Transcriptomic and Metabolomic Analyses of the Response of Resistant Peanut Seeds to Aspergillus flavus Infection
Abstract
1. Introduction
2. Results
2.1. Aflatoxin, Resveratrol Content and the Changes in Resistance-Related Enzyme Activities after Inoculation
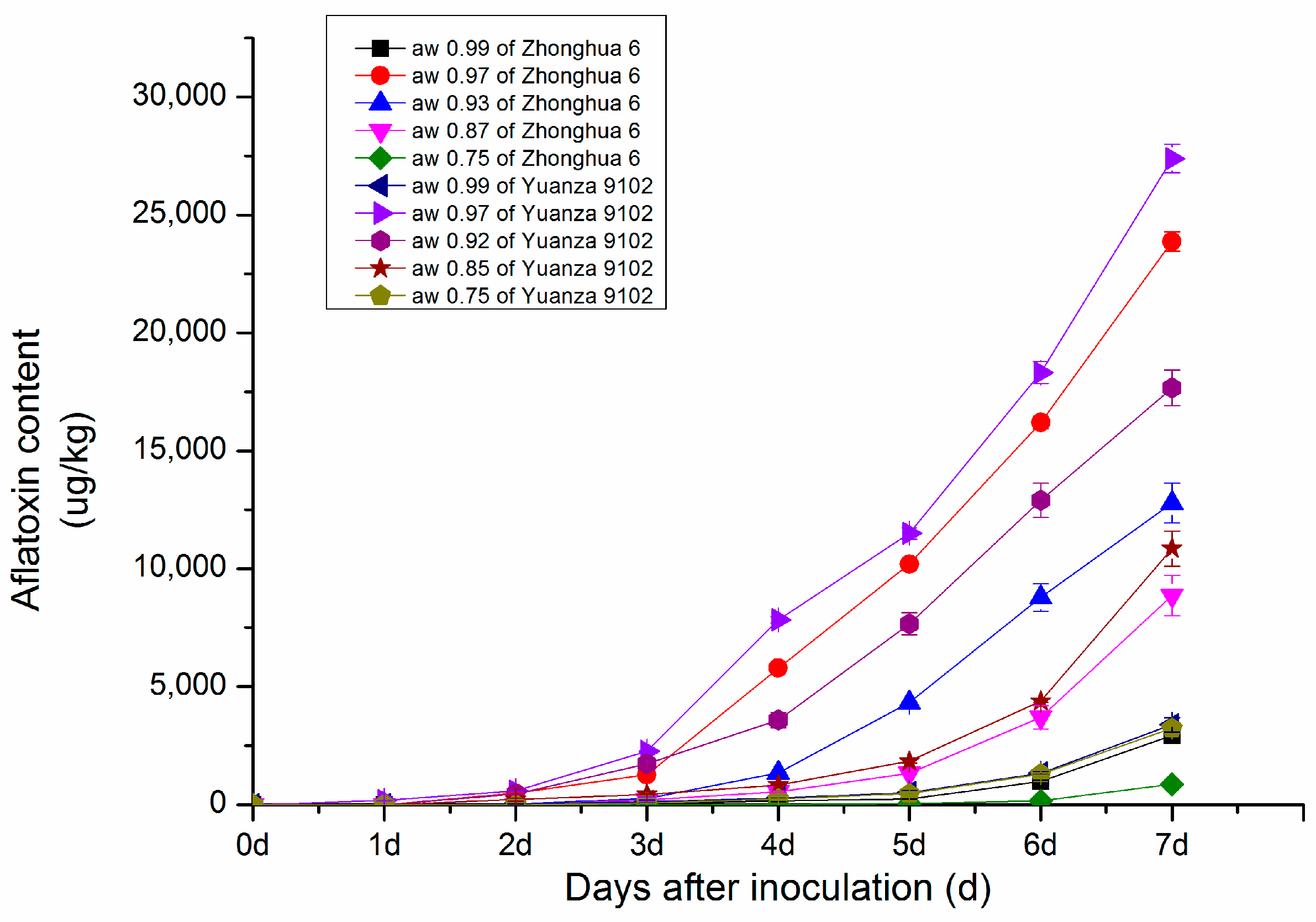
2.1.1. Changes in Aflatoxin Content in Peanut with Different Water Activity after Inoculation
2.1.2. Dynamic Changes in Resveratrol and Resistance-Related Enzyme (PAL, POD, LOX) Activities after Inoculation with A. flavus at a Suitable Water Activity Level
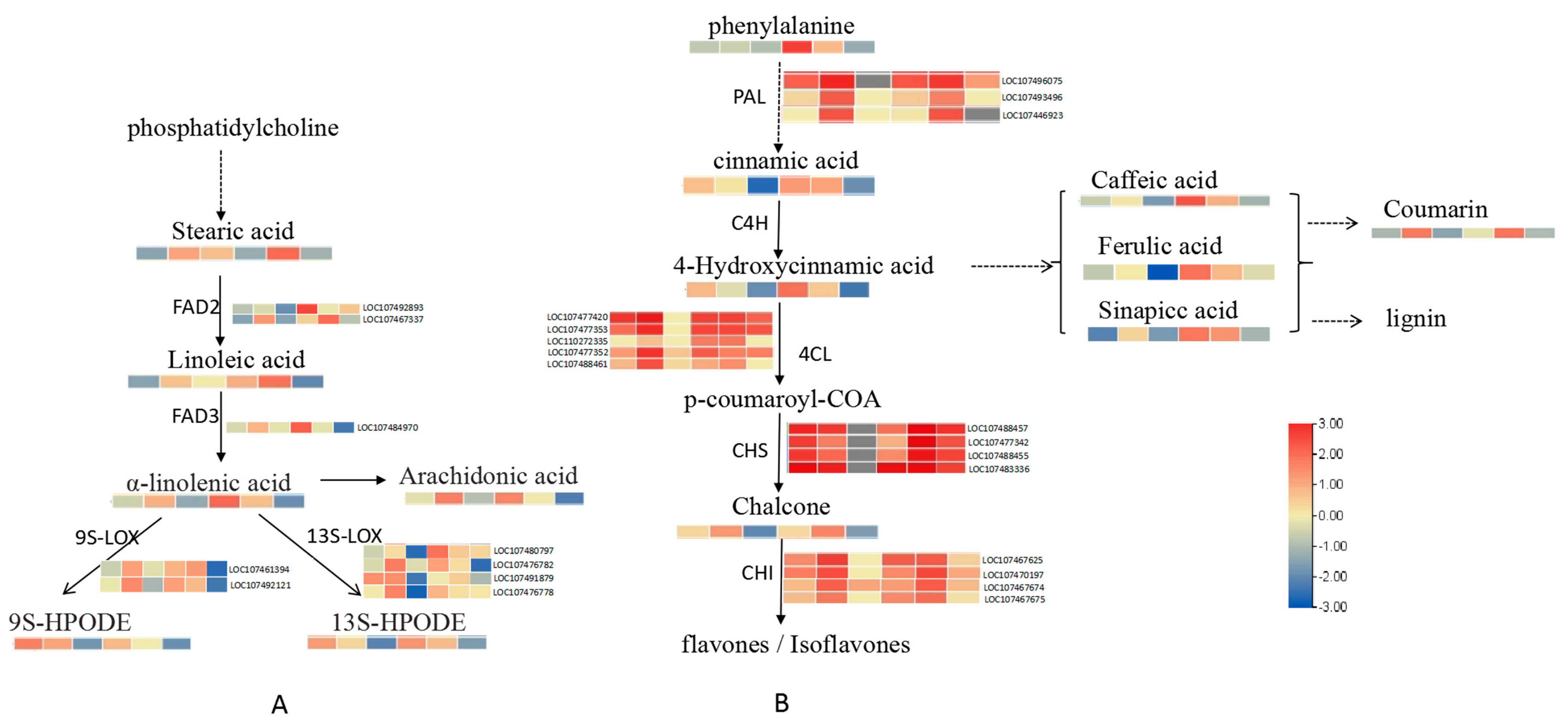
2.2. Transcriptomic and Metabolmic Profiles of A. flavus-Infected Peanut Seeds
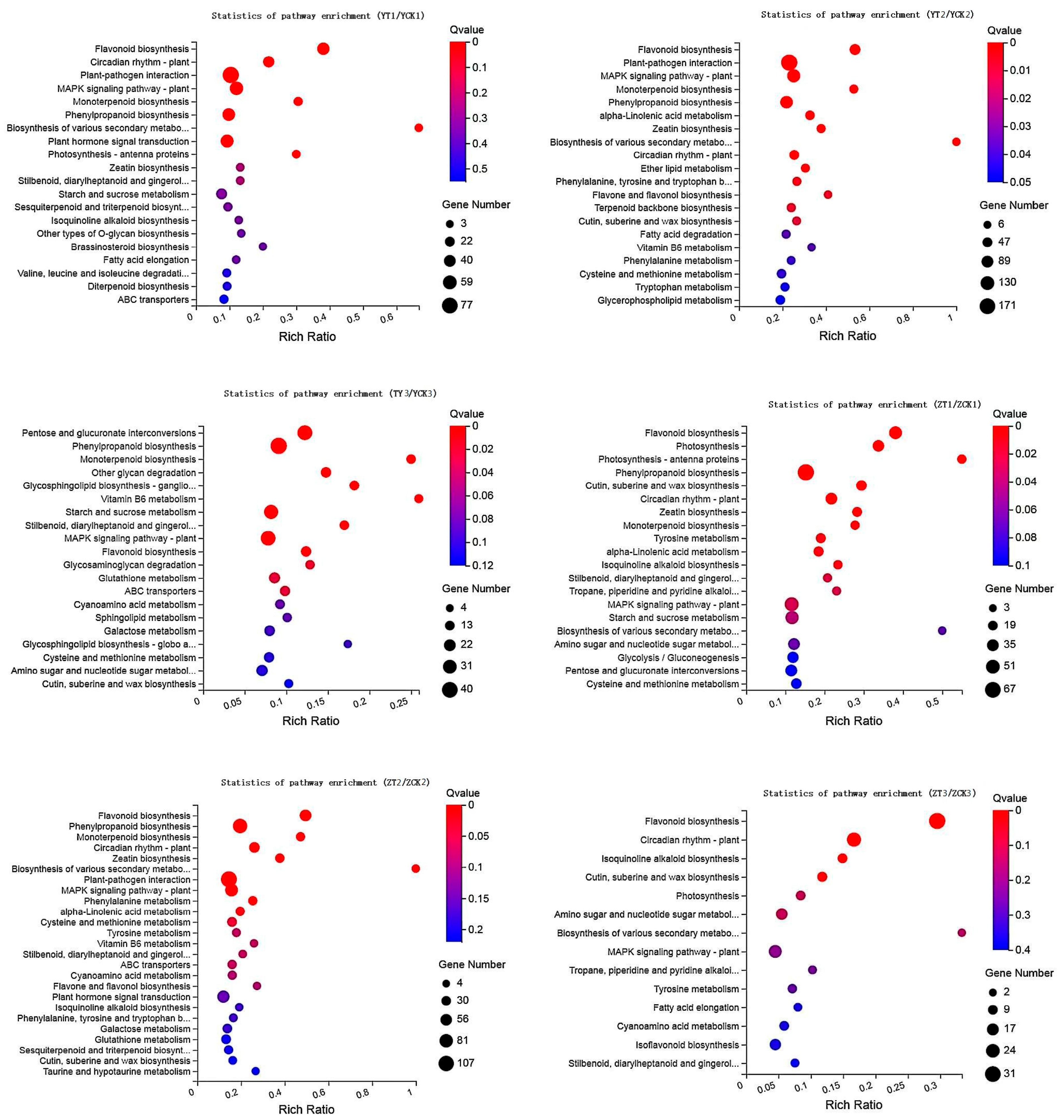
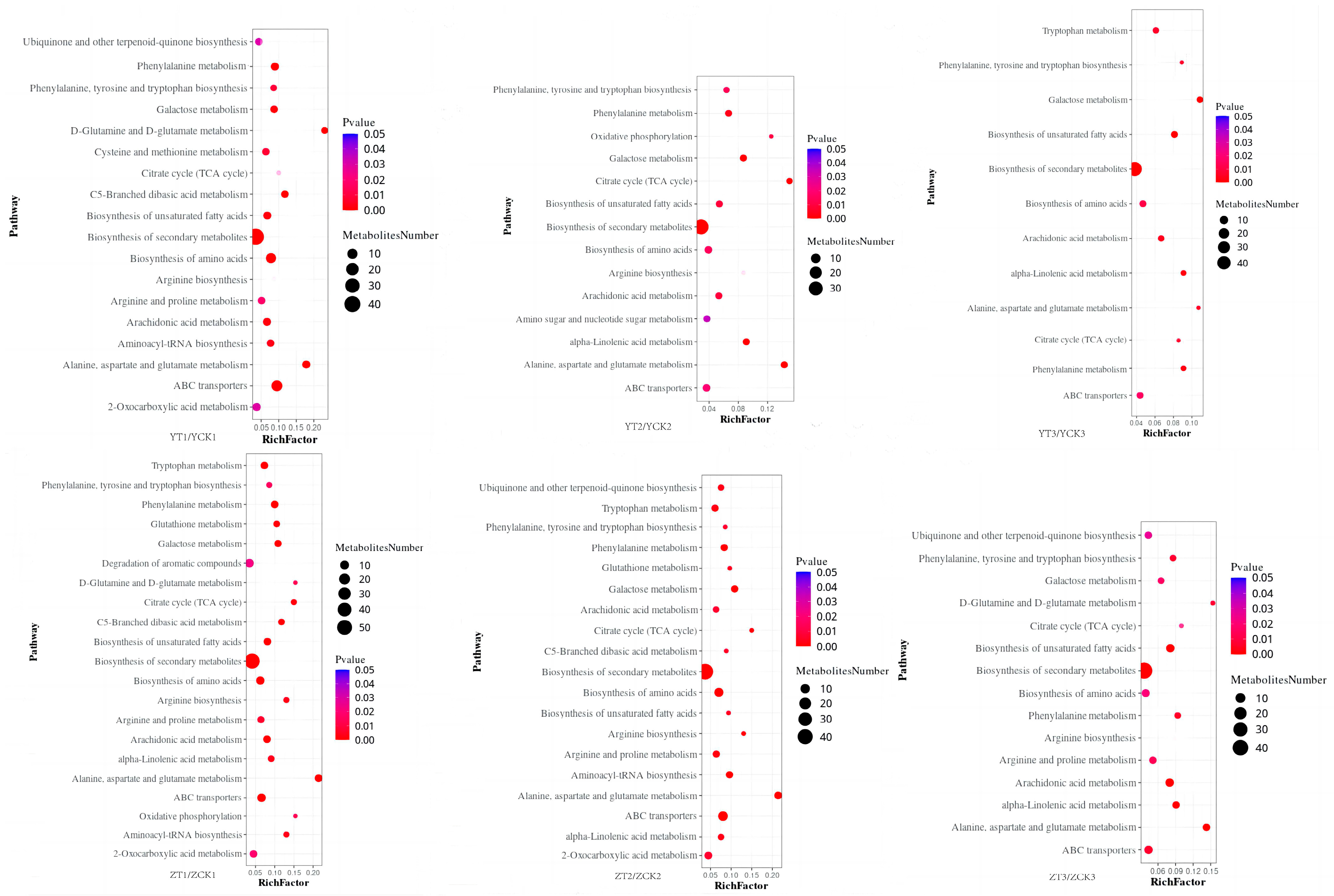
2.3. Functional Analysis of DEGs and DAMs
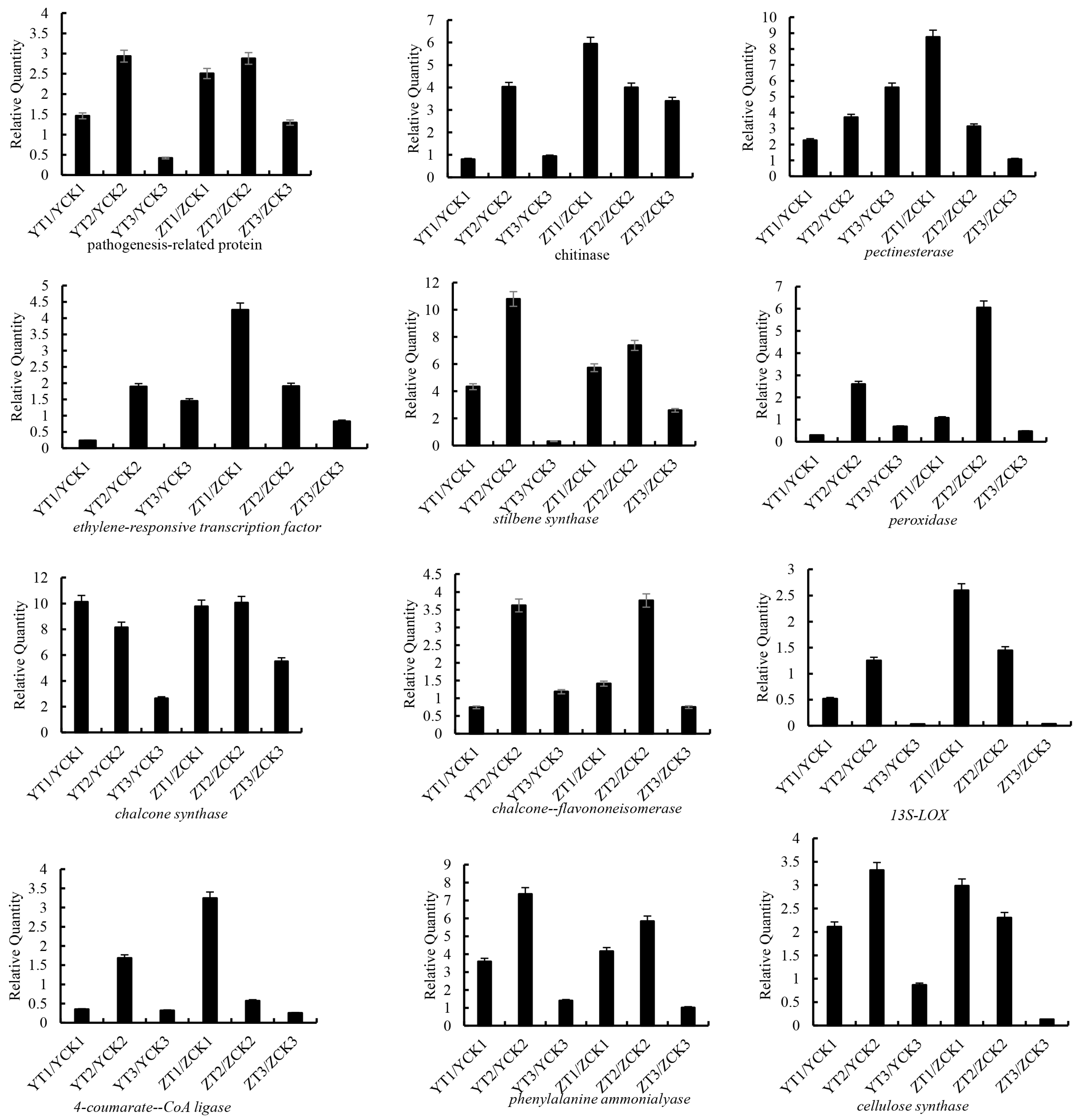
2.4. qRT–PCR Validation of Selected DEGs
2.5. Integrative Analysis of the Transcriptome and Metabolome
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Peanut Material and Pathogen Inoculation
5.2. Aflatoxin, Enzyme Activity and Resveratrol Content Assay
5.3. RNA Extraction and Transcriptome Sequencing
5.4. Alignment to the Reference Genome and Functional Annotation of DEGs
5.5. Validation of Transcriptome Data by RT–qPCR Assays
5.6. Metabolomic Sequencing Analysis
5.7. Joint Analysis of Transcriptome and Metabolome
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Settaluri, V.S.; Kandala, C.V.K.; Puppala, N.; Sundaram, J. Peanuts and their nutritional aspects-a review. Food Nutr. Sci. 2012, 3, 1644–1650. [Google Scholar] [CrossRef]
- Bonku, R.; Yu, J. Health aspects of peanuts as an outcome of its chemical composition. Food Sci. Hum. Wellness 2020, 9, 21–30. [Google Scholar] [CrossRef]
- Yao, Y.; Gao, S.; Ding, X.; Zhang, Q.; Li, P. Topography effect on Aspergillus flavus occurrence and aflatoxin B1 contamination associated with peanut. Curr. Res. Microb. Sci. 2021, 2, 100021. [Google Scholar] [CrossRef] [PubMed]
- Adetunji, M.C.; Akinola, S.A.; Nleya, N.; Mulunda, M. Nutrient composition and aflatoxin contamination of African sourced peanuts and cashew nuts: Its implications on health. In Nuts and Nut Products in Human Health and Nutrition; IntechOpen: London, UK, 2021. [Google Scholar]
