Biodegradation of Aflatoxin B1 in the Baijiu Brewing Process by Bacillus cereus
Abstract
1. Introduction
2. Results and Discussion
2.1. Isolation of Microbial Strains for AFB1 Degradation
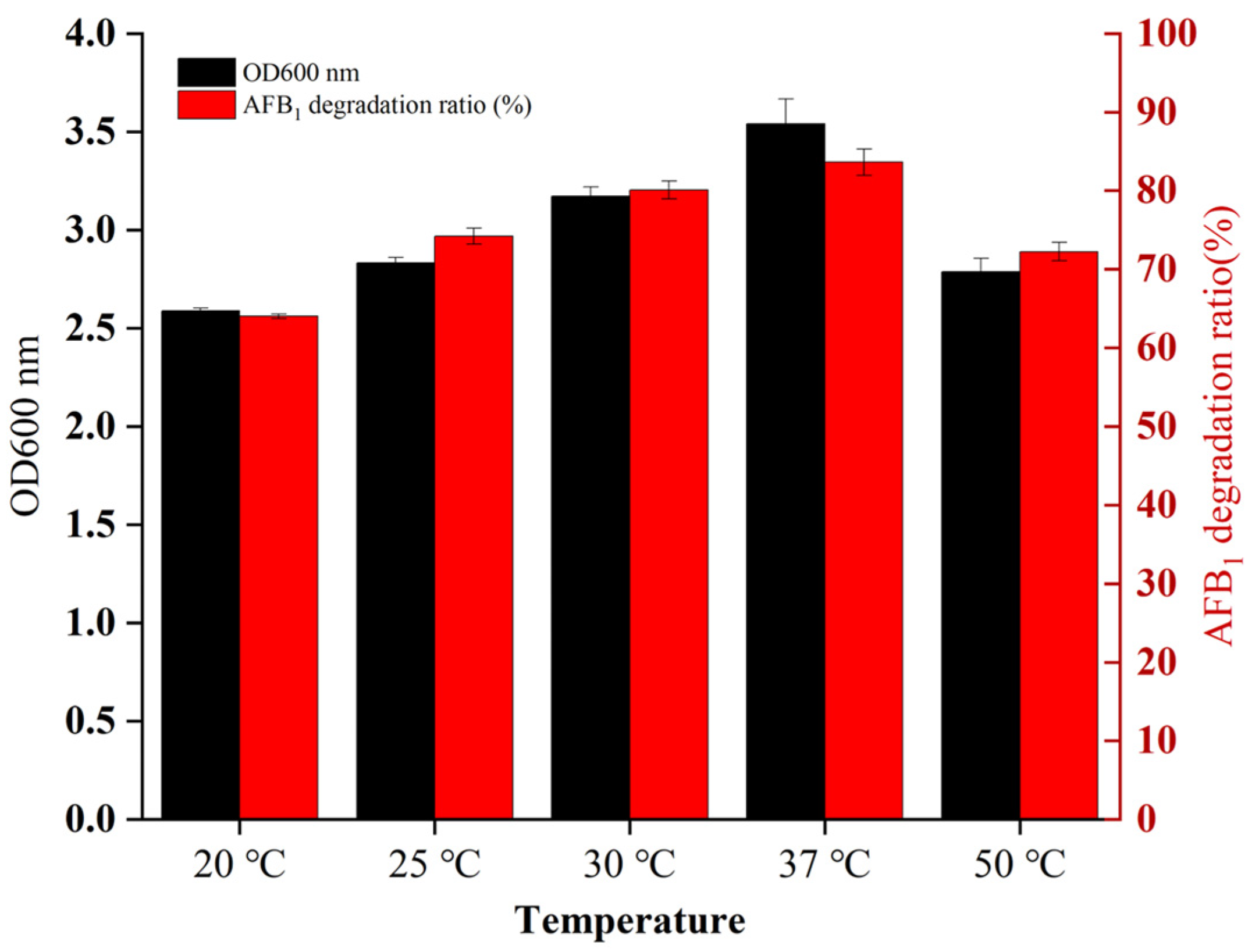
2.2. Adaptability of B. cereus XSSW9 to the Temperatures Found in Baijiu Fermentation Pits
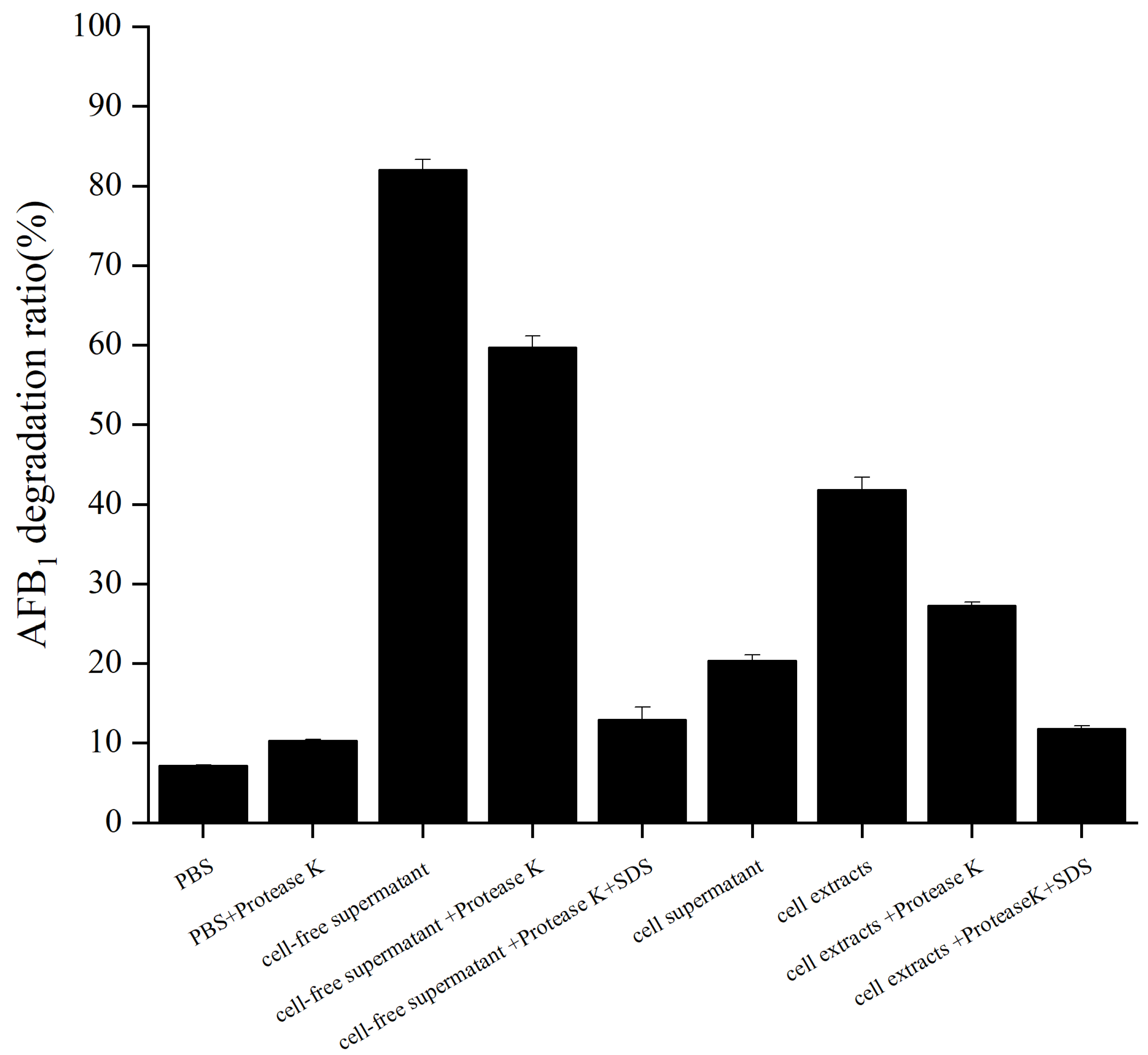
2.3. Degradation of AFB1 by the Cell-Free Supernatant, Cell Supernatant, and Cell Extracts of B. cereus XSWW9
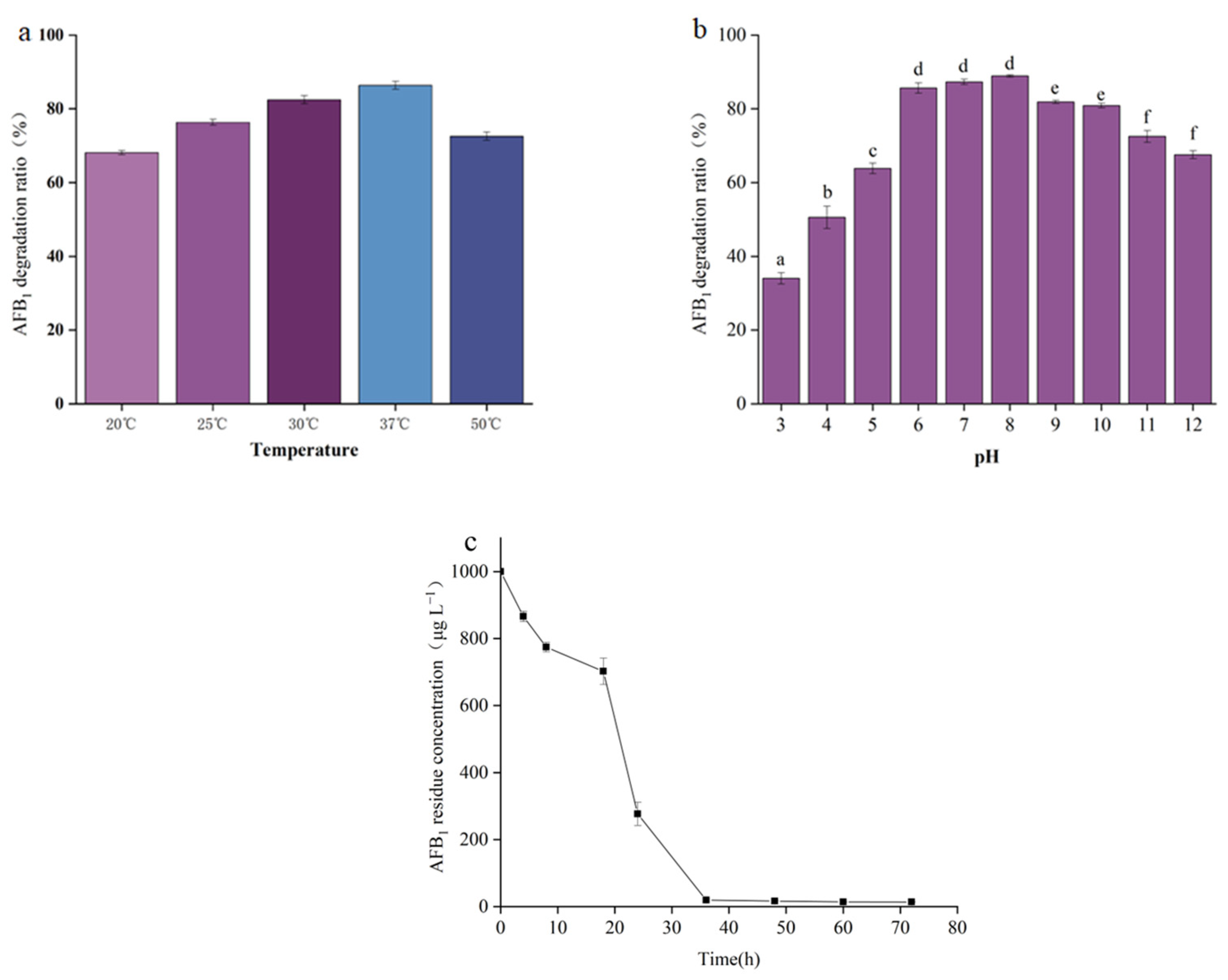
2.4. The Effect of Temperature and pH on AFB1 Degradation by the Cell-Free Supernatant of B. cereus XSSW9
2.5. Degradation of AFB1 by the Cell-Free Supernatant of XSSW9 during Grain Fermentation
