Characterization of Fusarium verticillioides Med1 LxxLL Motif Involved in Fumonisin Biosynthesis
Abstract
1. Introduction
2. Results
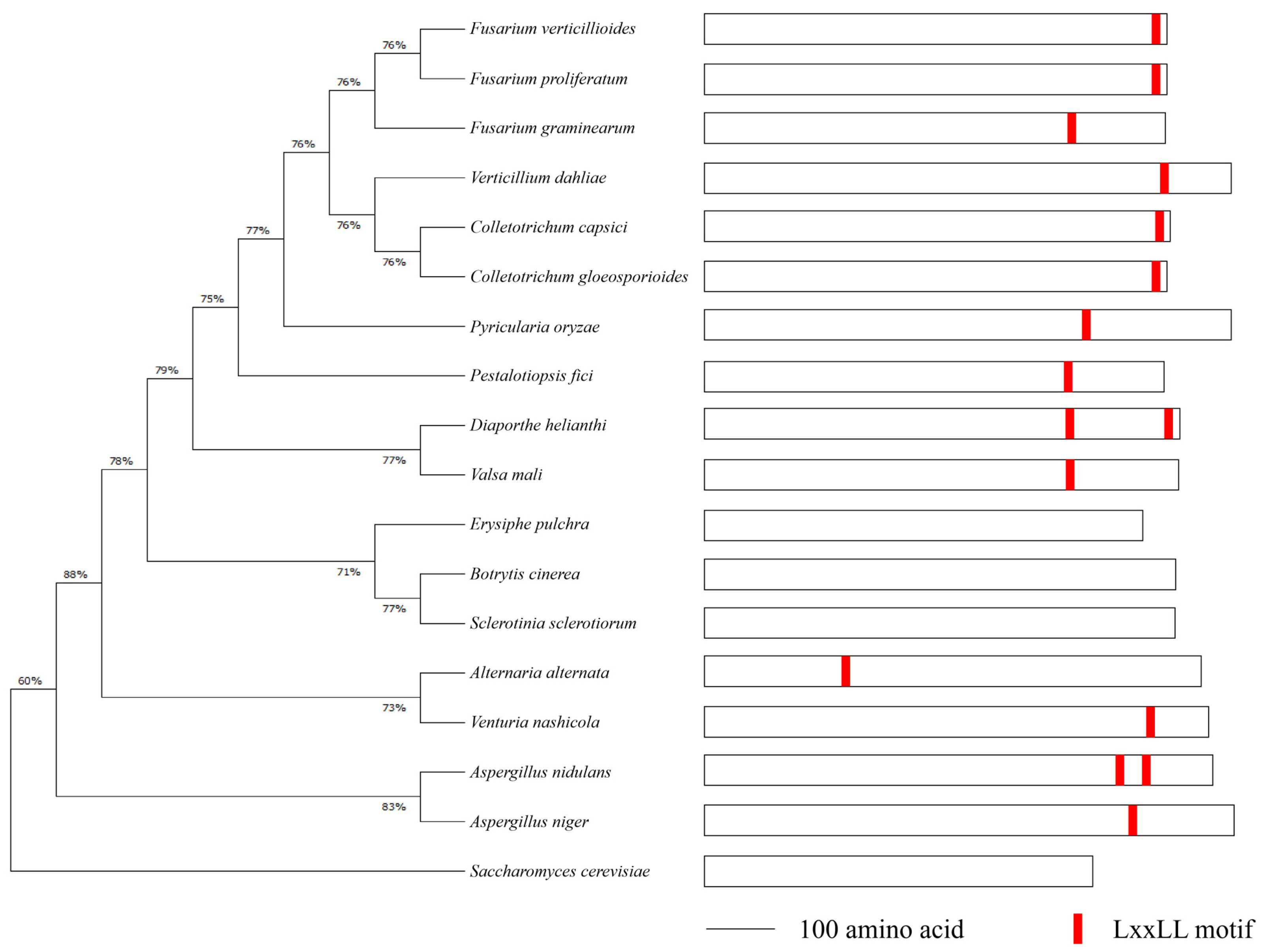
2.1. Identification of Med1 LxxLL Motif in F. verticillioides
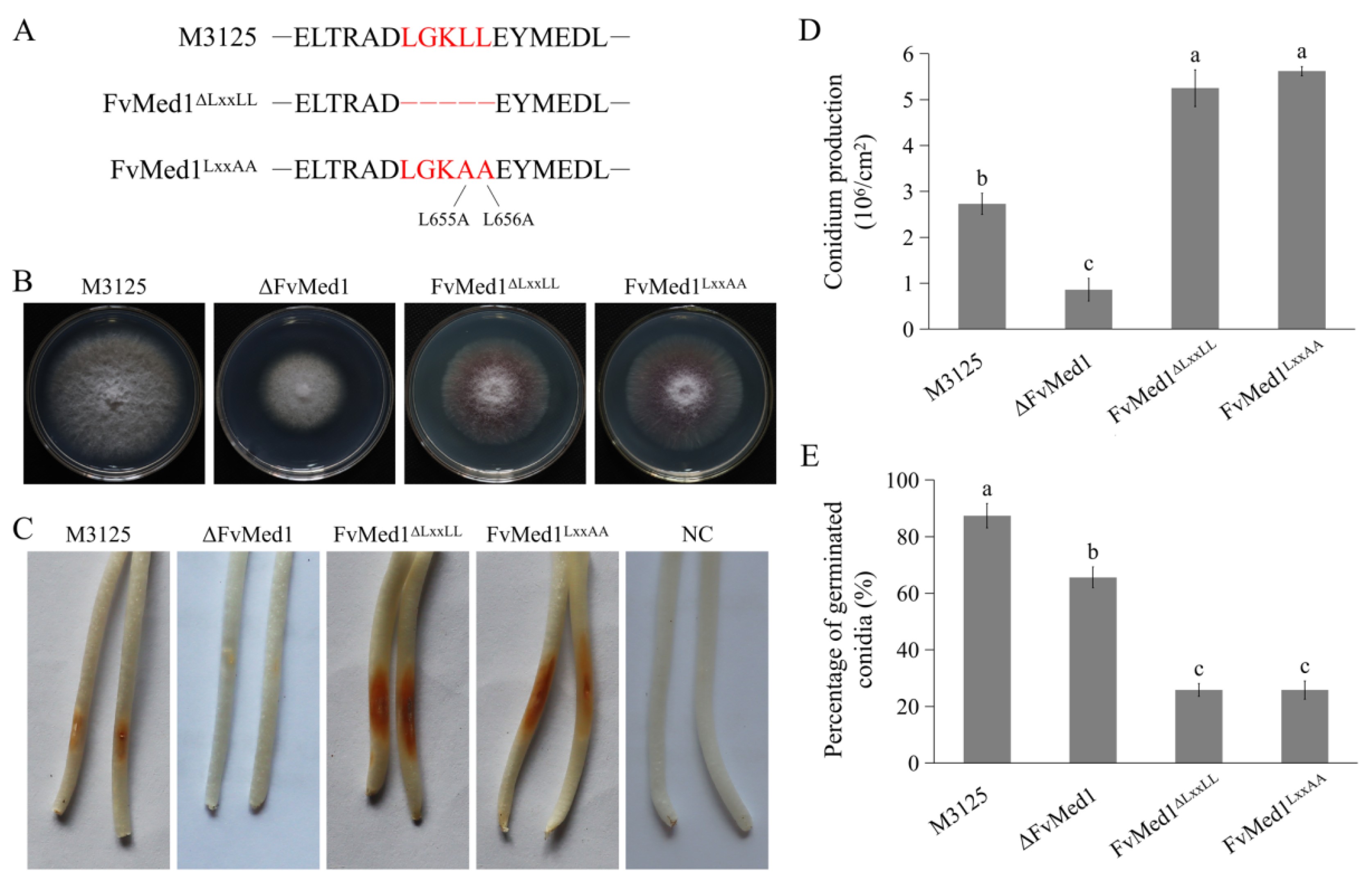
2.2. Med1 LxxLL Motif Plays a Role in F. verticillioides Development and Virulence
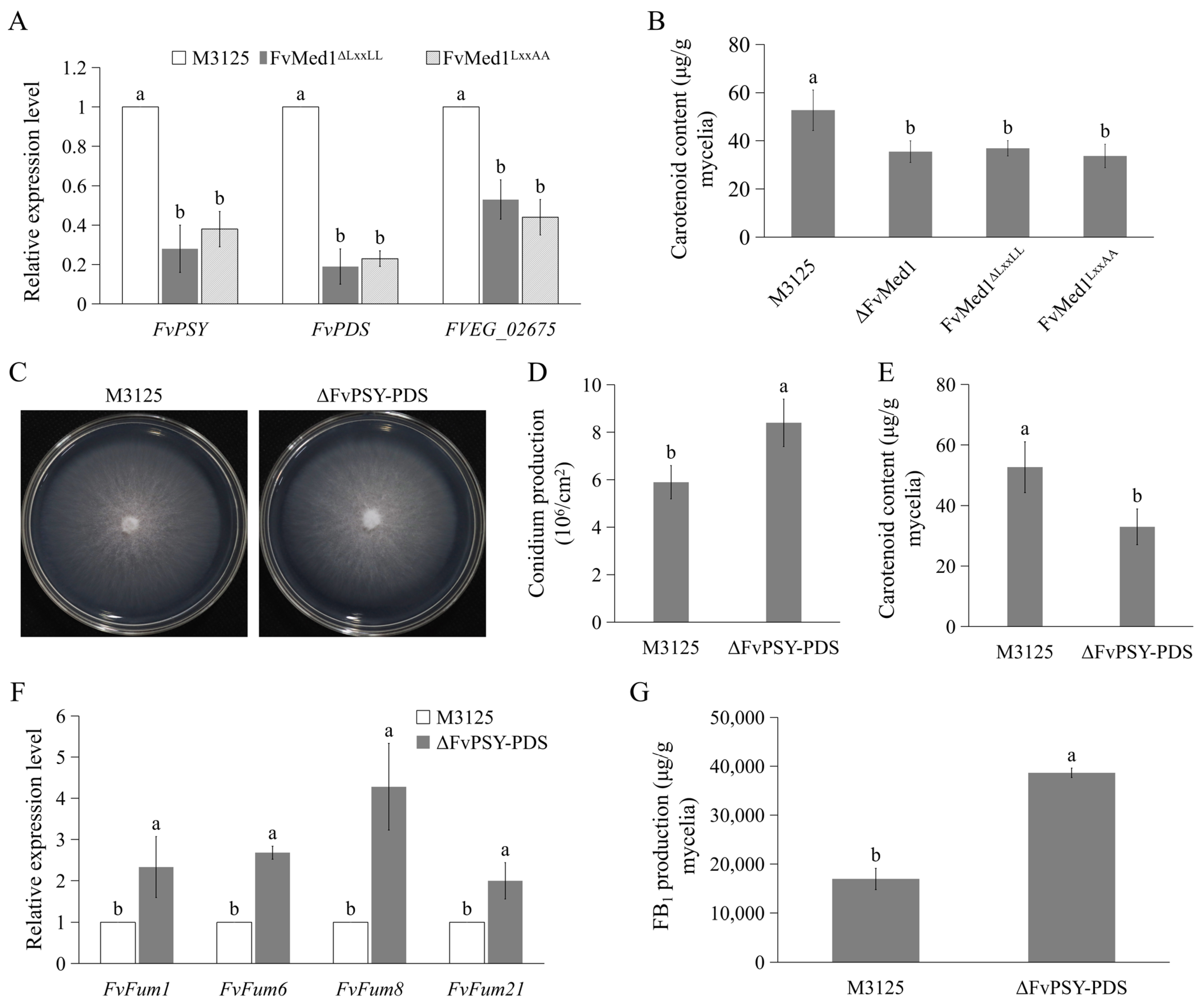
2.3. Med1 LxxLL Mutations Significantly Stimulate FB1 Biosynthesis and Upregulate Fum8 Gene Expression
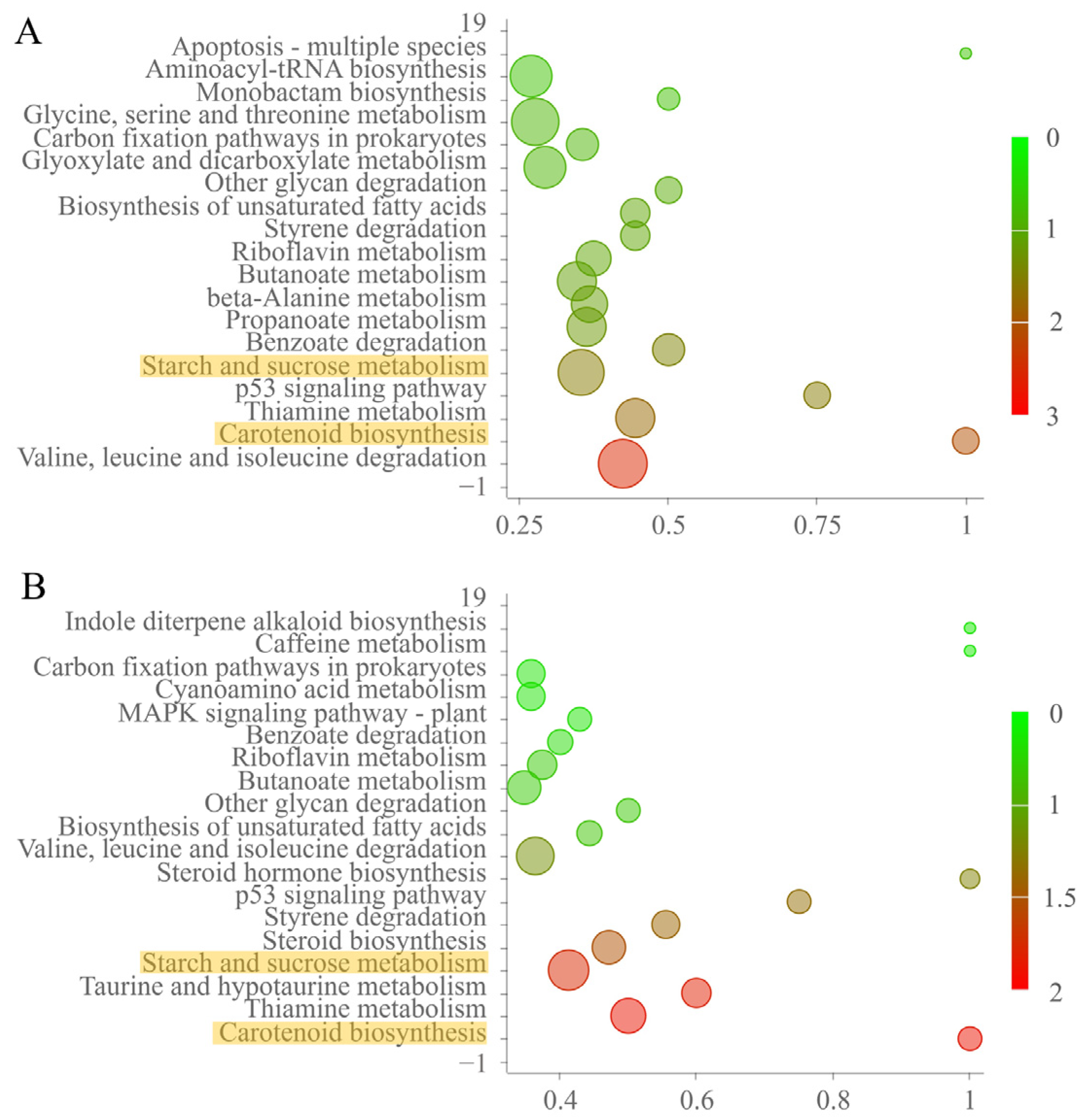
2.4. Proteomic Profiles of Med1 LxxLL Mutants Grown under Fumonisin-Inducing Conditions
2.5. Med1 LxxLL Motif Regulates Amylopectin Production in F. verticillioides
2.6. Suppressed Carotenoid Production Contributed to Enhanced FB1 Biosynthesis in Med1 LxxLL Mutants
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Strains and Culture Assays
5.2. Strain Construction
5.3. Analysis of FB1 Production
5.4. Microscopic Examinations
5.5. Proteomic Data and Bioinformatic Analysis
5.6. Quantification of Amylopectin Content and Carotenoid Production
5.7. Effect of Med1 LxxLL Mutations on the Expression of Key Genes in Starch Metabolism and Carotenoid Biosynthesis
5.8. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Allen, B.; Taatjes, D. The Mediator complex: A central integrator of transcription. Mol. Cell Biol. 2015, 16, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.; Trnka, M.; Bushnell, D.; Davis, R.; Mattei, P.; Burlingame, A.; Kornberg, R.D. Structure of a complete Mediator-RNA polymerase II pre-initiation complex. Cell 2016, 166, 1411–1422. [Google Scholar] [CrossRef] [PubMed]
- Richter, W.F.; Nayak, S.; Iwasa, J.; Taatjes, D.J. The Mediator complex is a master regulator of transcription by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2022, 23, 732–749. [Google Scholar] [CrossRef] [PubMed]
- Soutourina, J. Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 2018, 19, 262–274. [Google Scholar] [CrossRef]
- Paul, E.; Zhu, Z.I.; Landsman, D.; Morse, R.H. Genome-wide association of Mediator and RNA polymerase II in wild-type and Mediator mutant yeast. Mol. Cell Biol. 2015, 35, 331–342. [Google Scholar] [CrossRef]
