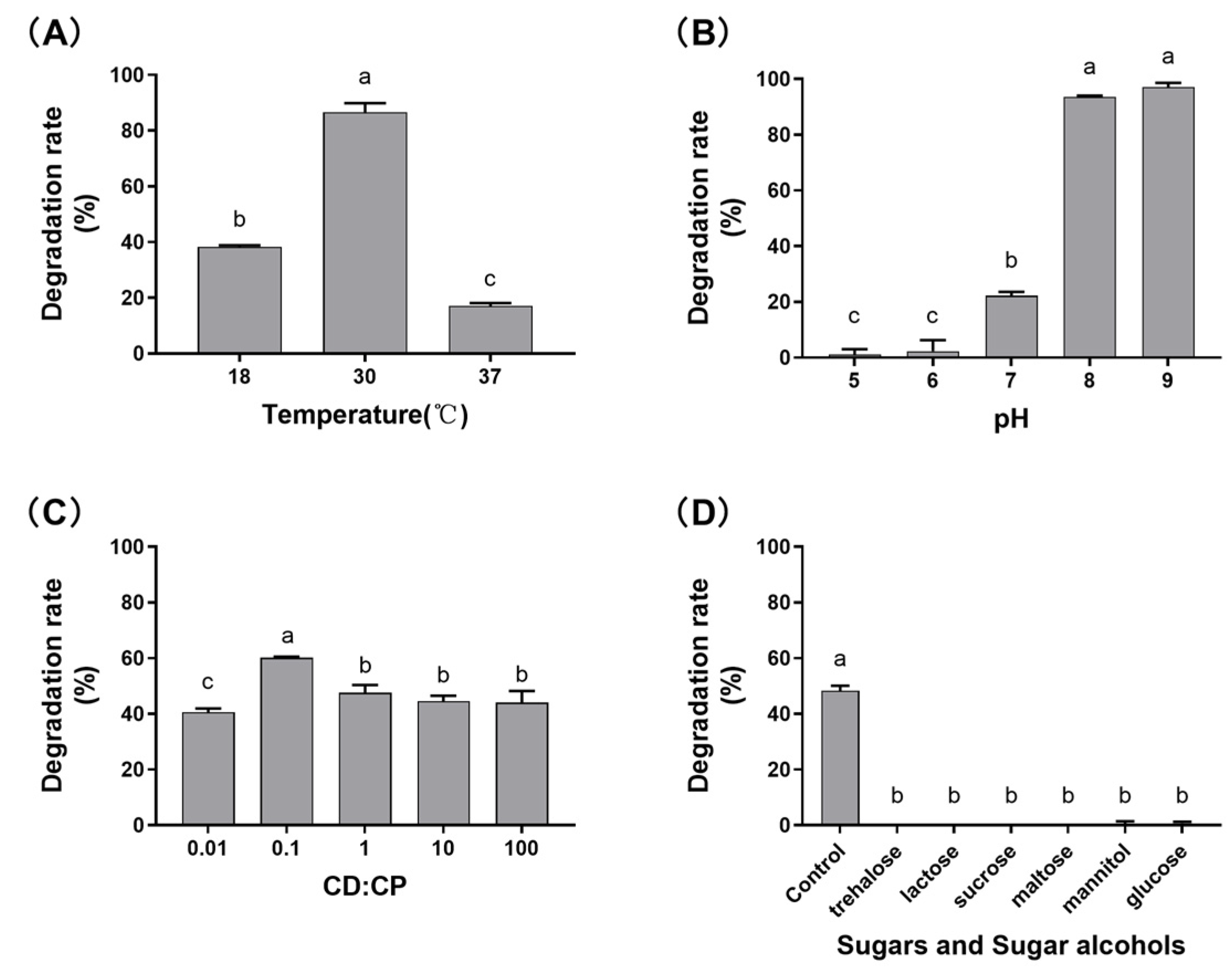
2.2. Characterization of the Effects of Different Factors on DON Transformation Activity
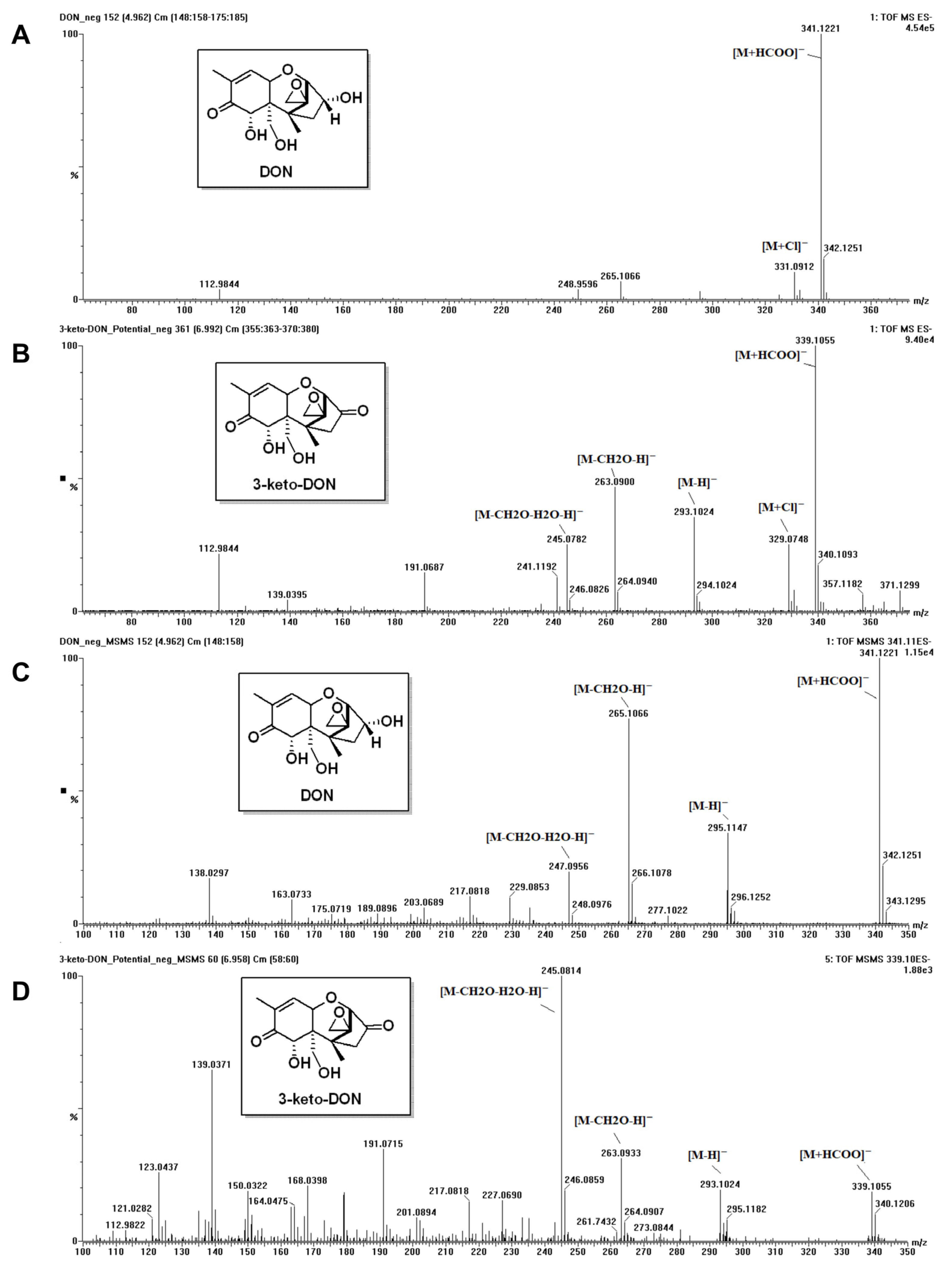
2.3. Determination of DON Metabolite’ Chemical Structure
2.4. Elucidation the Underlying Mechanisms of DON Biotransformation by the Microbial Consortium SD
2.4.1. Sequencing, Assembly, and Annotation of the Genomes of Strains SD17-1 and SD17-2
2.4.2. Homology Search for PQQ Biosynthetic Gene Cluster in Pseudomonas sp. 17-1 and PQQ-Dependent ADH Genes in Devosia sp. 17-2
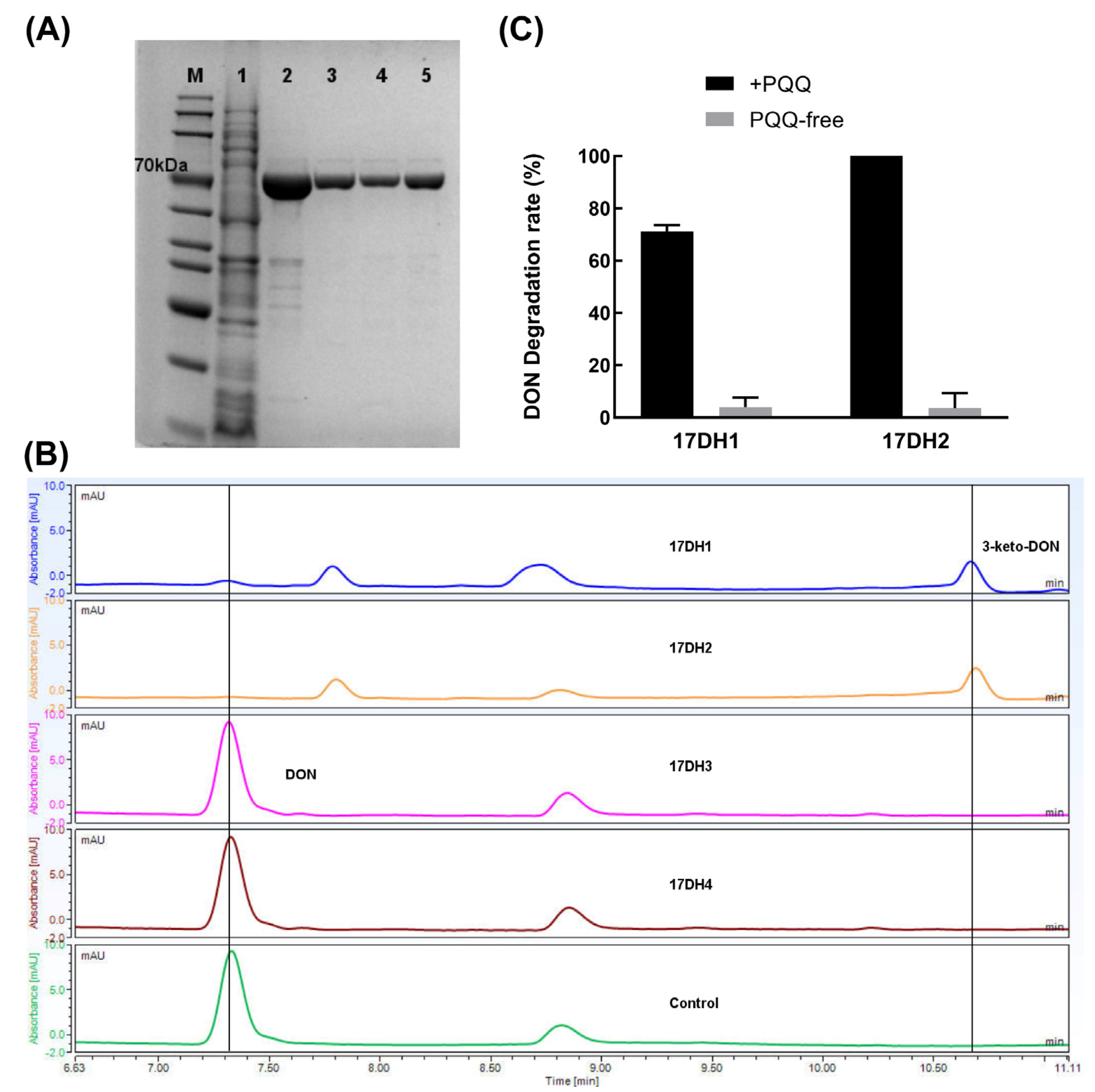
2.4.3. Biochemical Evidence for Pesudomonas and Devosia as Producer of PQQ and DON Dehydrogenase
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Reagent and Culture Medium
5.2. Screening and Identification of the DON-Transforming Bacterial Strains
5.3. Analysis of DON and Its Metabolite
5.3.1. Ultra-Performance Liquid Chromatography (UPLC) Analysis for DON
5.3.2. MS (Mass Spectrometry) Analysis of DON Metabolite
5.4. Effects of Various Factors on DON Transformation Rate
5.5. Elucidation of the Mechanism of DON Degradation by Microbial Community SD
5.5.1. Genomic Sequencing and Analysis
5.5.2. Identification of Two PQQ-Dependent DON Dehydrogenases from Devosia sp. SD17-2
5.5.3. NBT Assay for PQQ Produced by Pseudomonas sp. SD17-1
5.5.4. Activation of DON Oxidation Activity of Devosia sp. SD17-2 by PQQ Supplement
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mishra, S.; Srivastava, S.; Dewangan, J.; Divakar, A.; Kumar Rath, S. Global occurrence of deoxynivalenol in food commodities and exposure risk assessment in humans in the last decade: A survey. Crit. Rev. Food Sci. Nutr. 2020, 60, 1346–1374. [Google Scholar] [CrossRef]
- Tima, H.; Berkics, A.; Hannig, Z.; Ittzés, A.; Kecskésné Nagy, E.; Mohácsi-Farkas, C.; Kiskó, G. Deoxynivalenol in wheat, maize, wheat flour and pasta: Surveys in Hungary in 2008–2015. Food Addit. Contam. Part B Surveill. 2018, 11, 37–42. [Google Scholar] [CrossRef]