2.6. Dynamics of Microbial AFB1 Degradation during Grain Fermentation
2.7. Analysis of Volatile Aroma Compounds during AFB1 Degradation in Grain Fermentation
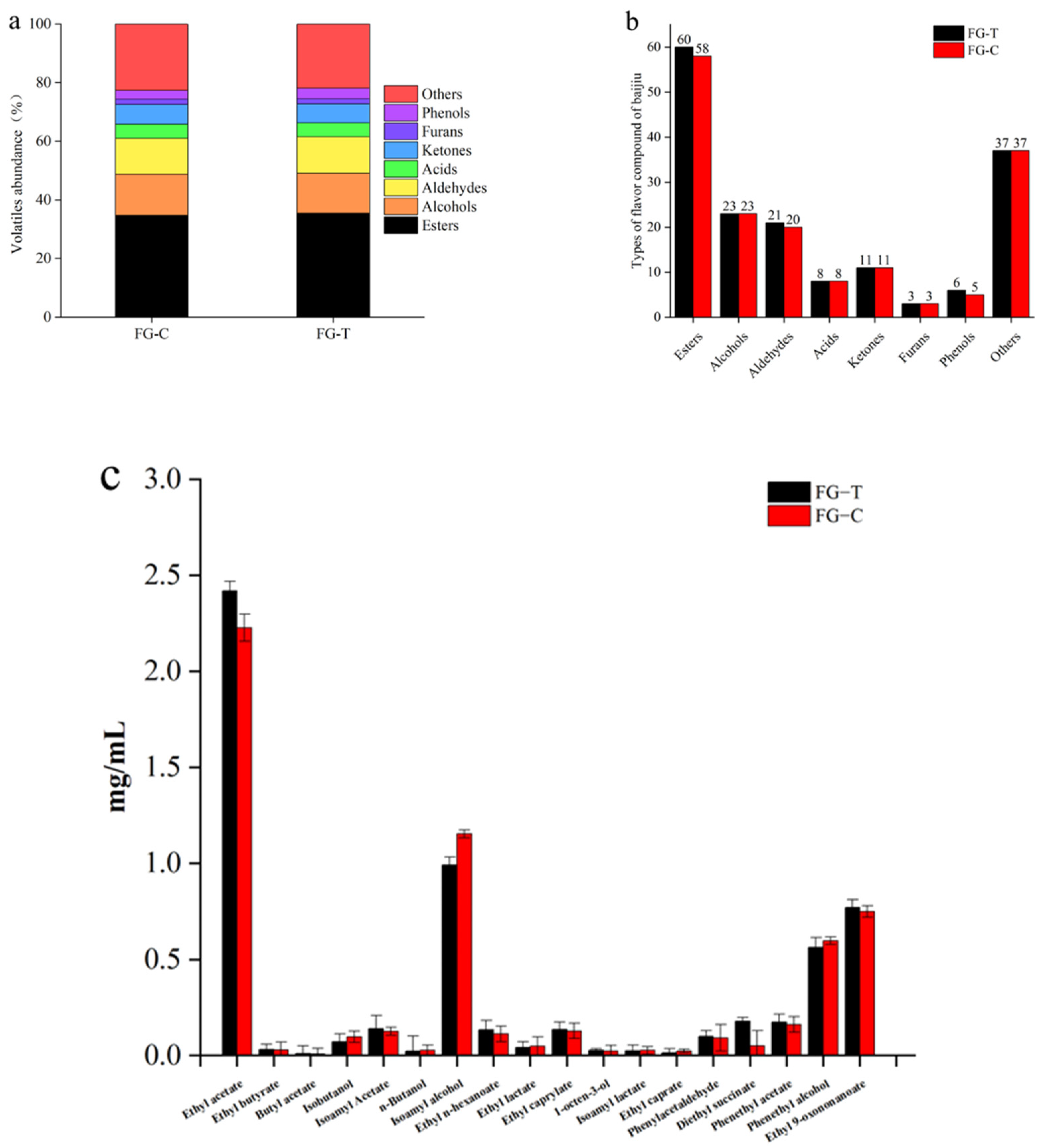
3. Conclusions
4. Materials and Methods
4.1. Chemicals and Culture Media
4.2. Preparation of Daqu Micro-Organism Suspensions
4.3. Enrichment and Isolation of Degrading Micro-Organism
4.4. Culture of Micro-Organisms
4.5. Degradation Test of AFB1 by B. cereus XSSW9
4.6. Detection of AFB1 Concentration
4.7. Identification of the AFB1 Degrading Strain
4.8. Preparation of Cell-Free Supernatant, Cell Supernatant, and Cell Extracts of B. cereus XSSW9