- Jeronimo, C.; Robert, F. The mediator complex: At the nexus of RNA polymerase II transcription. Trends Cell Biol. 2017, 27, 765–783. [Google Scholar] [CrossRef]
- Weber, H.; Garabedian, M.J. The mediator complex in genomic and non-genomic signaling in cancer. Steroids 2018, 133, 8–14. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Cevher, M.A.; Shi, Y.; Li, D.; Chait, B.T.; Malik, S.; Roeder, R.G. Reconstitution of active human core Mediator complex reveals a critical role of the MED14 subunit. Nat. Struct. Mol. Biol. 2014, 21, 1028–1034. [Google Scholar] [CrossRef]
- Tsai, K.L.; Tomomori-Sato, C.; Sato, S.; Conaway, R.C.; Conaway, J.W.; Asturias, F.J. Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 2014, 157, 1430–1444. [Google Scholar] [CrossRef]
- Plaschka, C.; Lariviere, L.; Wenzeck, L.; Seizl, M.; Hemann, M.; Tegunov, D.; Petrotchenko, E.V.; Borchers, C.H.; Baumeister, W.; Herzog, F.; et al. Architecture of the RNA polymerase II-mediator core initiation complex. Nature 2015, 518, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Holstege, F.C.P.; Jennings, E.G.; Wyrick, J.J.; Lee, T.I.; Hengartner, C.J.; Green, M.R.; Golub, T.R.; Lander, E.S.; Young, R.A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 1998, 95, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Andrau, J.C.; van de Pasch, L.; Lijnzaad, P.; Bijma, T.; Koerkamp, M.G.; van de Peppel, J.; Werner, M.; Holstege, F.C.P. Genome-wide location of the coactivator mediator: Binding without activation and transient Cdk8 interaction on DNA. Mol. Cell 2006, 22, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.L.; Sato, S.; Tomomori-Sato, C.; Conaway, R.C.; Conaway, J.W.; Asturias, F.J. A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat. Struct. Mol. Biol. 2013, 20, 611–619. [Google Scholar] [CrossRef]
- Zhu, Y.; Qi, C.; Jia, Y.; Nye, J.S.; Rao, M.S.; Reddy, J.K. Deletion of PBP/PPARBP, the gene for nuclear receptor coactivator peroxisome proliferator-activated receptor-binding protein, results in embryonic lethality. J. Biol. Chem. 2000, 275, 14779–14782. [Google Scholar] [CrossRef]