- Kumar, P.; Gupta, A.; Mahato, D.K.; Pandhi, S.; Pandey, A.K.; Kargwal, R.; Mishra, S.; Suhag, R.; Sharma, N.; Saurabh, V.; et al. Aflatoxins in Cereals and Cereal-Based Products: Occurrence, Toxicity, Impact on Human Health, and Their Detoxification and Management Strategies. Toxins 2022, 14, 687. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.N.; Zhang, Q.; Chen, Z.Y.; Liu, H.X.; Li, P.W. Investigation of Pseudomonas fluorescens strain 3JW1 on preventing and reducing aflatoxin contaminations in peanuts. PLoS ONE 2017, 12, e0178810. [Google Scholar] [CrossRef]
- Mauro, A.; Garcia-Cela, E.; Pietri, A.; Cotty, P.J.; Battilani, P. Biological control products for aflatoxin prevention in Italy: Commercial field evaluation of atoxigenic Aspergillus flavus active ingredients. Toxins 2018, 10, 30. [Google Scholar] [CrossRef]
- Ehrlich, K.C. Non-aflatoxigenic Aspergillus flavus to prevent aflatoxin contamination in crops: Advantages and limitations. Front. Microbiol. 2014, 5, 50. [Google Scholar] [CrossRef]
- Luo, M.; Brown, R.L.; Chen, Z.Y.; Cleveland, T.E. Host genes involved in the interaction between Aspergillus flavus and maize. Toxin Rev. 2009, 28, 118–128. [Google Scholar] [CrossRef]
- Torres, A.M.; Barros, G.G.; Palacios, S.A.; Chulze, S.N.; Battilani, P. Review on pre- and postharvest management of peanuts to minimize aflatoxin contamination. Food. Res. Int. 2014, 62, 11–19. [Google Scholar] [CrossRef]
- Wang, Z.; Yan, S.; Liu, C.; Chen, F.; Wang, T. Proteomic analysis reveals an aflatoxin-triggered immune response in cotyledons of Arachis hypogaea infected with Aspergillus flavus. J. Proteome Res. 2012, 11, 2739–2753. [Google Scholar] [CrossRef]
- Zhang, H.L.; Scharfenstein, L.; Zhang, D.H.; Chang, P.K.; Montalbano, B.G.; Guo, B.Z.; Meng, X.J.; Yu, J.J. Peanut Resistance Gene Expression in Response to Aspergillus flavus Infection During Seed Germination. J. Phytopathol. 2015, 163, 212–221. [Google Scholar] [CrossRef]