- Sobrova, P.; Adam, V.; Vasatkova, A.; Beklova, M.; Zeman, L.; Kizek, R. Deoxynivalenol and its toxicity. Interdiscip. Toxicol. 2010, 3, 94–99. [Google Scholar] [CrossRef]
- Liao, Y.; Peng, Z.; Chen, L.; Nüssler, A.K.; Liu, L.; Yang, W. Deoxynivalenol, gut microbiota and immunotoxicity: A potential approach? Food Chem. Toxicol. 2018, 112, 342–354. [Google Scholar] [CrossRef]
- Murata, H.; Yamaguchi, D.; Nagai, A.; Shimada, N. Reduction of deoxynivalenol contaminating corn silage by short-term ultraviolet irradiation: A pilot study. J. Vet. Med. Sci. 2011, 73, 1059–1060. [Google Scholar] [CrossRef]
- Bretz, M.; Beyer, M.; Cramer, B.; Knecht, A.; Humpf, H.-U. Thermal Degradation of the Fusarium Mycotoxin Deoxynivalenol. J. Agric. Food Chem. 2006, 54, 6445–6451. [Google Scholar] [CrossRef]
- Sabater-Vilar, M.; Malekinejad, H.; Selman, M.H.J.; van der Doelen, M.A.M.; Fink-Gremmels, J. In vitro assessment of adsorbents aiming to prevent deoxynivalenol and zearalenone mycotoxicoses. Mycopathologia 2007, 163, 81–90. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, Y.; Liu, S.; Wu, Y.; Zhou, Q.; Zhang, Y.; Zheng, X.; Han, Y.; Xie, C.; Liu, N. Adsorption of deoxynivalenol by pillared montmorillonite. Food Chem. 2021, 343, 128391. [Google Scholar] [CrossRef]
- Wang, L.; Shao, H.; Luo, X.; Wang, R.; Li, Y.; Li, Y.; Luo, Y.; Chen, Z. Effect of Ozone Treatment on Deoxynivalenol and Wheat Quality. PLoS ONE 2016, 11, e0147613. [Google Scholar] [CrossRef]
- Hassan, Y.I.; He, J.W.; Perilla, N.; Tang, K.; Karlovsky, P.; Zhou, T. The enzymatic epimerization of deoxynivalenol by Devosia mutans proceeds through the formation of 3-keto-DON intermediate. Sci. Rep. 2017, 7, 6929. [Google Scholar] [CrossRef]
- Wang, G.; Wang, Y.; Ji, F.; Xu, L.; Yu, M.; Shi, J.; Xu, J. Biodegradation of deoxynivalenol and its derivatives by Devosia insulae A16. Food Chem. 2019, 276, 436–442. [Google Scholar] [CrossRef]
