First Identification of Amphidinols from Mexican Strains and New Analogs
Abstract
:1. Introduction
2. Results
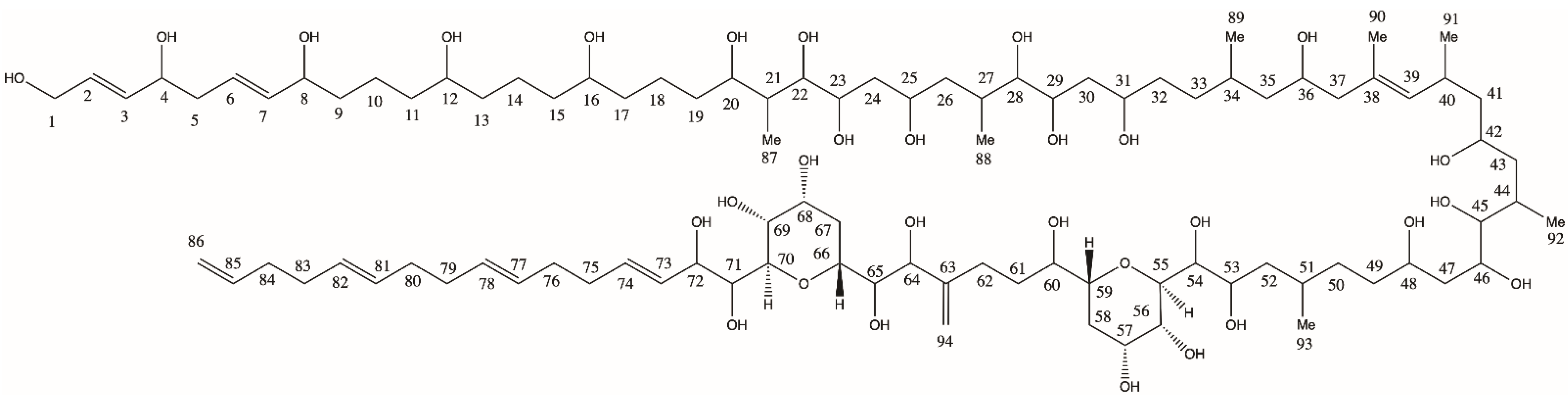
2.1. New Amphidinols
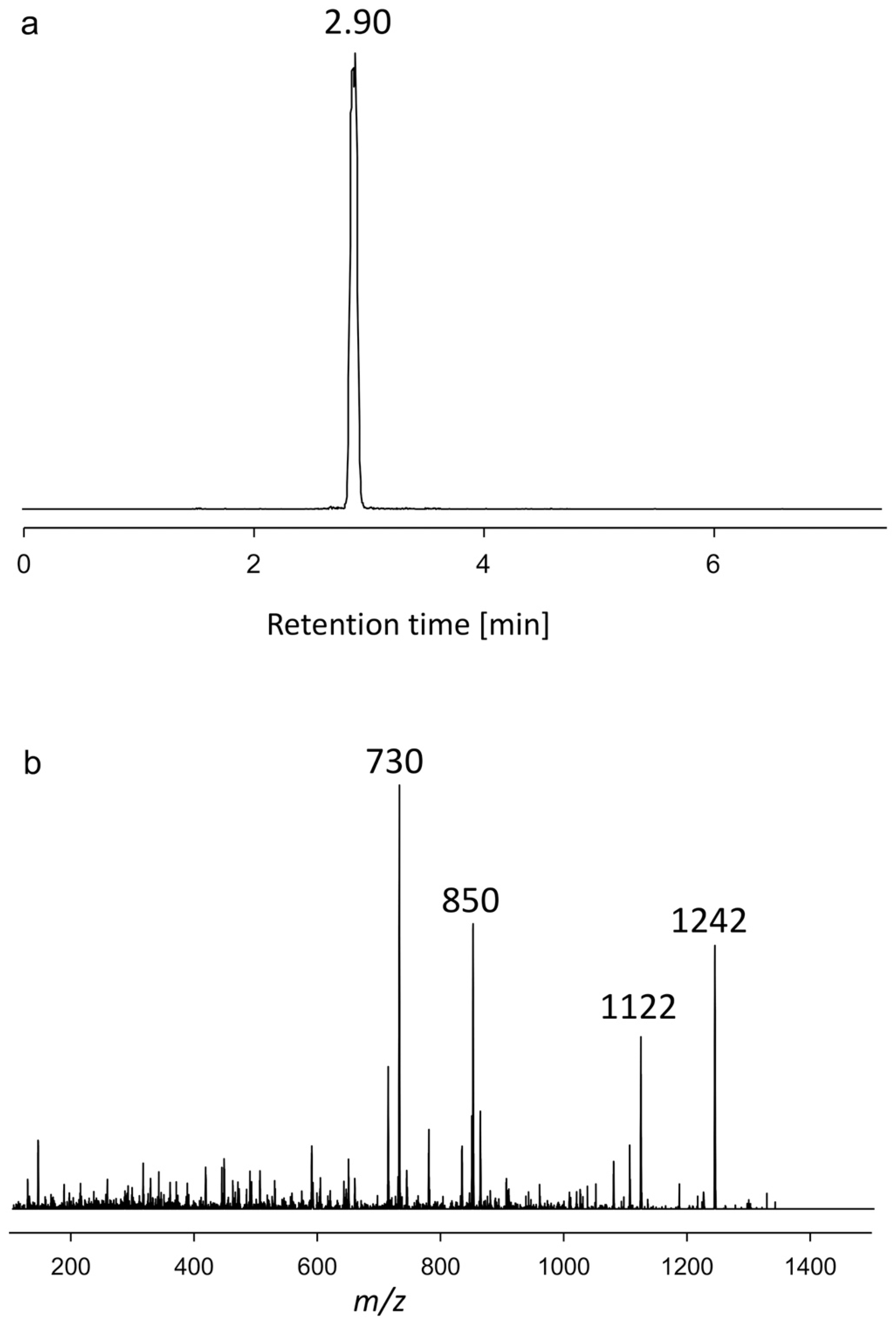
2.1.1. Variant U1
2.1.2. Variant U2
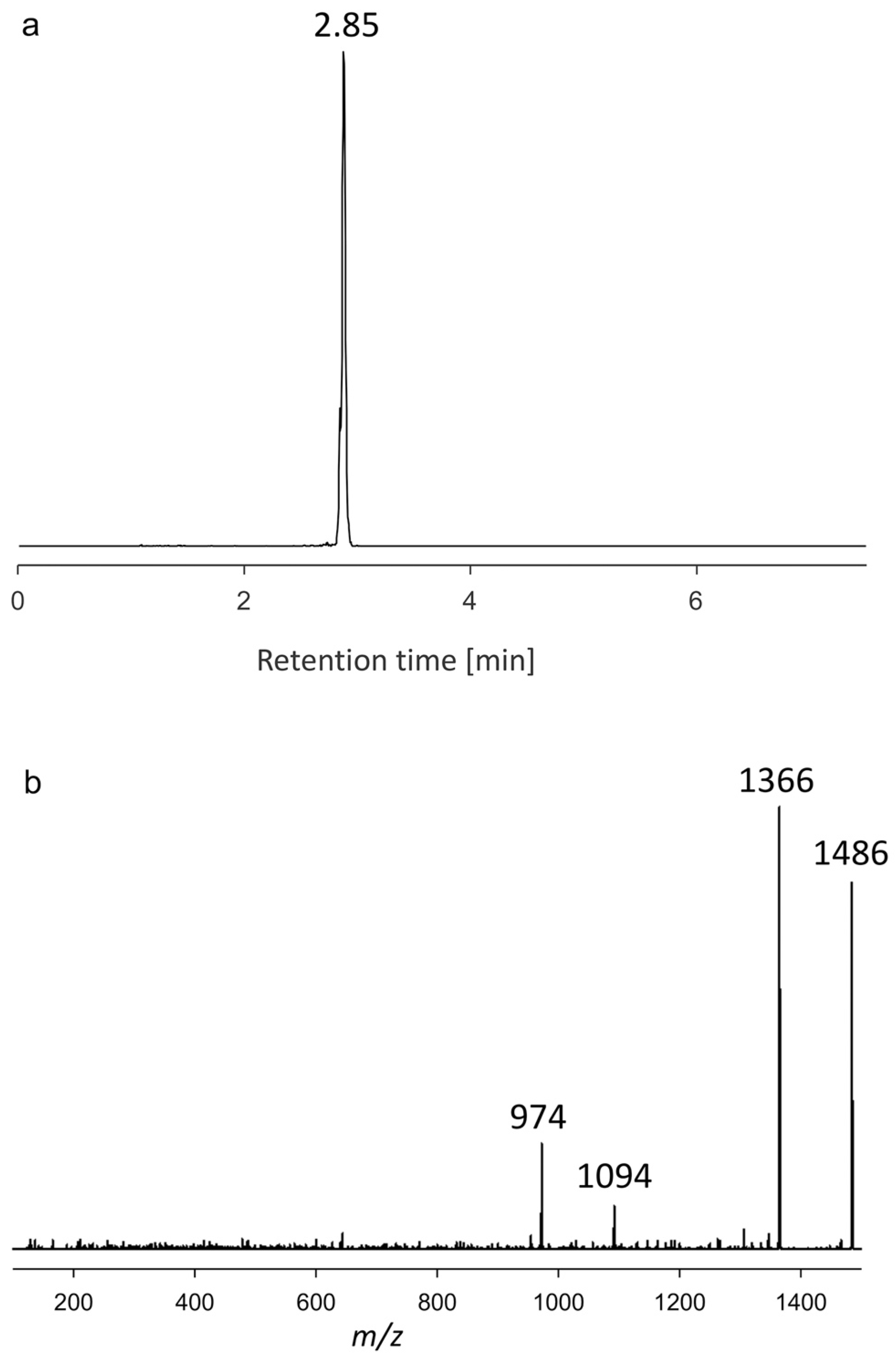
2.1.3. Variant U3
2.2. Amphidinol Profiles
3. Discussion
3.1. Novel Amphidinols
3.2. Confirmation of Detected Amphidinols
3.3. Amphidinol Cell Quotas, Profiles, and Geographical Patterns