4.9. The Effect of Temperature and pH on AFB1 Degradation by the Cell-Free Supernatant of B. cereus XSSW9
4.10. Grain Fermentation (Baijiu Brewing)
4.11. Determination of Volatile Compounds
4.12. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guengerich, F.P.; Johnson, W.W.; Ueng, Y.F.; Yamazaki, H.; Shimada, T. Involvement of cytochrome P450, glutathione S-transferase, and epoxide hydrolase in the metabolism of aflatoxin B1 and relevance to risk of human liver cancer. Environ. Health Perspect. 1996, 104, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Hussein, H.S.; Brasel, J.M. Toxicity, metabolism, and impact of mycotoxins on humans and animals. Toxicology 2001, 167, 101–134. [Google Scholar] [CrossRef] [PubMed]
- Kabak, B.; Dobson, A.D.; Var, I. Strategies to prevent mycotoxin contamination of food and animal feed: A review. Crit. Rev. Food Sci. Nutr. 2006, 46, 593–619. [Google Scholar] [CrossRef] [PubMed]
- Mishra, H.N.; Das, C. A review on biological control and metabolism of aflatoxin. Crit. Rev. Food Sci. Nutr. 2003, 43, 245–264. [Google Scholar] [CrossRef]
- Zhu, M.; Zheng, J.; Xie, J.; Zhao, D.; Qiao, Z.; Huang, D.; Luo, H. Effects of environmental factors on the microbial community changes during medium-high temperature Daqu manufacturing. Food Res. Int. 2022, 153, 110955. [Google Scholar] [CrossRef]
- Ratnavathi, C.V.; Sashidhar, R.B. Substrate suitability of different geno-types of sorghum in relation to Aspergillus infection and aflatoxin production. J. Agric. Food Chem. 2003, 51, 3482–3492. [Google Scholar] [CrossRef]
- Wang, Y.; Ning, B.; Peng, Y.; Bai, J.; Liu, M.; Fan, X.; Sun, Z. Application of suspension array for simultaneous detection of four different mycotoxins in corn and peanut. Biosens. Bioelectron. 2013, 41, 391–396. [Google Scholar] [CrossRef]
- Nagatomi, Y.; Inoue, T.; Uyama, A.; Mochizuki, N. The Fate of Mycotoxins during the Distillation Process of Barley Shochu, a Distilled Alcoholic Beverage. Biosci. Biotechnol. Biochem. 2012, 76, 202–204. [Google Scholar] [CrossRef]
- GB2761-2017; Limits of Fungal Toxins in Food. China Standards Press: Beijing, China, 2017.