- Jia, Y.Z.; Guo, G.L.; Surapureddi, S.; Sarkar, J.; Qi, C.; Guo, D.S.; Xia, J.; Kashireddi, P.; Yu, S.T.; Cho, Y.W.; et al. Transcription coactivator peroxisome proliferator-activated receptor-binding protein/mediator 1 deficiency abrogates acetaminophen hepatotoxicity. Proc. Natl. Acad. Sci. USA 2015, 102, 12531–12536. [Google Scholar] [CrossRef]
- Zhou, J.; Singh, B.K.; Ho, J.P.; Lim, A.; Bruinstroop, E.; Ohba, K.; Sinha, R.A.; Yen, P.M. MED1 mediator subunit is a key regulator of hepatic autophagy and lipid metabolism. Autophagy 2021, 17, 4043–4061. [Google Scholar] [CrossRef]
- Kuriyan, J.; Cowburn, D. Modular peptide recognition domains in eukaryotic signaling. Annu. Rev. Biophys. Biomol. Struct. 1997, 26, 259–288. [Google Scholar] [CrossRef]
- Hoeflich, K.P.; Ikura, M. Calmodulin in action: Diversity in target recognition and activation mechanisms. Cell 2002, 108, 739–742. [Google Scholar] [CrossRef]
- Heery, D.M.; Kalkhoven, E.; Hoare, S.; Parker, M.G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 1997, 387, 733–736. [Google Scholar] [CrossRef]
- Torchia, J.; Rose, D.W.; Inostroza, J.; Kamei, Y.; Westin, S.; Glass, C.K.; Rosenfeld, M.G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 1997, 387, 677–684. [Google Scholar] [CrossRef]
- Martinez-Zapien, D.; Ruiz, F.X.; Poirson, J.; Mitschler, A.; Ramirez, J.; Forster, A.; Cousido-Siah, A.; Masson, M.; Vande Pol, S.; Podjarny, A.; et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature 2016, 529, 541–545. [Google Scholar] [CrossRef]
- Qian, H.; He, P.; Lv, F.; Wu, W. Genome-wide analysis of LXXLL-mediated DAX1/SHP-nuclear receptor interaction network and rational design of stapled LXXLL-based peptides to target the specific network profile. Int. J. Biol. Macromol. 2019, 129, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.X.; Ito, M.; Fondell, J.D.; Fu, Z.Y.; Roeder, R.G. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl. Acad. Sci. USA 1998, 95, 7939–7944. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Roeder, R.G. The Mediator subunit MED1/TRAP220 is required for optimal glucocorticoid receptor-mediated transcription activation. Nucleic Acids Res. 2007, 35, 6161–6169. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, X.; Birsoy, K.; Roeder, R.G. A muscle-specific knockout implicates nuclear receptor coactivator MED1 in the regulation of glucose and energy metabolism. Proc. Natl. Acad. Sci. USA 2010, 107, 10196–101201. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.P.; Hu, Q.P.; Ito, M.; Meyer, S.; Waltz, S.; Khan, S.; Roeder, R.G.; Zhang, X.T. Key roles for MED1 LxxLL motifs in pubertal mammary gland development and luminal-cell differentiation. Proc. Natl. Acad. Sci. USA 2010, 107, 6765–6770. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wen, J.; Tang, Y.; Shi, J.; Mu, G.; Yan, R.; Cai, J.; Long, M. Research progress on fumonisin B1 contamination and toxicity: A review. Molecules 2021, 26, 5238. [Google Scholar] [CrossRef]
- Howard, P.C.; Eppley, R.M.; Stack, M.E.; Warbritton, A.; Voss, K.A.; Lorentzen, R.J.; Kovach, R.M.; Bucci, T.J. Fumonisin b1 carcinogenicity in a two-year feeding study using F344 rats and B6C3F1 mice. Environ. Health Perspect. 2001, 109, 277–282. [Google Scholar] [CrossRef]
- Chen, J.; Wei, Z.; Wang, Y.; Long, M.; Wu, W.; Kuca, K. Fumonisin B1: Mechanisms of toxicity and biological detoxification progress in animals. Food Chem. Toxicol. 2021, 149, 111977. [Google Scholar] [CrossRef]
- Woloshuk, C.P.; Shim, W.B. Aflatoxins, fumonisins, and trichothecenes: A convergence of knowledge. FEMS Microbiol. Rev. 2013, 37, 94–109. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Groopman, J.D.; Pestka, J.J. Public health impacts of foodborne mycotoxins. Annu. Rev. Food Sci. Technol. 2014, 5, 351–372. [Google Scholar] [CrossRef]
- Alexander, N.J.; Proctor, R.H.; McCormick, S.P. Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Rev. 2009, 28, 198–215. [Google Scholar] [CrossRef]
- Zhou, Z.H.; Yan, H.J.; Shim, W.B. Distinct function of Mediator subunits in fungal development, stress response and secondary metabolism in maize pathogen Fusarium verticillioides. Phytopathology 2022, 112, 1730–1738. [Google Scholar] [CrossRef]