4. Conclusions
5. Materials and Methods
5.1. Isolation, Culture, and Harvest of Amphidinium Strains
5.2. Amphidinol Extraction
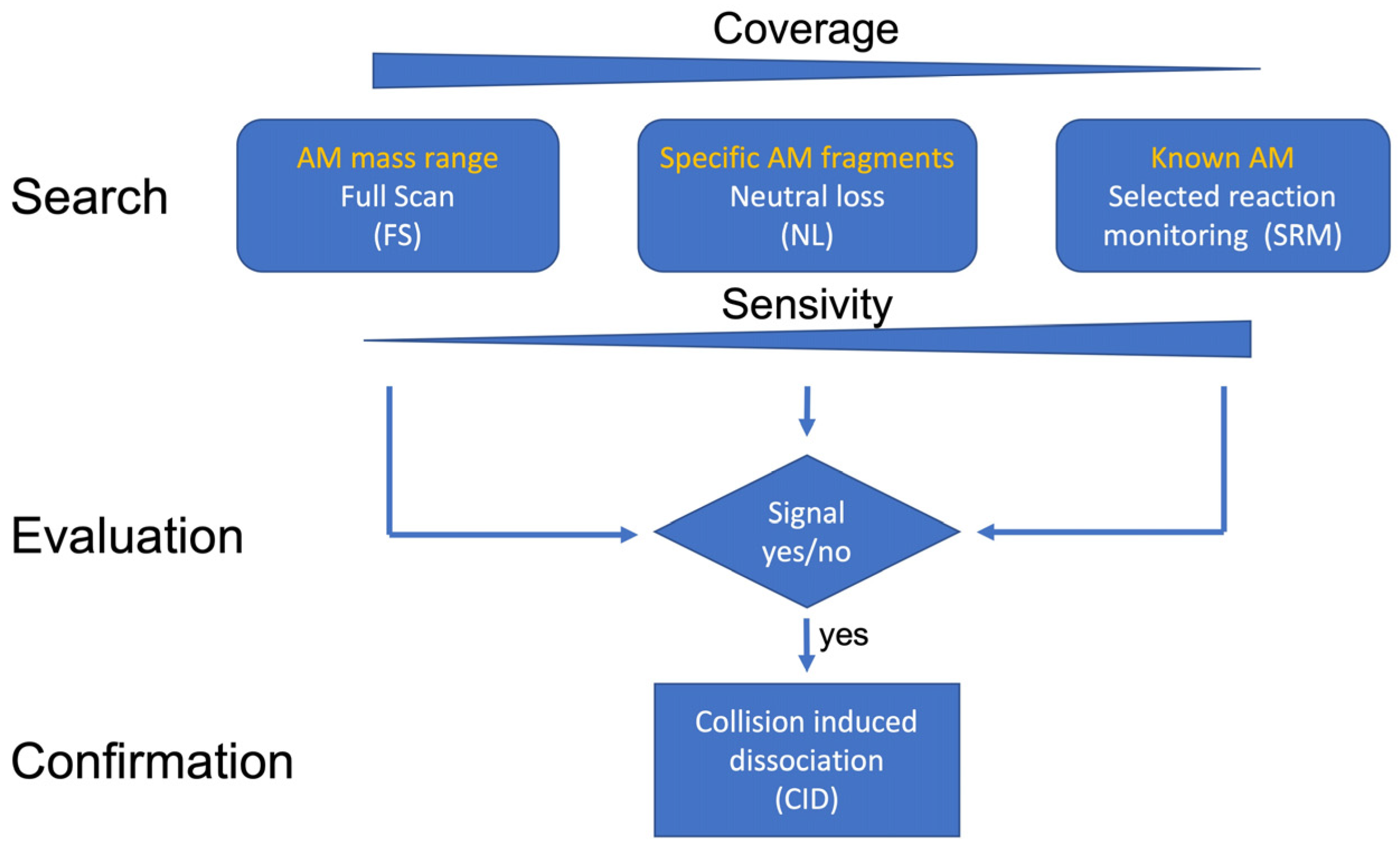
5.3. Analysis via Ultra-Performance Liquid Chromatography Coupled with Tandem Mass Spectrometry
5.3.1. Selected Reaction Monitoring (SRM) Mode
5.3.2. Neutral Loss (NL) Measurement Mode
5.3.3. Full-Scan (FS) Measurement Mode
5.3.4. Collision-Induced Dissociation (CID) Measurement Mode
5.3.5. Selection of Neutral Loss Scans
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flø Jørgensen, M.; Murray, S.; Daugbjerg, N. Amphidinium revisited. I. Redefinition of Amphidinium (Dinophyceae) based on cladistic and molecular phylogenetic analyses. J. Phycol. 2004, 40, 351–365. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M. World-Wide Electronic Publication; National University of Ireland: Galway, Ireland, 2022; Available online: https://www.algaebase.org (accessed on 5 January 2022).