- Boeira, L.S.; Bryce, J.H.; Stewart, G.G.; Flannigan, B. Inhibitory effect of Fusarium mycotoxins on growth of brewing yeast. 1. Zearalenone and Fumonisin B1. J. Inst. Brew. 1996, 105, 366–375. [Google Scholar] [CrossRef]
- Boeira, L.S.; Bryce, J.H.; Stewart, G.G.; Flannigan, B. Inhibitory effect of Fusarium mycotoxins on growth of brewing yeast. 1. Zearalenone and Fumonisin B1. J. Inst. Brew. 1996, 105, 376–381. [Google Scholar] [CrossRef]
- Boeira, L.S.; Bryce, J.H.; Stewart, G.G.; Flannigan, B. Influence of cultural conditions on sensitivity of brewing yeasts growth to Fusarium mycotoxins zearalenone, deoxynivalenol and fumonisin B1. Int. Biodeterior. Biodegrad. 2002, 50, 69–81. [Google Scholar] [CrossRef]
- Cundliffe, E.; Davies, C.J. Mechanism of Inhibition of Eukaryotic Protein Synthesis by Trichothecene Fungal Toxins. Proc. Natl. Acad. Sci. USA 1974, 71, 30–34. [Google Scholar] [CrossRef]
- Cundliffe, E.; Davies, J.E. Inhibition of Initiation, Elongation, and Termination of Eukaryotic Protein Synthesis by Trichothecene Fungal Toxins. Antimicrob. Agents Chemother. 1977, 11, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Line, J.E.; Brackett, R.E.; Wilkinson, R.E. Evidence for degradation of aflatoxin B1 by Flavobacterium aurantiacum. J. Food Prot. 1994, 57, 788–791. [Google Scholar] [CrossRef]
- Guan, S.; Ji, C.; Zhou, T.; Li, J.; Ma, Q.; Niu, T. Aflatoxin B 1 degradation by Stenotrophomonas maltophilia and other microbes selected using coumarin medium. Int. J. Mol. Sci. 2008, 9, 1489–1503. [Google Scholar] [CrossRef] [PubMed]
- Foszczyn’ska, B.; Dziuba, E. Physiological status of brewing yeast during fermentation of worts contaminated with mycotoxins, P. 1: T-2 and ZEA. Acta Sci. Pol. Biotechnol. 2007, 6, 3–12. [Google Scholar]
- Foszczyn´ska, B.; Dziuba, E. Physiological status of brewing yeast during fermentation of worts contaminated with mycotoxins, P. 2: DAS and OTA. Acta Sci. Pol. Biotechnol. 2007, 6, 25–34. [Google Scholar]
- Kłosowski, G.; Błajet-Kosicka, A. Mechanisms of pyrazine compounds formation and validation of raw material thermal processing during technological process based on the presence of pyrazine in raw spirits. Biotechnologia 2010, 1, 140–153. [Google Scholar]
- Kłosowski, G.; Mikulski, D.; Grajewski, J.; Błajet-Kosicka, A. The influence of raw material contamination with mycotoxins on alcoholic fermentation indicators. Bioresour. Technol. 2010, 101, 3147–3152. [Google Scholar] [CrossRef]
- Womack, E.D.; Brown, A.E.; Sparks, D.L. A recent review of non-biological remediation of aflatoxin-contaminated crops. J. Sci. Food Agric. 2014, 94, 1706–1714. [Google Scholar] [CrossRef]
- Halasz, A.; Lasztity, R.; Abonyi, T. Decontamination of Mycotoxin-Containing Food and Feed by Biodegradation. Food Rev. Int. 2009, 25, 284–298. [Google Scholar] [CrossRef]
- Fuchs, S.; Sontag, G.; Stidl, R.; Ehrlich, V.; Kundi, M. Detoxification of patulin and ochratoxin A, two abundant mycotoxins, by lactic acid bacteria. Food Chem. Toxicol. 2008, 46, 1398–1407. [Google Scholar] [CrossRef] [PubMed]