- Ikunaga, Y.; Sato, I.; Grond, S.; Numaziri, N.; Yoshida, S.; Yamaya, H.; Hiradate, S.; Hasegawa, M.; Toshima, H.; Koitabashi, M.; et al. Nocardioides sp. strain WSN05-2, isolated from a wheat field, degrades deoxynivalenol, producing the novel intermediate 3-epi-deoxynivalenol. Appl. Microbiol. Biotechnol. 2011, 89, 419–427. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, H.; Qin, X.; Wang, X.; Wang, Y.; Bin, Y.; Xie, X.; Zheng, F.; Luo, H. Biodegradation of Deoxynivalenol by Nocardioides sp. ZHH-013: 3-keto-Deoxynivalenol and 3-epi-Deoxynivalenol as Intermediate Products. Front. Microbiol. 2021, 12, 658421. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, H.H.; Zhao, C.; Han, Y.T.; Liu, Y.C.; Zhang, X.L. Isolation and characterization of a novel deoxynivalenol-transforming strain Paradevosia shaoguanensis DDB001 from wheat field soil. Lett. Appl. Microbiol. 2017, 65, 414–422. [Google Scholar] [CrossRef]
- Shima, J.; Takase, S.; Takahashi, Y.; Iwai, Y.; Fujimoto, H.; Yamazaki, M.; Ochi, K. Novel detoxification of the trichothecene mycotoxin deoxynivalenol by a soil bacterium isolated by enrichment culture. Appl. Environ. Microbiol. 1997, 63, 3825–3830. [Google Scholar] [CrossRef]
- Zhang, J.; Qin, X.; Guo, Y.; Zhang, Q.; Ma, Q.; Ji, C.; Zhao, L. Enzymatic degradation of deoxynivalenol by a novel bacterium, Pelagibacterium halotolerans ANSP101. Food Chem. Toxicol. 2020, 140, 111276. [Google Scholar] [CrossRef]
- Ito, M.; Sato, I.; Ishizaka, M.; Yoshida, S.; Koitabashi, M.; Yoshida, S.; Tsushima, S. Bacterial Cytochrome P450 System Catabolizing the Fusarium Toxin Deoxynivalenol. Appl. Environ. Microbiol. 2013, 79, 1619–1628. [Google Scholar] [CrossRef]
- He, W.-J.; Zhang, L.; Yi, S.-Y.; Tang, X.-L.; Yuan, Q.-S.; Guo, M.-W.; Wu, A.-B.; Qu, B.; Li, H.-P.; Liao, Y.-C. An aldo-keto reductase is responsible for Fusarium toxin-degrading activity in a soil Sphingomonas strain. Sci. Rep. 2017, 7, 9549. [Google Scholar] [CrossRef]