- Murray, S.; Patterson, D. The benthic dinoflagellate genus Amphidinium in south-eastern Australian waters, including three new species. Eur. J. Phycol. 2002, 37, 279–298. [Google Scholar] [CrossRef]
- Murray, S.; Flø Jørgensen, M.; Daugbjerg, N.; Rhodes, L. Amphidinium revisited. II. Resolving species boundaries in the Amphidinium operculatum species complex (Dinophyceae), including the descriptions of Amphidinium trulla sp. nov. and Amphidinium gibbosum. comb. nov. J. Phycol. 2004, 40, 366–382. [Google Scholar] [CrossRef] [Green Version]
- Murray, S.A.; Garby, T.; Hoppenrath, M.; Neilan, B.A. Genetic diversity, morphological uniformity and polyketide production in dinoflagellates (Amphidinium, Dinoflagellata). PLoS ONE 2012, 7, e38253. [Google Scholar] [CrossRef] [Green Version]
- Pagliara, P.; Caroppo, C. Toxicity assessment of Amphidinium carterae, Coolia cfr. monotis and Ostreopsis cfr. ovata (Dinophyta) isolated from the northern Ionian Sea (Mediterranean Sea). Toxicon 2012, 60, 1203–1214. [Google Scholar] [CrossRef]
- Baig, H.; Saifullah, S.; Dar, A. Occurrence and toxicity of Amphidinium carterae Hulburt in the North Arabian Sea. Harmful Algae 2006, 5, 133–140. [Google Scholar] [CrossRef]
- Murray, S.A.; Kohli, G.S.; Farrell, H.; Spiers, Z.B.; Place, A.R.; Dorantes-Aranda, J.J.; Ruszczyk, J. A fish kill associated with a bloom of Amphidinium carterae in a coastal lagoon in Sydney, Australia. Harmful Algae 2015, 49, 19–28. [Google Scholar] [CrossRef]
- Sampayo, M.A. Encystment and excystment of a Portugese isolate of Amphidinium carterae in cultures. In Toxic Dinoflagellates; Anderson, D.M., White, A.W., Baden, D.G., Eds.; Elsevier: Amsterdam, The Netherlands, 1985; pp. 125–130. [Google Scholar]
- Cousseau, A.; Siano, R.; Probert, I.; Bach, S.; Mehiri, M. Marine dinoflagellates as a source of new bioactive structures. Stud. Nat. Prod. Chem. 2020, 65, 125–171. [Google Scholar]
- Mooney, B.D.; Dorantes-Aranda, J.J.; Place, A.R.; Hallegraeff, G.M. Ichthyotoxicity of gymnodinioid dinoflagellates: PUFA and superoxide effects in sheepshead minnow larvae and rainbow trout gill cells. Mar. Ecol. Prog. Ser. 2011, 426, 213–224. [Google Scholar] [CrossRef] [Green Version]
- Barone, M.E.; Murphy, E.; Parkes, R.; Fleming, G.T.; Campanile, F.; Thomas, O.P.; Touzet, N. Antibacterial activity and amphidinol profiling of the marine dinoflagellate Amphidinium carterae (Subclade III). Int. J. Mol. Sci. 2021, 22, 12196. [Google Scholar] [CrossRef]