- Nayak, S.N.; Agarwal, G.; Pandey, M.K.; Sudini, H.K.; Jayale, A.S.; Purohit, S.; Desai, A.; Wan, L.Y.; Guo, B.Z.; Liao, B.S.; et al. Aspergillus flavus infection triggered immune responses and host-pathogen cross-talks in groundnut during in-vitro seed colonization. Sci. Rep. 2017, 7, 9659. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.A.; Luo, M.; Guo, B.Z. Resistance Mechanisms to Aspergillus flavu infection and aflatoxin contaminatin in peanut (Arachis hypogaea). Plant Pathol. J. 2006, 5, 115–124. [Google Scholar] [CrossRef]
- Liang, X.Q.; Holbrook, C.C.; Lynch, R.E.; Guo, B.Z. β-1, 3-Glucanase activity in peanut seed (Arachis hypogaea) is induced by inoculation with Aspergillus flavus and copurifies with a conglutin-like protein. Phytopathology 2005, 95, 506–511. [Google Scholar] [CrossRef]
- Müller, V.; Amé, M.V.; Carrari, V.; Gieco, J.; Asis, R. Lipoxygenase Activation in Peanut Seed Cultivars Resistant and Susceptible to Aspergillus parasiticus Colonization. Phytopathology 2014, 104, 1340–1348. [Google Scholar] [CrossRef]
- Mierziak, J.; Kostyn, K.; Kulma, A. Flavonoids as Important Molecules of Plant Interactions with the Environment. Molecules 2014, 19, 16240–16265. [Google Scholar] [CrossRef]
- Yan, S.; Liang, Y.; Zhang, J.; Chen, Z.; Liu, C.M. Autoxidated linolenic acid inhibits aflatoxin biosynthesis in Aspergillus flavus via oxylipin species. Fungal Genet. Biol. 2015, 81, 229–237. [Google Scholar] [CrossRef]
- Korani, W.; Chu, Y.; Holbrook, C.C.; Ozias-Akins, P. Insight into genes regulating postharvest aflatoxin contamination of tetraploid peanut from transcriptional profiling. Genetics 2018, 209, 143–156. [Google Scholar] [CrossRef]
- Prasad, K.; Bhatnagar-Mathur, P.; Waliyar, F.; Sharma, K.K. Overexpression of a chitinase gene in transgenic peanut confers enhanced resistance to major soil borne and foliar fungal pathogens. J. Plant Biochem. Biotechnol. 2013, 22, 222–233. [Google Scholar] [CrossRef]
- Xie, C.; Wen, S.; Liu, H.; Chen, X.; Li, H.; Hong, Y.; Liang, X. Overexpression of ARAhPR10, a member of the PR10 family, decreases levels of Aspergillus flavus infection in peanut seeds. Am. J. Plant Sci. 2013, 4, 602–607. [Google Scholar] [CrossRef]
