Evaluation of the Synthetic Multifunctional Peptide Hp-MAP3 Derivative of Temporin-PTa
Abstract
1. Introduction
2. Results
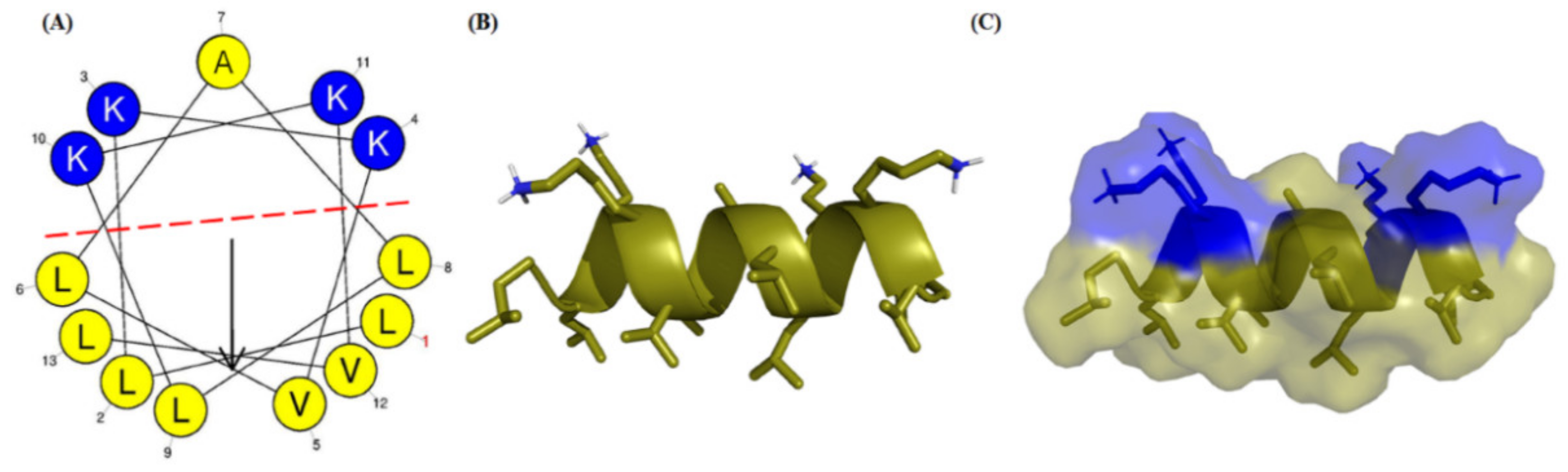
2.1. Rational Design and Synthesis