- Satake, M.; Murata, M.; Yasumoto, T.; Fujita, T.; Naoki, H. Amphidinol, a polyhydroxy-polyene antifungal agent with an unprecedented structure, from a marine dinoflagellate, Amphidinium klebsii. J. Am. Chem. Soc. 1991, 113, 9859–9861. [Google Scholar] [CrossRef]
- Morsy, N.; Houdai, T.; Matsuoka, S.; Matsumori, N.; Adachi, S.; Oishi, T.; Murata, M.; Iwashita, T.; Fujita, T. Structures of new amphidinols with truncated polyhydroxyl chain and their membrane-permeabilizing activities. Bioorg. Med. Chem. 2006, 14, 6548–6554. [Google Scholar] [CrossRef] [PubMed]
- Kellmann, R.; Stüken, A.; Orr, R.J.; Svendsen, H.M.; Jakobsen, K.S. Biosynthesis and molecular genetics of polyketides in marine dinoflagellates. Mar. Drugs 2010, 8, 1011–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satake, M.; Cornelio, K.; Hanashima, S.; Malabed, R.; Murata, M.; Matsumori, N.; Zhang, H.; Hayashi, F.; Mori, S.; Kim, J.S. Structures of the largest amphidinol homologues from the dinoflagellate Amphidinium carterae and structure–activity relationships. J. Nat. Prod. 2017, 80, 2883–2888. [Google Scholar] [CrossRef]
- Wellkamp, M.; García-Camacho, F.; Durán-Riveroll, L.M.; Tebben, J.; Tillmann, U.; Krock, B. LC-MS/MS method development for the discovery and identification of amphidinols produced by Amphidinium. Mar. Drugs 2020, 18, 497. [Google Scholar] [CrossRef]
- Holmes, M.J.; Brust, A.; Lewis, R.J. Dinoflagellate toxins: An overview. In Seafood and Freshwater Toxins: Pharmacology, Physiology, and Detection; Botana, L., Ed.; CRC Press: Boca Raton, FL, USA, 2014; pp. 3–38. [Google Scholar]
- Echigoya, R.; Rhodes, L.; Oshima, Y.; Satake, M. The structures of five new antifungal and hemolytic amphidinol analogs from Amphidinium carterae collected in New Zealand. Harmful Algae 2005, 4, 383–389. [Google Scholar] [CrossRef]
- Kobayashi, J.I.; Kubota, T. Bioactive macrolides and polyketides from marine dinoflagellates of the genus Amphidinium. J. Nat. Prod. 2007, 70, 451–460. [Google Scholar] [CrossRef]
- Meng, Y.; Van Wagoner, R.M.; Misner, I.; Tomas, C.; Wright, J.L. Structure and biosynthesis of amphidinol 17, a hemolytic compound from Amphidinium carterae. J. Nat. Prod. 2010, 73, 409–415. [Google Scholar] [CrossRef]
- Durán-Riveroll, L.D.; Cembella, A.D.; Okolodkov, Y.B. A review on the biodiversity and biogeography of toxigenic benthic marine dinoflagellates of the coasts of Latin America. Front. Mar. Sci. 2019, 6, 148. [Google Scholar] [CrossRef] [Green Version]