- Jayaprakash, A.; Roy, A.; Thanmalagan, R.R.; Arunachalam, A.; Lakshmi, P.T.V. Immune response gene coexpression network analysis of Arachis hypogaea infected with Aspergillus flavus. Genomics 2021, 113, 2977–2988. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.M.; Lei, Y.; Wan, L.Y.; Yan, L.Y.; Lv, J.W.; Dai, X.F.; Ren, X.P.; Guo, W.; Jiang, H.F.; Liao, B.S. Comparative transcript profiling of resistant and susceptible peanut post-harvest seeds in response to aflatoxin production by Aspergillus flavus. BMC Plant Biol. 2016, 16, 54. [Google Scholar] [CrossRef] [PubMed]
- Coppola, M.; Diretto, G.; Digilio, M.C.; Woo, S.L.; Giuliano, G.; Molisso, D.; Pennacchio, F.; Lorito, M.; Rao, R. Transcriptome and metabolome reprogramming in tomato plants by Trichoderma harzianum strain T22 primes and enhances defense responses against aphids. Front. Physiol. 2019, 10, 745. [Google Scholar] [CrossRef]
- Li, Q.Q.; Liu, C.B.; Huang, C.Y.; Wang, M.F.; Long, T.; Liu, J.J.; Shi, J.L.; Li, L.; He, Y.Q.; Xu, D.L. Transcriptome and metabolomics analysis revealed the molecular mechanism of differential metabolite production of Dendrobium mobile under different epiphytic patterns. Front. Plant Sci. 2022, 13, 1193. [Google Scholar]
- Cui, M.J.; Han, S.Y.; Wang, D.; Haider, M.S.; Guo, J.J.; Zhao, Q.; Du, P.; Sun, Z.Q.; Qi, F.Y.; Zheng, Z.; et al. Gene co-expression network analysis of the comparative transcriptome identifies hub genes associated with resistance to Aspergillus flavus L. in cultivated peanut (Arachis hypogea L.). Front. Plant Sci. 2022, 13, 899177. [Google Scholar] [CrossRef]
- Zhao, X.; Li, C.; Yan, C.; Wang, J.; Yuan, C.; Zhang, H.; Shan, S. Transcriptome and proteome analyses of resistant preharvest peanut seed coat in response to Aspergillus flavus infection. Electron. J. Biotechnol. 2019, 39, 82–90. [Google Scholar] [CrossRef]
- Soni, P.; Nayak, S.N.; Kumar, R.; Pandey, M.K.; Singh, N.; Sudini, H.K.; Bajaj, P.; Fountain, J.C.; Singam, P.; Hong, Y.B.; et al. Transcriptome analysis identified coordinated control of key pathways regulating cellular physiology and metabolism upon Aspergillus flavus infection resulting in reduced aflatoxin production in groundnut. J. Fungi 2020, 6, 370. [Google Scholar] [CrossRef]
- Abdel-Hadi, A.; Schmidt-Heydt, M.; Parra, R.; Geisen, R.; Magan, N. A systems approach to model the relationship between aflatoxin gene cluster expression, environmental factors, growth and toxin production by Aspergillus flavus. J. R. Soc. Interface 2012, 9, 757–767. [Google Scholar] [CrossRef]