- Gratz, S.; Mykkanen, H.; El-Nezami, H. Aflatoxin B1 binding by a mixture of Lactobacillus and Propionibacterium: In vitro versus ex vivo. J. Food Prot. 2005, 68, 2470. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Zhang, H.; Yang, Q.; Ren, R. Efficacy of Pichia caribbica in controlling blue mold rot and patulin degradation in apples. Int. J. Food Microbiol. 2013, 162, 167–173. [Google Scholar] [CrossRef]
- Castoria, R.; Mannina, L.; Duran-Patron, R.; Maffei, F.; Sobolev, A.P. Conversion of the Mycotoxin Patulin to the Less Toxic Desoxypatulinic Acid by the Biocontrol Yeast Rhodosporidium kratochvilovae Strain LS11. J. Agric. Food Chem. 2011, 59, 11571. [Google Scholar] [CrossRef]
- Motomura, M.; Toyomasu, T.; Mizuno, K.; Shinozawa, T. Purification and characterization of an aflatoxin degradation enzyme from Pleurotus ostreatus. Microbiol. Res. 2003, 158, 237–242. [Google Scholar] [CrossRef]
- Xu, D.; Wang, H.; Zhang, Y.; Yang, Z.; Sun, X. Inhibition of non-toxigenic Aspergillus Niger FS10 isolated from Chinese fermented soybean on growth and aflatoxin B1 production by Aspergillus flavus. Food Control 2013, 32, 359–365. [Google Scholar] [CrossRef]
- Ahlberg, S.H.; Joutsjoki, V.; Korhonen, H.J. Potential of lactic acid bacteria in aflatoxin risk mitigation. Int. J. Food Microbiol. 2015, 207, 87–102. [Google Scholar] [CrossRef]
- Haskard, C.A.; El-Nezami, H.S.; Kankaanpaa, P.E. Surface Binding of Aflatoxin B1 by Lactic Acid Bacteria. Appl. Environ. Microbiol. 2001, 67, 3086–3091. [Google Scholar] [CrossRef]
- Silosuh, L.A.; Lethbridge, B.J.; Raffel, S.J. Biological activities of two fungistatic antibiotics produced by Bacillus cereus UW85. Appl. Environ. Microbiol. 1994, 60, 2023–2030. [Google Scholar] [CrossRef]
- Lei, S.; Liang, Z.; Li, J. Ochratoxin A biocontrol and biodegradation by Bacillus subtilis CW 14. J. Sci. Food Agric. 2014, 94, 1879–1885. [Google Scholar]
- Xu, J.; Wang, H.; Zhu, Z. Isolation and characterization of Bacillus amyloliquefaciens ZDS-1: Exploring the degradation of Zearalenone by Bacillus spp. Food Control 2016, 68, 244–250. [Google Scholar] [CrossRef]
- Smiley, R.; Draughon, F. Preliminary evidence that degradation of aflatoxin B1 by Flavobacterium aurantiacum is enzymatic. J. Food Prot. 2000, 63, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Farzaneh, M.; Shi, Z.Q.; Ahmadzadeh, M. Inhibition of the Aspergillus flavus Growth and Aflatoxin B1 Contamination on Pistachio Nut by Fengycin and Surfactin-Producing Bacillus subtilis UTBSP. Plant Pathol. J. 2016, 32, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Zhang, Y.; Li, M.; Garba, B.; Zhang, Q.; Wang, Y.; Zhang, H.; Li, P. Isolation and characterization of a Bacillus subtilis strain with aflatoxin B1 biodegradation capability. Food Control 2017, 75, 92–98. [Google Scholar] [CrossRef]
- Gu, X.; Sun, J.; Cui, Y.; Wang, X.; Sang, Y. Biological degradation of aflatoxin M1 by Bacillus pumilus E-1-1-1. MicrobiologyOpen 2019, 8, e00663. [Google Scholar] [CrossRef]