- Krock, B.; Busch, J.A.; Tillmann, U.; García-Camacho, F.; Sánchez-Mirón, A.; Gallardo-Rodríguez, J.J.; López-Rosales, L.; Andree, K.B.; Fernández-Tejedor, M.; Witt, M.; et al. LC-MS/MS detection of karlotoxins reveals new variants in strains of the marine dinoflagellate Karlodinium veneficum from the Ebro Delta (NW Mediterranean). Mar. Drugs 2017, 15, 391. [Google Scholar] [CrossRef] [Green Version]
- Krock, B.; Tillmann, U.; Wen, Y.; Hansen, P.J.; Larsen, T.O.; Andersen, A.J. Development of a LC-MS/MS method for the quantification of goniodomins A and B and its application to Alexandrium pseudogonyaulax strains and plankton field samples of Danish coastal waters. Toxicon 2018, 155, 51–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Place, A.R.; Bowers, H.A.; Bachvaroff, T.R.; Adolf, J.E.; Deeds, J.R.; Sheng, J. Karlodinium veneficum—The little dinoflagellate with a big bite. Harmful Algae 2012, 14, 179–195. [Google Scholar] [CrossRef]
- Blackburn, S.I.; Bolch, C.J.S.; Haskard, K.A.; Hallegraeff, G.M. Reproductive compatibility among four global populations of the toxic dinoflagellate Gymnodinium catenatum (Dinophyceae). Phycologia 2001, 40, 78–87. [Google Scholar] [CrossRef]
- Molina-Miras, A.; Morales-Amador, A.; de Vera, C.; López-Rosales, L.; Sánchez-Mirón, A.; Souto, M.; Fernández, J.; Norte, M.; García-Camacho, F.; Molina-Grima, E. A pilot-scale bioprocess to produce amphidinols from the marine microalga Amphidinium carterae: Isolation of a novel analogue. Algal Res. 2018, 31, 87–98. [Google Scholar] [CrossRef]




| Number of Neutral Losses | Neutral Loss (m/z) | C Atoms | Hydroxyl Groups | Double Bonds |
|---|---|---|---|---|
| Nonsulfated | ||||
| 1 | 392.13 | 22 | 5 | 5 |
| 2 | 426.23 | 22 | 7 | 4 |
| 3 | 418.24 | 24 | 5 | 6 |
| 4 | 398.28 | 22 | 5 | 2 |
| 5 | 440.25 | 23 | 7 | 4 |
| 6 | 442.23 | 22 | 8 | 4 |
| 7 | 338.18 | 18 | 5 | 4 |
| Sulfated | ||||
| 1.1 | 512.13 | 22 | 5 | 5 |
| 2.1 | 546.23 | 22 | 7 | 4 |
| 3.1 | 538.24 | 24 | 5 | 6 |
| 4.1 | 518.28 | 22 | 5 | 2 |
| 5.1 | 560.25 | 23 | 7 | 4 |
| 6.1 | 562.12 | 22 | 8 | 4 |
| 7.1 | 458.18 | 18 | 5 | 4 |
| Strain | tR (min) | Q1-Mass (m/z) | Neutral Loss |
|---|---|---|---|
| AxLT113 | 3.75 | 1114 | 518 |
| AxLT111 | 2.93 | 1138 | 338 |
| AxLT111 | 3.90 | 1140 | 392 |
| AtLPZ38 | 2.03 | 1238 | 440 |
| AxSQ175 | 2.90 | 1242 | 392/512 |
| AxSQ175 | 3.80 | 1264 | 392 |
| AA39 | 2.90 | 1296 | 426 |
| AeSQ172 | 3.79 | 1305 | 442 |
| AA60 | 3.75 | 1384 | 392 |
| AxSQ175 | 3.90 | 1392 | 442 |
| AxSQ175 | 3.90 | 1396 | 392 |
| AA60 | 2.92 | 1418 | 426 |
| AA60 | 2.85 | 1486 | 512 |