- Xin, M.M.; Wang, X.F.; Peng, H.R.; Yao, Y.Y.; Xie, C.J.; Han, Y.; Ni, Z.F.; Sun, Q.X. Transcriptome comparison of susceptible and resistant wheat in response to powdery mildew infection. Genom. Proteom. Bioinform. 2012, 10, 94–106. [Google Scholar] [CrossRef]
- Sels, J.; Mathys, J.; Barbara, M.A.; DeConinck, B.M.A.; Cammue, B.P.A.; De Bolle, M.F.C. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950. [Google Scholar] [CrossRef]
- Agarwal, P.; Agarwal, P.K. Pathogenesis related-10 proteins are small, structurally similar but with diverse role in stress signaling. Mol. Biol. Rep. 2014, 41, 599–611. [Google Scholar] [CrossRef]
- Chen, Z.Y.; Brown, R.L.; Damann, K.E.; Cleveland, T.E. PR10 expression in maize and its effect on host resistance against Aspergillus flavus infection and aflatoxin production. Mol. Plant Pathol. 2010, 11, 69–81. [Google Scholar] [CrossRef]
- Chen, Z.Y.; Brown, R.L.; Rajasekaran, K.; Damann, K.E.; Cleveland, T.E. Identification of a maize kernel pathogenesis-related protein and evidence for its involvement in resistance to Aspergillus flavus infection and aflatoxin production. Phytopathology 2006, 96, 87–95. [Google Scholar] [CrossRef]
- Poria, V.; Rana, A.; Kumari, A.; Grewal, J.; Pranaw, K.; Singh, S. Current Perspectives on Chitinolytic Enzymes and Their Agro-Industrial Applications. Biology 2021, 10, 1319. [Google Scholar] [CrossRef]
- Qiao, L.X.; Ding, X.; Wang, H.C.; Sui, J.M.; Wang, J.S. Characterization of the β-1,3-glucanase gene in peanut (Arachis hypogaea L.) by cloning and genetic transformation. Genet. Mol. Res. 2014, 13, 1893–1904. [Google Scholar] [CrossRef] [PubMed]
- Alaux, P.L.; Naveau, F.; Declerck, S.; Cranenbrouck, S. Common mycorrhizal network induced JA/ET genes expression in healthy potato plants connected to potato plants infected by Phytophthora infestans. Front. Plant Sci. 2020, 11, 602. [Google Scholar] [CrossRef]
- Fan, P.M.; Wan, W.D.; Kang, Y.P.; Wan, L.Y.; Lei, Y.; Huai, D.X.; Chen, Y.N.; Wang, X.; Jiang, H.F.; Yan, L.Y.; et al. Phenotypic identification of peanut germplasms for resistance to southern stem rot. Oil Crop Sci. 2020, 5, 174–179. [Google Scholar] [CrossRef]