- Ding, J.L.; Peng, Y.J.; Chu, X.L.; Feng, M.G.; Ying, S.H. Autophagy-related gene BbATG11 is indispensable for pexophagy and mitophagy, and contributes to stress response, conidiation and virulence in the insect mycopathogen Beauveria bassiana. Environ. Microbiol. 2018, 20, 3309–3324. [Google Scholar] [CrossRef]
- Yang, W.; Wu, H.; Wang, Z.; Sun, Q.; Qiao, L.; Huang, B. The APSES gene MrStuA regulates sporulation in Metarhizium robertsii. Front. Microbiol. 2018, 9, 1208. [Google Scholar] [CrossRef]
- Lin, H.Y.; Wang, J.J.; Feng, M.G.; Ying, S.H. Autophagy-related gene ATG7 participates in the asexual development, stress response and virulence of filamentous insect pathogenic fungus Beauveria bassiana. Curr. Genet. 2019, 65, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.J.; Shim, W.B. Characterization of non-canonical G beta-like protein FvGbb2 and its relationship with heterotrimeric G proteins in Fusarium verticillioides. Environ. Microbiol. 2020, 22, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.J.; Zhou, Z.H.; Zhang, H.; Shim, W.B. Vacuole proteins with optimized microtubule assembly is required for Fum1 protein localization and fumonisin biosynthesis in mycotoxigenic fungus Fusarium verticillioides. J. Fungi 2023, 9, 268. [Google Scholar] [CrossRef]
- Shim, W.B.; Flaherty, J.E.; Woloshuk, C.P. Comparison of fumonisin B1 biosynthesis in maize germ and degermed kernels by Fusarium verticillioides. J. Food Prot. 2003, 66, 2116–2122. [Google Scholar] [CrossRef]
- Bluhm, B.H.; Woloshuk, C.P. Amylopectin induces fumonisin B1 production by Fusarium verticillioides during colonization of maize kernels. Mol. Plant Microbe Interact. 2005, 18, 1333–1339. [Google Scholar] [CrossRef]
- Ansari, S.A.; Morse, R.H. Mechanisms of Mediator complex action in transcriptional activation. Cell Mol. Life Sci. 2013, 70, 2743–2756. [Google Scholar] [CrossRef] [PubMed]
- Poss, Z.C.; Ebmeier, C.C.; Taatjes, D.J. The Mediator complex and transcription regulation. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 575–608. [Google Scholar] [CrossRef] [PubMed]