- Wang, S.; Hou, Q.; Guo, Q.-Q.; Zhang, J.; Sun, Y.; Wei, H.; Shen, L. Isolation and Characterization of a Deoxynivalenol-Degrading Bacterium Bacillus licheniformis YB9 with the Capability of Modulating Intestinal Microbial Flora of Mice. Toxins 2020, 12, 184. [Google Scholar] [CrossRef]
- Li, B.; Duan, J.; Ren, J.; Francis, F.; Li, G. Isolation and Characterization of Two New Deoxynivalenol-Degrading Strains, Bacillus sp. HN117 and Bacillus sp. N22. Toxins 2022, 14, 781. [Google Scholar] [CrossRef]
- Ito, M.; Sato, I.; Koitabashi, M.; Yoshida, S.; Imai, M.; Tsushima, S. A novel actinomycete derived from wheat heads degrades deoxynivalenol in the grain of wheat and barley affected by Fusarium head blight. Appl. Microbiol. Biotechnol. 2012, 96, 1059–1070. [Google Scholar] [CrossRef]
- Qu, R.; Jiang, C.; Wu, W.; Pang, B.; Lei, S.; Lian, Z.; Shao, D.; Jin, M.; Shi, J. Conversion of DON to 3-epi-DON in vitro and toxicity reduction of DON in vivo by Lactobacillus rhamnosus. Food Funct. 2019, 10, 2785–2796. [Google Scholar] [CrossRef]
- Fuchs, E.; Binder, E.M.; Heidler, D.; Krska, R. Structural characterization of metabolites after the microbial degradation of type A trichothecenes by the bacterial strain BBSH 797. Food Addit. Contam. 2002, 19, 379–386. [Google Scholar] [CrossRef]
- Gao, X.; Mu, P.; Zhu, X.; Chen, X.; Tang, S.; Wu, Y.; Miao, X.; Wang, X.; Wen, J.; Deng, Y. Dual Function of a Novel Bacterium, Slackia sp. D-G6: Detoxifying Deoxynivalenol and Producing the Natural Estrogen Analogue, Equol. Toxins 2020, 12, 85. [Google Scholar] [CrossRef]
- He, W.-J.; Shi, M.-M.; Yang, P.; Huang, T.; Yuan, Q.-S.; Yi, S.-Y.; Wu, A.-B.; Li, H.-P.; Gao, C.-B.; Zhang, J.-B.; et al. Novel Soil Bacterium Strain Desulfitobacterium sp. PGC-3-9 Detoxifies Trichothecene Mycotoxins in Wheat via De-Epoxidation under Aerobic and Anaerobic Conditions. Toxins 2020, 12, 363. [Google Scholar] [CrossRef]