- Lee, J.; Yang, K.; Lee, M.; Kim, S.; Kim, J.; Lim, S.; Kang, G.; Min, A.J.; Park, A.U.; Jang, Y.S.; et al. Differentiated cuticular wax content and expression patterns of cuticular wax biosynthetic genes in bloomed and bloomless broccoli (Brassica oleracea var.italica). Process Biochem. 2015, 50, 456–462. [Google Scholar] [CrossRef]
- Nandini, D.; Bariya, H. Induction of systemic acquired resistance in Arachis hypogaea L. by aspergillus flavus derived elicitors. J. Cell Tissue Res. 2014, 14, 4395–4403. [Google Scholar]
- Jain, D.; Khurana, J.P. Role of Pathogenesis-Related (PR) Proteins in Plant Defense Mechanism. Mol. Asp. Plant-Pathog. Interact. 2018, 12, 265–281. [Google Scholar]
- Li, X.M.; Li, Z.Y.; Wang, Y.D.; Wang, J.Q.; Yang, P.L. Quercetin inhibits the proliferation and aflatoxins biosynthesis of Aspergillus flavus. Toxins 2019, 11, 154–167. [Google Scholar] [CrossRef]
- Norton, R.A. Inhibition of aflatoxin B1 biosynthesis in Aspergillus flavus by anthocyanidins and related flavonoids. J. Agric. Food Chem. 1999, 47, 1230–1235. [Google Scholar] [CrossRef] [PubMed]
- Firozi, P.F.; Bhattacharya, R.K. Effects of natural polyphenols on aflatoxin B1 activation in a reconstituted microsomal monooxygenase system. J. Biochem.Toxicol. 1995, 10, 25–31. [Google Scholar] [PubMed]
- Wang, H.; Huang, J.Q.; Lei, Y.; Yan, L.Y.; Wang, S.Y.; Jiang, H.F.; Ren, X.P.; Lou, Q.R.; Liao, B.S. Relationship of resveratrol content and resistance to aflatoxin accumulation caused by Aspergillus flavus in peanut seeds. Acta Agron. Sin. 2012, 38, 1875–1883. [Google Scholar] [CrossRef]
- Chong, J.; Poutaraud, A.; Hugueney, P. Metabolism and roles of stilbenes in plants. Plant Sci. 2009, 177, 143–155. [Google Scholar] [CrossRef]
- Abouraïcha, E.F.; El Alaoui-Talibi, Z.; Tadlaoui-Ouafi, A.; El Boutachfaiti, R.; Petit, E.; Douira, A.; Courtois, B.; Courtois, J.; El Modafar, C. Glucuronan and oligoglucuronans isolated from green algae activate natural defense responses in apple fruit and reduce postharvest blue and gray mold decay. J. Appl. Phys. 2017, 29, 471–480. [Google Scholar] [CrossRef]
- Jiang, L.; Jin, P.; Wang, L.; Yu, X.; Wang, H.; Zheng, Y. Methyl jasmonate primes defense responses against Botrytis cinerea and reduces disease development in harvested table grapes. Sci. Hortic. 2015, 192, 218–223. [Google Scholar] [CrossRef]