2.2. Evaluation of Antimicrobial Activity
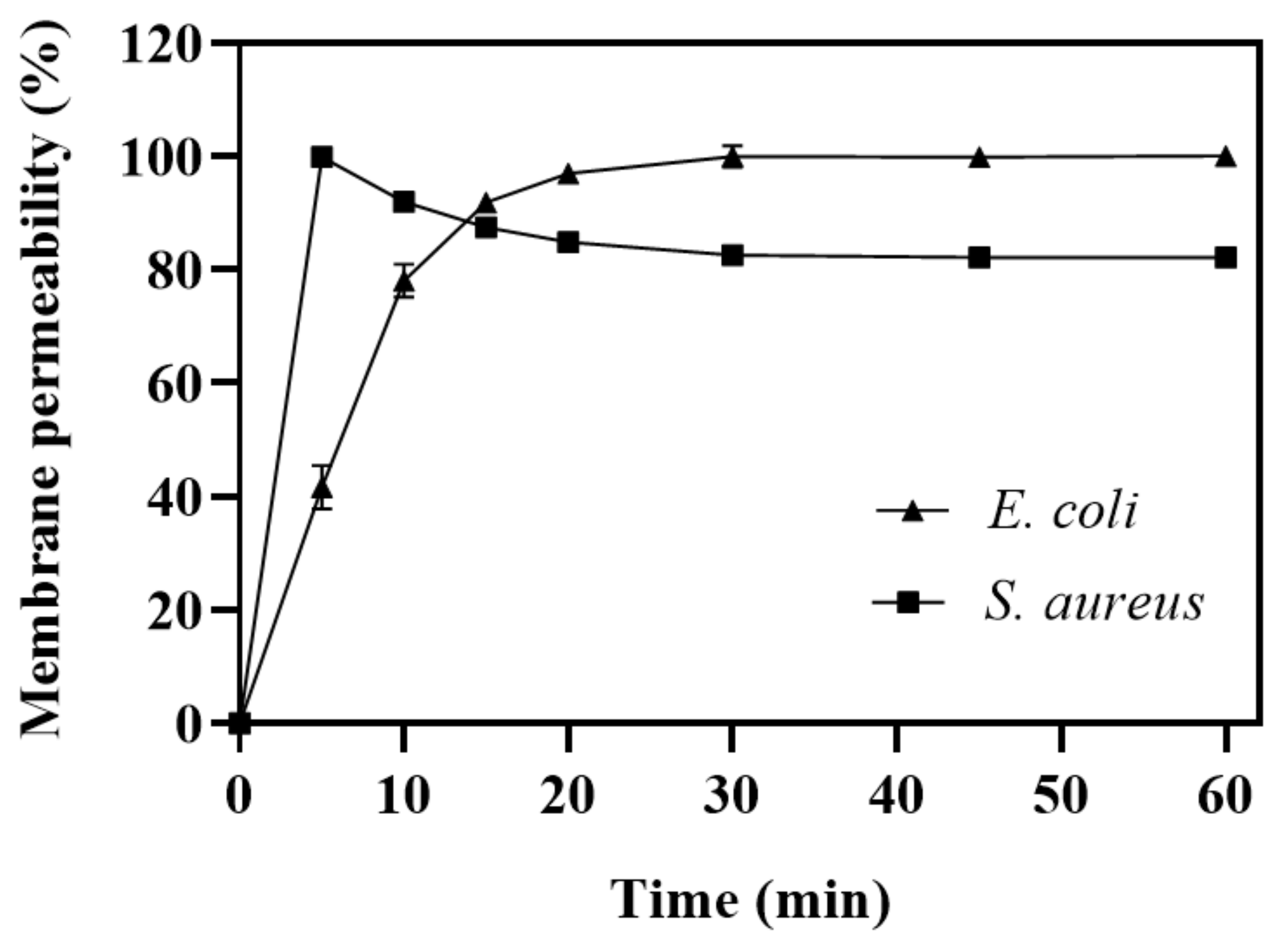
2.3. Evaluation of Damage to Bacterial Membrane
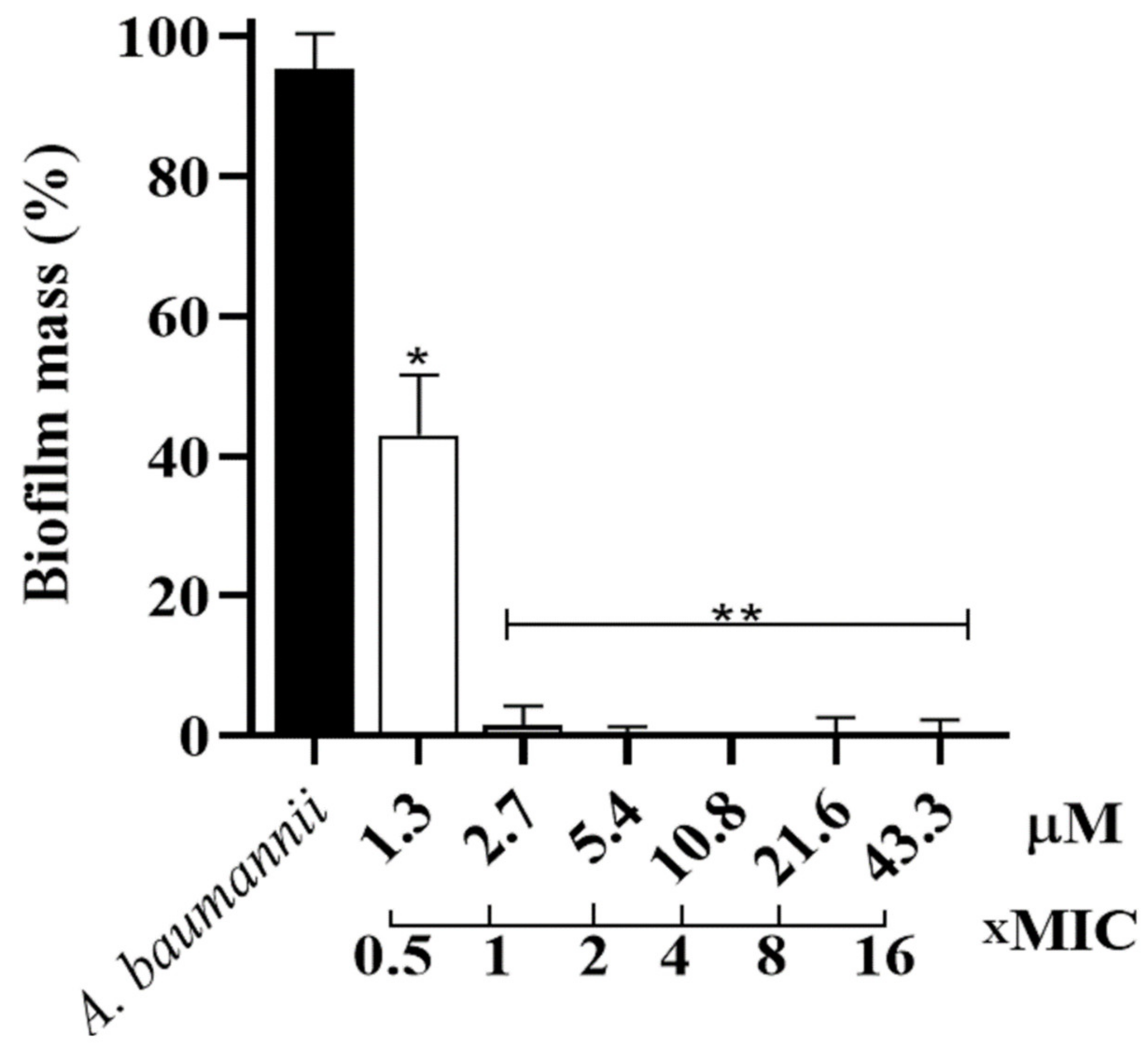