- Ahad, R.; Zhou, T.; Lepp, D.; Pauls, K.P. Microbial detoxification of eleven food and feed contaminating trichothecene mycotoxins. BMC Biotechnol. 2017, 17, 30. [Google Scholar] [CrossRef]
- He, W.; Yuan, Q.-S.; Zhang, Y.; Guo, M.; Gong, A.; Zhang, J.; Wu, A.; Huang, T.; Qu, B.; Li, H.; et al. Aerobic De-Epoxydation of Trichothecene Mycotoxins by a Soil Bacterial Consortium Isolated Using In Situ Soil Enrichment. Toxins 2016, 8, 277. [Google Scholar] [CrossRef]
- Zhai, Y.; Zhong, L.; Gao, H.; Lu, Z.; Bie, X.; Zhao, H.; Zhang, C.; Lu, F. Detoxification of Deoxynivalenol by a Mixed Culture of Soil Bacteria With 3-epi-Deoxynivalenol as the Main Intermediate. Front Microbiol. 2019, 10, 2172. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, J.; Dai, Y.; Wang, Y.; Shi, J.; Wang, G.; Xu, J.; De Saeger, S. Design and characterization of an artificial two-strain bacterial consortium for the efficient biodegradation of deoxynivalenol. Biol. Control. 2023, 179, 105172. [Google Scholar] [CrossRef]
- Gao, H.; Niu, J.; Yang, H.; Lu, Z.; Zhou, L.; Meng, F.; Lu, F.; Chen, M. Epimerization of Deoxynivalenol by the Devosia Strain A6-243 Assisted by Pyrroloquinoline Quinone. Toxins 2022, 14, 16. [Google Scholar] [CrossRef]
- Carere, J.; Hassan, Y.I.; Lepp, D.; Zhou, T. The enzymatic detoxification of the mycotoxin deoxynivalenol: Identification of DepA from the DON epimerization pathway. Microb. Biotechnol. 2018, 11, 1106–1111. [Google Scholar] [CrossRef]
- Carere, J.; Hassan, Y.I.; Lepp, D.; Zhou, T. The Identification of DepB: An Enzyme Responsible for the Final Detoxification Step in the Deoxynivalenol Epimerization Pathway in Devosia mutans 17-2-E-8. Front. Microbiol. 2018, 9, 1573. [Google Scholar] [CrossRef]
- Yang, S.; Wu, W.; Brandon, A.M.; Fan, H.; Receveur, J.P.; Li, Y.; Wang, Z.; Fan, R.; McClellan, R.L.; Gao, S.-H.; et al. Ubiquity of polystyrene digestion and biodegradation within yellow mealworms, larvae of Tenebrio molitor Linnaeus (Coleoptera: Tenebrionidae). Chemosphere 2018, 212, 262–271. [Google Scholar] [CrossRef]