- Shao, X.; Wang, H.; Xu, F.; Cheng, S. Effects and possible mechanisms of tea tree oil vapor treatment on the main disease in postharvest strawberry fruit. Postharvest Biol. Technol. 2013, 77, 94–101. [Google Scholar] [CrossRef]
- MacDonald, M.J.; D’Cunha, G.B. A modern view of phenylalanine ammonialyase. Biochem. Cell Biol. 2007, 85, 273–282. [Google Scholar] [CrossRef]
- Mohammadi, M.; Kazemi, H. Changes in peroxidase and polyphenol oxidase activities in susceptible and resistant wheat heads inoculated with Fusarium graminearum and induced resistance. Plant Sci. 2002, 162, 491–498. [Google Scholar] [CrossRef]
- Koramutla, M.K.; Kaur, A.; Negi, M.; Venkatachalam, P.; Bhattacharya, R. Elicitation of jasmonate-mediated host defense in Brassica juncea (L.) attenuates population growth of mustard aphid Lipaphis erysimi (Kalt.). Planta 2014, 240, 177–194. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, B.L.; Sun, S.; Xing, G.M.; Wang, F.; Li, M.Y.; Tian, Y.S.; Xiong, A.S. AP2/ERF Transcription Factors Involved in Response to Tomato Yellow Leaf Curly Virus in Tomato. Plant Genome 2016, 9, 2. [Google Scholar] [CrossRef]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef]
- Nakano, Y.; Yamaguchi, M.; Endo, H.; Rejab, N.A.; Ohtani, M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 2015, 6, 288. [Google Scholar] [CrossRef]
- Sun, L.Y.; Zhang, H.J.; Li, D.Y.; Huang, L.; Hong, Y.B.; Ding, X.S.; Nelson, R.S.; Zhou, X.P.; Song, F.M. Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea. Plant Mol. Biol. 2013, 81, 41–56. [Google Scholar] [CrossRef]
- Ogunola, O.F.; Hawkins, L.K.; Mylroie, E.; Kolomiets, M.V.; Borrego, E.; Tang, J.D.; Williams, W.P.; Warburton, M.L. Character-ization of the maize lipoxygenase gene family in relation to aflatoxin accumulation resistance. PLoS ONE 2017, 12, e0181265. [Google Scholar] [CrossRef] [PubMed]
- Burow, G.B.; Gardner, H.W.; Keller, N.P. A peanut seed lipoxygenase responsive to Aspergillus colonization. Plant Mol. Biol. 2000, 42, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.M.; Sucharitha, A.; Devi, P.U.M. Impact of 13-lipoxygenase on induction of resistance in peanut against aflatoxigenic Aspergillus. Int. J. Biol. Sci. 2012, 5, 4–13. [Google Scholar]