| AeSQ172 | 3.77 | 1764 | 440 |
| Strain | (min) | Q1-Mass (m/z) |
|---|---|---|
| AxLT113 | 4.16 | 1028 |
| AxLT113 | 4.16 | 1044 |
| AxRoq139 | 3.73 | 1074 |
| AxLT113 | 3.78 | 1134 |
| AxSQ175 | 4.04 | 1290 |
| AA60 | 3.03 | 1334 |
| Strain | (min) | Q1-Mass (m/z) | Detection Mode | Variant Name |
|---|---|---|---|---|
| AxSQ175 | 2.90 | 1242 | Neutral loss | U1 |
| AA60 | 3.75 | 1384 | Neutral loss | U2 |
| AA60 | 2.85 | 1486 | Neutral loss | U3 |
| AM | (min) | Strain/Species | ||||||
|---|---|---|---|---|---|---|---|---|
| AA39 A. massartii | AA60 A. operculatum | AxLT111 * | AeSQ 172 A. eilatiensis | AxSQ175 * | AeSQ177 A. eilatiensis | AeSQ181 A. eilatiensis | ||
| AM02 | 3.78 | 4 | 21 | 200 | 22 | 231 | 632 | |
| AM04 | 3.87 | 441 | 547 | 2 | ||||
| AM05 | 4.07 | 317 | 406 | 1 | ||||
| AM06 | 3.18 | 22 | ||||||
| AM07 | 3.3 | 45 | 79 | 250 | ||||
| AM09 | 4.10 | 1 | 19 | 796 | ||||
| AM11 | 3.43 | 2 | 5 | 5 | ||||
| AM14 | 2.61 | 2 | 10 | 2 | ||||
| AM15 | 3.23 | 1 | ||||||
| AM17 | 4.17 | 2 | 5 | 2 | ||||
| N7 | 4.20 | 2 | ||||||
| N8/N9 | 4.17 | 2 | ||||||
| N12 | 3.88 | 14 | 15 | |||||
| N13 | 3.52 | 1 | 2 | |||||
| U1 | 2.90 | 985 | ||||||
| U2 | 3.75 | 32 | ||||||
| U3 | 2.85 | 79 | ||||||
| Total AM content | 1 | 139 | 23 | 1021 | 1012 | 1314 | 1694 | |
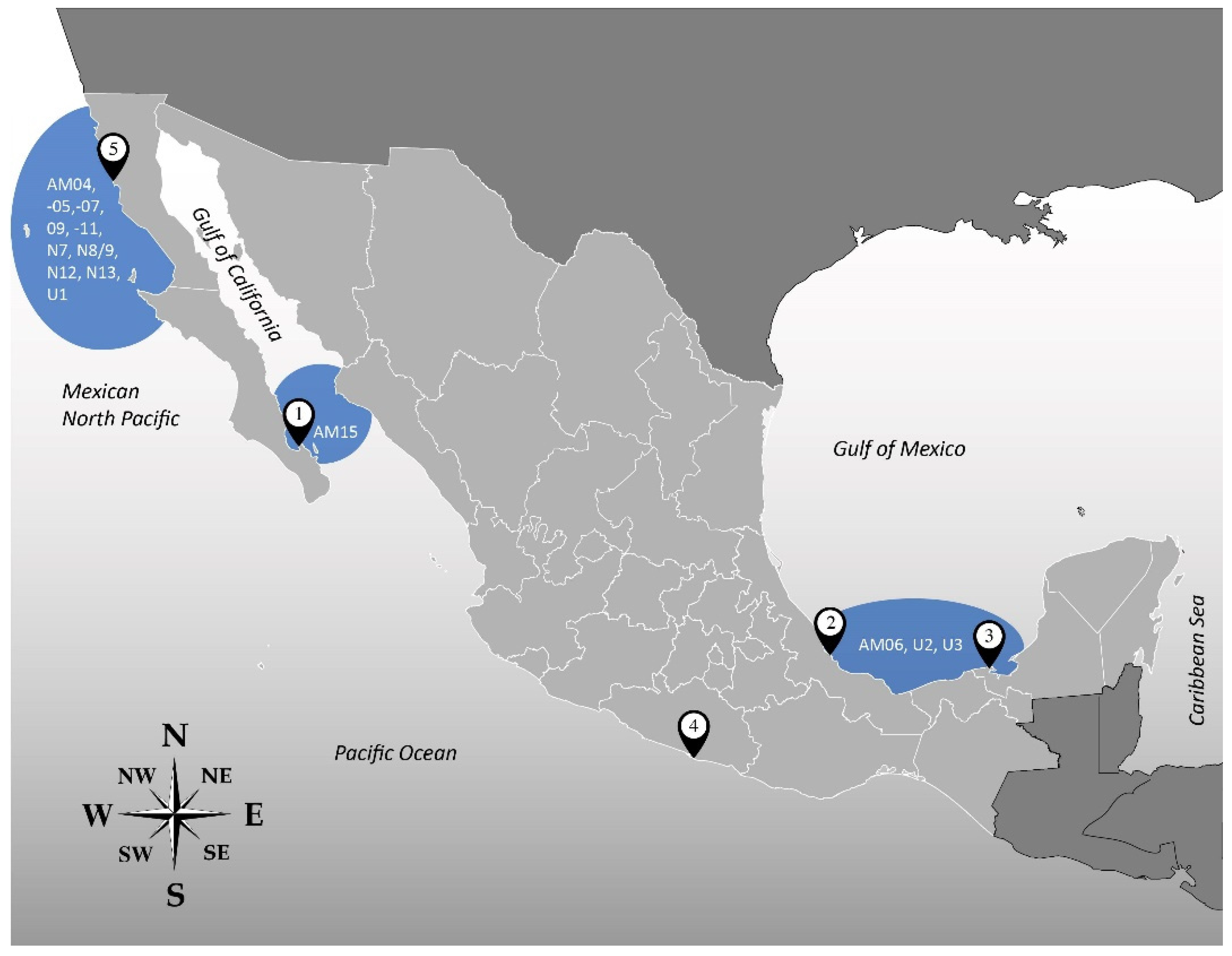
| Isolate | Locality of Origin * | Geographical Location | Date of Sampling | Species ** |
|---|---|---|---|---|
| AtLPZ38 | Balandra Bay, Baja California Sur, (1) | Gulf of California | January 2018 | A. theodorei |
| AA39 | A. massartii | |||
| AA40 | A. theodorei | |||
| AA60 | Veracruz Reef System, Veracruz (2) | Gulf of Mexico | May 2018 | A. operculatum |
| AxLT111 | Laguna de Términos, Campeche (3) | January 2019 | Unidentified | |
| AxLT112 | A. massartii | |||
| AxLT113 | Unidentified | |||
| AxRoq139 | Roqueta Island, Acapulco, Guerrero (4) | Mexican South Pacific | June 2019 | Unidentified |
| AeSQ172 | San Quintín, Baja California (5) | Mexican North Pacific | December 2019 | A. eilatiensis |
| AxSQ175 | Unidentified | |||
| AeSQ177 | A. eilatiensis | |||
| AeSQ181 | A. eilatiensis |
| Chromatographic Parameters | |
| Eluent composition | A: 500 mL ultrapure water + 252.5 µL NH4OH (25%) B: 450 mL acetonitrile + 50 mL ultrapure water + 252.5 µL NH4OH (25%) |
| Eluent gradient | 80% Eluent A to 20% Eluent B |
| Total duration (min) | 5 |
| Flow rate (mL min−1) | 0.2 |
| Injection volume (µL) | 0.5 |
| Collision energy (eV) | 75 85 for AMs over 1500 m/z during product ion scans |