- Brai, A.; Poggialini, F.; Trivisani, C.I.; Vagaggini, C.; Tarchi, F.; Francardi, V.; Dreassi, E. Efficient use of agricultural waste to naturally fortify Tenebrio molitor mealworms and evaluation of their nutraceutical properties. J. Insects Food Feed. 2022, 9, 599–610. [Google Scholar] [CrossRef]
- Brai, A.; Poggialini, F.; Vagaggini, C.; Pasqualini, C.; Simoni, S.; Francardi, V.; Dreassi, E. Tenebrio molitor as a Simple and Cheap Preclinical Pharmacokinetic and Toxicity Model. Int. J. Mol. Sci. 2023, 24, 2296. [Google Scholar] [CrossRef]
- Niermans, K.; Woyzichovski, J.; Kröncke, N.; Benning, R.; Maul, R. Feeding study for the mycotoxin zearalenone in yellow mealworm (Tenebrio molitor) larvae—Investigation of biological impact and metabolic conversion. Mycotoxin Res. 2019, 35, 231–242. [Google Scholar] [CrossRef]
- Dilshaan, D. Yellow Mealworm Larvae (Tenebrio molitor) Grown on Deoxynivalenol-Contaminated Wheat as a Feed Ingredient for Broiler Chickens. PhD Thesis, University of Saskatchewan, Saskatoon, SK, Canada, 2021. [Google Scholar]
- Zhao, D.; Xie, H.; Gao, L.; Zhang, J.; Li, Y.; Mao, G.; Zhang, H.; Wang, F.; Lam, S.S.; Song, A. Detoxication and bioconversion of aflatoxin B1 by yellow mealworms (Tenebrio molitor): A sustainable approach for valuable larval protein production from contaminated grain. Ecotoxicol. Environ. Saf. 2022, 242, 113935. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, D.; Zhang, W.; Wang, S.; Wu, Y.; Wang, S.; Yang, Y.; Guo, B. Four PQQ-Dependent Alcohol Dehydrogenases Responsible for the Oxidative Detoxification of Deoxynivalenol in a Novel Bacterium Ketogulonicigenium vulgare D3_3 Originated from the Feces of Tenebrio molitor Larvae. Toxins 2023, 15, 367. [Google Scholar] [CrossRef]