- Tudor, M.; Murray, P.J.; Onufryk, C.; Jaenisch, R.; Young, R.A. Ubiquitous expression and embryonic requirement for RNA polymerase II coactivator subunit Srb7 in mice. Genes Dev. 1999, 13, 2365–2368. [Google Scholar] [CrossRef][Green Version]
- Thompson, C.M.; Young, R.A. General requirement for RNA polymerase II holoenzymes in vivo. Proc. Natl. Acad. Sci. USA 1995, 92, 4587–4590. [Google Scholar] [CrossRef]
- Lindsay, A.K.; Morales, D.K.; Liu, Z.L.; Grahl, N.; Zhang, A.; Willger, S.D.; Myers, L.C.; Hogan, D.A. Analysis of Candida albicans mutants defective in the Cdk8 module of mediator reveal links between metabolism and biofilm formation. PLoS Genet. 2014, 10, e1004567. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, W.; Chen, Q.; Fang, M.; Zheng, S.; Scheres, B.; Li, C.Y. Mediator subunit MED31 is required for radial patterning of Arabidopsis roots. Proc. Natl. Acad. Sci. USA 2018, 115, E5624–E5633. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Roeder, R.G. Mediator-dependent nuclear receptor function. Semin. Cell Dev. Biol. 2011, 22, 749–758. [Google Scholar] [CrossRef]
- Fondell, J.D.; Ge, H.; Roeder, R.G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. USA 1996, 93, 8329–8333. [Google Scholar] [CrossRef]
- Malik, S.; Guermah, M.; Yuan, C.X.; Wu, W.; Yamamura, S.; Roeder, R.G. Structural and functional organization of TRAP220, the TRAP/mediator subunit that is targeted by nuclear receptors. Mol. Cell Biol. 2004, 24, 8244–8254. [Google Scholar] [CrossRef]
- Ge, K.; Cho, Y.W.; Guo, H.; Hong, T.B.; Guermah, M.; Ito, M.; Yu, H.; Kalkum, M.; Roeder, R.G. Alternative mechanisms by which mediator subunit MED1/TRAP220 regulates peroxisome proliferator-activated receptor gamma-stimulated adipogenesis and target gene expression. Mol. Cell. Biol. 2008, 28, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Lefterova, M.I.; Zhang, Y.; Steger, D.J.; Schupp, M.; Schug, J.; Cristancho, A.; Feng, D.; Zhuo, D.; Stoeckert, C.J., Jr.; Liu, X.S.; et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008, 22, 2941–2952. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Gade, P.; Nallar, S.C.; Raha, A.; Roy, S.K.; Karra, S.; Reddy, J.K.; Reddy, S.P.; Kalvakolanu, D.V. The Med1 subunit of transcriptional mediator plays a central role in regulating CCAAT/enhancer-binding protein-beta-driven transcription in response to interferon-gamma. J. Biol. Chem. 2008, 283, 13077–13086. [Google Scholar] [CrossRef] [PubMed]
- Tomaru, T.; Steger, D.J.; Lefterova, M.I.; Schupp, M.; Lazar, M.A. Adipocyte-specific expression of murine resistin is mediated by synergism between peroxisome proliferator-activated receptor gamma and CCAAT/enhancer-binding proteins. J. Biol. Chem. 2009, 284, 6116–6125. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Huang, Y.; Huang, R.; Wu, Y.; Wang, W. The genetic architecture of amylose biosynthesis in maize kernel. Plant Biotechnol. J. 2018, 16, 688–695. [Google Scholar] [CrossRef]
- Nakamura, Y. A model for the reproduction of amylopectin cluster by coordinated actions of starch branching enzyme isoforms. Plant Mol. Biol. 2023, 112, 199–212. [Google Scholar] [CrossRef] [PubMed]
- Ponts, N.; Pinson-Gadais, L.; Barreau, C.; Richard-Forget, F.; Ouellet, T. Exogenous H2O2 and catalase treatments interfere with Tri genes expression in liquid cultures of Fusarium graminearum. FEBS Lett. 2007, 581, 443–447. [Google Scholar] [CrossRef]
- Audenaert, K.; Callewaert, E.; Holfe, M.; De Saeger, S.; Haesaert, G. Hydrogen peroxide induced by the fungicide prothioconazole triggers deoxynivalenol (DON) production by Fusarium graminearum. BMC Microbiol. 2010, 10, 112. [Google Scholar] [CrossRef]
- Reverberi, M.; Ricelli, A.; Zjalic, S.; Fabbri, A.A.; Fanelli, C. Natural functions of mycotoxins and control of their biosynthesis in fungi. Appl. Environ. Microbiol. 2010, 87, 899–911. [Google Scholar] [CrossRef]
- Ponts, N.; Pinson-Gadais, L.; Verdal-Bonnin, M.N.; Barreau, C.; Richard-Forget, F. Accumulation of deoxynivalenol and its 15-acetylated form is significantly modulated by oxidative stress in liquid cultures of Fusarium graminearum. FEMS Microbiol. Lett. 2006, 258, 102–107. [Google Scholar] [CrossRef]