| Scanning time (sec) | 0.133 |
| Scanning rate (points per peak) | 12 |
| Spectrometric Parameters | |
| Ion source | |
| Capillary voltage (kV) Cone voltage (V) Desolvation temperature (°C) | 3.00 40 600 |
| Gas flow | |
| Desolvation gas flow (L h−1) Cone gas flow (L h−1) Nebulizer gas flow (bar) | 1000 150 7.0 |
| Further settings | |
| Autosampler temperature (°C) Column temperature (°C) Electrospray Full-scan mass range (m/z) | 10 40 ES+ 1000–1800 |
| Toxin | Q1-Mass (m/z) | Q3-Mass (m/z) | Toxin | Q1-Mass (m/z) | Q3-Mass (m/z) |
|---|---|---|---|---|---|
| AM1 | 1512 | 974 | LP-A | 1278 | 754 |
| AM2 | 1398 | 1006 | LP-B | 1344 | 904 |
| AM3 | 1350 | 932 | LP-C | 1344 | 904 |
| AM4 | 1324 | 932 | LP-D | 1330 | 904 |
| AM5 | 1394 | 976 | LS | 1374 | 976 |
| AM6 | 1368 | 976 | LS-A | 1296 | 904 |
| AM7 | 1254 | 742 | LS-B | 1266 | 754 |
| AM9 | 1350 | 932 | SP | 1266 | 754 |
| AM10 | 1296 | 904 | N1 | 1268 | 876 |
| AM11 | 1500 | 988 | N2 | 1430 | 1038 |
| AM12 | 1426 | 914 | N3 | 1458 | 1066 |
| AM13 | 1452 | 914 | N4 | 1316 | 876 |
| AM14 | 1288 | 742 | N5 | 1346 | 946 |
| AM15 | 1186 | 760 | N6 | 1346 | 946 |
| AM17 | 1306 | 816 | N7 | 1346 | 928 |
| AM18 | 1382 | 964 | N8 | 1364 | 946 |
| AM19 | 1484 | 946 | N9 | 1364 | 946 |
| AM20 (M) * | 1346 | 904 | N10 | 1364 | 946 |
| AM20 (S) ** | 1653 | 1260 | N11 | 1364 | 946 |
| AM21 | 1798 | 1406 | N12 | 1326 | 946 |
| AM22 | 1668 | 1330 | N13 | 1326 | 928 |
| AM-A | 1362 | 964 | N14 | 1344 | 946 |
| AM-B | 1464 | 946 | N15 | 1344 | 946 |
| AMD-G | 1300 | 768 | N16 | 1344 | 946 |
| CAR-E | 1422 | 1030 | U1 | 1242 | 730 |
| KAR-A | 1480 | 1082 | U2 | 1384 | 992 |
| KAR-B | 1462 | 1064 | U3 | 1486 | 974 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Durán-Riveroll, L.M.; Weber, J.; Krock, B. First Identification of Amphidinols from Mexican Strains and New Analogs. Toxins 2023, 15, 163. https://doi.org/10.3390/toxins15020163
Durán-Riveroll LM, Weber J, Krock B. First Identification of Amphidinols from Mexican Strains and New Analogs. Toxins. 2023; 15(2):163. https://doi.org/10.3390/toxins15020163
Chicago/Turabian StyleDurán-Riveroll, Lorena M., Jannik Weber, and Bernd Krock. 2023. "First Identification of Amphidinols from Mexican Strains and New Analogs" Toxins 15, no. 2: 163. https://doi.org/10.3390/toxins15020163
APA StyleDurán-Riveroll, L. M., Weber, J., & Krock, B. (2023). First Identification of Amphidinols from Mexican Strains and New Analogs. Toxins, 15(2), 163. https://doi.org/10.3390/toxins15020163