- Ge, X.; Wang, W.; Du, B.; Wang, J.; Xiong, X.; Zhang, W. Multiple pqqA genes respond differently to environment and one contributes dominantly to pyrroloquinoline quinone synthesis. J. Basic Microbiol. 2015, 55, 312–323. [Google Scholar] [CrossRef]
- He, W.-J.; Shi, M.-M.; Yang, P.; Huang, T.; Zhao, Y.; Wu, A.-B.; Dong, W.-B.; Li, H.-P.; Zhang, J.-B.; Liao, Y.-C. A quinone-dependent dehydrogenase and two NADPH-dependent aldo/keto reductases detoxify deoxynivalenol in wheat via epimerization in a Devosia strain. Food Chem. 2020, 321, 126703. [Google Scholar] [CrossRef]
- Yu, H.; Zhou, T.; Gong, J.; Young, C.; Su, X.; Li, X.-Z.; Zhu, H.; Tsao, R.; Yang, R. Isolation of deoxynivalenol-transforming bacteria from the chicken intestines using the approach of PCR-DGGE guided microbial selection. BMC Microbiol. 2010, 10, 182. [Google Scholar] [CrossRef]
- He, J.W. Detoxification of Deoxynivalenol by a Soil Bacterium Devosia mutans 17-2-E-8. PhD Thesis, University of Guelph, Guelph, ON, Canada, 2015. [Google Scholar]
- Li, X.-Z.; Hassan, Y.I.; Lepp, D.; Zhu, Y.; Zhou, T. 3-keto-DON, but Not 3-epi-DON, Retains the in Planta Toxicological Potential after the Enzymatic Biotransformation of Deoxynivalenol. Int. J. Mol. Sci. 2022, 23, 7230. [Google Scholar] [CrossRef]
- Sarmiento-Pavía, P.D.; Sosa-Torres, M.E. Bioinorganic insights of the PQQ-dependent alcohol dehydrogenases. J. Biol. Inorg. Chem. 2021, 26, 177–203. [Google Scholar] [CrossRef]
- Studier, F.W. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, S. Complete Genome Sequence of Paradevosia shaoguanensis Type Strain J5-3, Obtained Using Nanopore and Illumina Sequencing Technologies. Microbiol. Resour. Announc. 2021, 10, e0009921. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Xie, Y.; Li, H.; Luo, X.; Li, H.; Gao, Q.; Zhang, L.; Teng, Y.; Zhao, Q.; Zuo, Z.; Ren, J. IBS 2.0: An upgraded illustrator for the visualization of biological sequences. Nucleic Acids Res. 2022, 50, W420–W426. [Google Scholar] [CrossRef]