- Jayashree, T.; Subramanyam, C. Oxidative stress as a prerequisite for aflatoxin production by Aspergillus parasiticus. Free Radic. Biol. Med. 2000, 29, 981–985. [Google Scholar] [CrossRef] [PubMed]
- Reverberi, M.; Zjalic, S.; Ricelli, A.; Fabbri, A.A.; Fanelli, C. Oxidant/antioxidant balance in Aspergillus parasiticus affects aflatoxin biosynthesis. Mycotoxin Res. 2006, 22, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Stahl, W.; Sies, H. Antioxidant activity of carotenoids. Mol. Asp. Med. 2003, 24, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Gomez, J.; Marin, S.; Nogareda, C.; Sanchis, V.; Ramos, A.J. The effect of enhanced carotenoid content of transgenic maize grain on fungal colonization and mycotoxin content. Mycotoxin Res. 2016, 32, 221–228. [Google Scholar] [CrossRef]
- Zhou, Z.H.; Duan, Y.B.; Zhou, M.G. Carbendazim-resistance associated β2-tubulin substitutions increase deoxynivalenol biosynthesis by reducing the interaction between β2-tubulin and IDH3 in Fusarium graminearum. Environ. Microbiol. 2020, 22, 598–614. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Z.; Liu, J.; Zhang, J.; Yan, H.; Yi, T.; Shim, W.B. Characterization of Fusarium verticillioides Med1 LxxLL Motif Involved in Fumonisin Biosynthesis. Toxins 2023, 15, 652. https://doi.org/10.3390/toxins15110652
Zhou Z, Liu J, Zhang J, Yan H, Yi T, Shim WB. Characterization of Fusarium verticillioides Med1 LxxLL Motif Involved in Fumonisin Biosynthesis. Toxins. 2023; 15(11):652. https://doi.org/10.3390/toxins15110652
Chicago/Turabian StyleZhou, Zehua, Jie Liu, Jie Zhang, Huijuan Yan, Tuyong Yi, and Won Bo Shim. 2023. "Characterization of Fusarium verticillioides Med1 LxxLL Motif Involved in Fumonisin Biosynthesis" Toxins 15, no. 11: 652. https://doi.org/10.3390/toxins15110652
APA StyleZhou, Z., Liu, J., Zhang, J., Yan, H., Yi, T., & Shim, W. B. (2023). Characterization of Fusarium verticillioides Med1 LxxLL Motif Involved in Fumonisin Biosynthesis. Toxins, 15(11), 652. https://doi.org/10.3390/toxins15110652