- Rao, K.R.; Vipin, A.V.; Hariprasad, P. Biological detoxification of Aflatoxin B1 by Bacillus licheniformis CFR1. Food Control 2016, 71, 234–241. [Google Scholar]
- Müller, R.; Antranikian, G.; Maloney, S.; Sharp, R. Thermophilic degradation of environmental pollutants. In Biotechnology of Extremophiles. Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2007; Volume 61, pp. 155–169. [Google Scholar]
- Yi, W.; Zhao, C.; Zhang, D. Effective degradation of aflatoxin B1 using a novel thermophilic microbial consortium TADC7. Bioresour. Technol. 2017, 224, 166–173. [Google Scholar]
- Sangare, L.; Zhao, Y.; Folly, Y.E. Aflatoxin B1 Degradation by a Pseudomonas Strain. Toxins 2014, 6, 3028. [Google Scholar] [CrossRef]
- Yu, Y.; Qiu, L.; Wu, H.; Tang, Y.; Lai, F. Oxidation of zearalenone by extracellular enzymes from Acinetobactersp. SM04 into smaller estrogenic products. World J. Microbiol. Biotechnol 2011, 27, 2675–2681. [Google Scholar] [CrossRef]
- Adebo, O.A.; Njobeh, P.B.; Gbashi, S.; Nwinyi, O.C.; Mavumengwana, V. Review on microbial degradation of aflatoxins. Crit. Rev. Food. Sci. Nutr 2017, 57, 3208–3217. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Jin, H.; Lan, J.; Zhang, R.; Ren, H.; Zhang, X. Detoxification of zearalenone by three strains of Lactobacillus Plantarumfrom fermented food in vitro. Food Control 2015, 54, 158–164. [Google Scholar] [CrossRef]
- Verheecke, C.; Liboz, T.; Mathieu, F. Microbial degradation of aflatoxin B1: Current status and future advances. Int. J. Food Microbiol 2016, 237, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Alberts, J.F.; Engelbrecht, Y.; Steyn, P.S. Biological degradation of aflatoxin B1 by Rhodococcus erythropolis cultures. Int. J. Food Microbiol. 2006, 109, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Le, V.; Zheng, X.; Chen, J. Characterization of volatile compounds in Fen-Daqu—A traditional Chinese liquor fermentation starter. J. Inst. Brew. 2012, 118, 107–113. [Google Scholar] [CrossRef]


| Samples | AFB1 Content (μg/kg) | AFB1 Degradation Ratio (%) |
|---|---|---|
| Top layer of grains | <0.1 | >99% |
| Middle layer of grains | <0.1 | >99% |
| Lower layer of grains | <0.1 | >99% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, G.; Qu, Y.; Wu, D.; Huang, S.; Che, Y.; Yu, J.; Song, P. Biodegradation of Aflatoxin B1 in the Baijiu Brewing Process by Bacillus cereus. Toxins 2023, 15, 65. https://doi.org/10.3390/toxins15010065
Xue G, Qu Y, Wu D, Huang S, Che Y, Yu J, Song P. Biodegradation of Aflatoxin B1 in the Baijiu Brewing Process by Bacillus cereus. Toxins. 2023; 15(1):65. https://doi.org/10.3390/toxins15010065
Chicago/Turabian StyleXue, Guoli, Yanjun Qu, Dan Wu, Shuyuan Huang, Yuqing Che, Jing Yu, and Ping Song. 2023. "Biodegradation of Aflatoxin B1 in the Baijiu Brewing Process by Bacillus cereus" Toxins 15, no. 1: 65. https://doi.org/10.3390/toxins15010065
APA StyleXue, G., Qu, Y., Wu, D., Huang, S., Che, Y., Yu, J., & Song, P. (2023). Biodegradation of Aflatoxin B1 in the Baijiu Brewing Process by Bacillus cereus. Toxins, 15(1), 65. https://doi.org/10.3390/toxins15010065