- Xia, Y.; Li, K.; Li, J.; Wang, T.; Gu, L.; Xun, L. T5 exonuclease-dependent assembly offers a low-cost method for efficient cloning and site-directed mutagenesis. Nucleic Acids Res. 2019, 47, e15. [Google Scholar] [CrossRef]
- Yang, H.; Yan, R.; Li, Y.; Lu, Z.; Bie, X.; Zhao, H.; Lu, F.; Chen, M. Structure-Function Analysis of a Quinone-Dependent Dehydrogenase Capable of Deoxynivalenol Detoxification. J. Agric. Food Chem. 2022, 70, 6764–6774. [Google Scholar] [CrossRef]



| Pseudomonas sp. SD17-1 | Devosia sp. SD17-2 | |
|---|---|---|
| Genome coverage | 180.0× | 261.0× |
| Contig Length (bp) | 5,727,087 | 4,231,113 |
| Type | chromosome | chromosome |
| Topology | circular | circular |
| Genes | 5249 | 4152 |
| Protein-coding | 5170 | 4004 |
| GC content (%) | 64.25 | 62.11 |
| rRNA number | 19 | 12 |
| tRNA number | 73 | 58 |
| Assembly accession number | GCA_029201585.1 | GCF_029201565.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhao, D.; Zhang, W.; Wang, S.; Huang, K.; Guo, B. Biotransformation of Deoxynivalenol by a Dual-Member Bacterial Consortium Isolated from Tenebrio molitor Larval Feces. Toxins 2023, 15, 492. https://doi.org/10.3390/toxins15080492
Wang Y, Zhao D, Zhang W, Wang S, Huang K, Guo B. Biotransformation of Deoxynivalenol by a Dual-Member Bacterial Consortium Isolated from Tenebrio molitor Larval Feces. Toxins. 2023; 15(8):492. https://doi.org/10.3390/toxins15080492
Chicago/Turabian StyleWang, Yang, Donglei Zhao, Wei Zhang, Songxue Wang, Kai Huang, and Baoyuan Guo. 2023. "Biotransformation of Deoxynivalenol by a Dual-Member Bacterial Consortium Isolated from Tenebrio molitor Larval Feces" Toxins 15, no. 8: 492. https://doi.org/10.3390/toxins15080492
APA StyleWang, Y., Zhao, D., Zhang, W., Wang, S., Huang, K., & Guo, B. (2023). Biotransformation of Deoxynivalenol by a Dual-Member Bacterial Consortium Isolated from Tenebrio molitor Larval Feces. Toxins, 15(8), 492. https://doi.org/10.3390/toxins15080492