- Tang, J.D.; Perkins, A.; Williams, W.P.; Warburton, M.L. Using genome- wide associations to identify metabolic pathways involved in maize aflatoxin accumulation resistance. BMC Genom. 2015, 16, 673. [Google Scholar] [CrossRef]
- Wang, Q.F.; Wang, M.; Li, L.P.; Gao, L.; Ying, D.S.; Zhang, R. Research on Correlation between Synthesis of Stilbenes and Aflatoxin Accumulation Caused by Aspergillus flavus in Peanut Seeds. Agric. Sci. Technol. 2016, 17, 265–272+278. [Google Scholar]
- Limmongkon, A.; Janhom, P.; Amthong, M.; Kawpanuk, M.; Nopprang, P.; Poohadsuan, J.; Somboon, T.; Saijeen, S.; Surangkul, D.; Srikummool, M.; et al. Antioxidant activity, total phenolic, and resveratrol content in five cultivars of peanut sprouts. Asian Pac. J. Trop. Biomed. 2017, 7, 332–338. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, X.L.; Zhang, X.; He, X.; Li, H.; Cui, D.Q.; Yin, D. ITRAQ-based proteomic analysis of the metabolic mechanisms behind lipid accumulation and degradation during peanut seed development and postgermination. J. Proteome Res. 2016, 15, 4277–4289. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Liu, D.; Yin, H.; Wang, H.; Cao, C.; Wang, J.; Zheng, J.; Liu, J. Transcriptomic and Metabolomic Analyses of the Response of Resistant Peanut Seeds to Aspergillus flavus Infection. Toxins 2023, 15, 414. https://doi.org/10.3390/toxins15070414
Wang Y, Liu D, Yin H, Wang H, Cao C, Wang J, Zheng J, Liu J. Transcriptomic and Metabolomic Analyses of the Response of Resistant Peanut Seeds to Aspergillus flavus Infection. Toxins. 2023; 15(7):414. https://doi.org/10.3390/toxins15070414
Chicago/Turabian StyleWang, Yun, Dongmei Liu, Haiyan Yin, Hongqi Wang, Cheng Cao, Junyan Wang, Jia Zheng, and Jihong Liu. 2023. "Transcriptomic and Metabolomic Analyses of the Response of Resistant Peanut Seeds to Aspergillus flavus Infection" Toxins 15, no. 7: 414. https://doi.org/10.3390/toxins15070414
APA StyleWang, Y., Liu, D., Yin, H., Wang, H., Cao, C., Wang, J., Zheng, J., & Liu, J. (2023). Transcriptomic and Metabolomic Analyses of the Response of Resistant Peanut Seeds to Aspergillus flavus Infection. Toxins, 15(7), 414. https://doi.org/10.3390/toxins15070414