2.4. Evaluation of Antibiofilm Activity
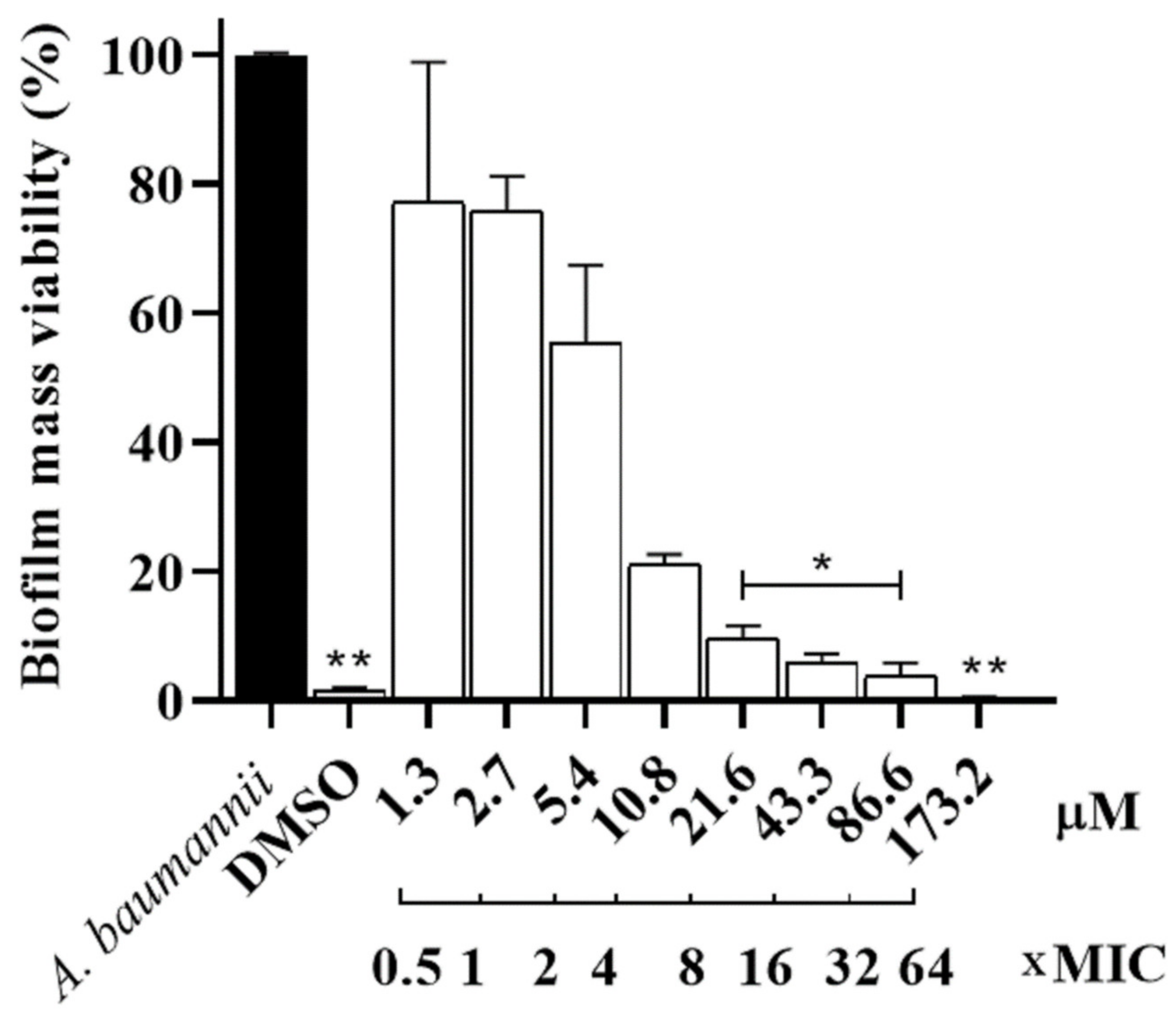
2.5. Evaluation of Pre-Formed Biofilm Activity
2.6. Evaluation of Antifungal Activity
2.7. Evaluation of Hp-MAP3 Cytotoxicity
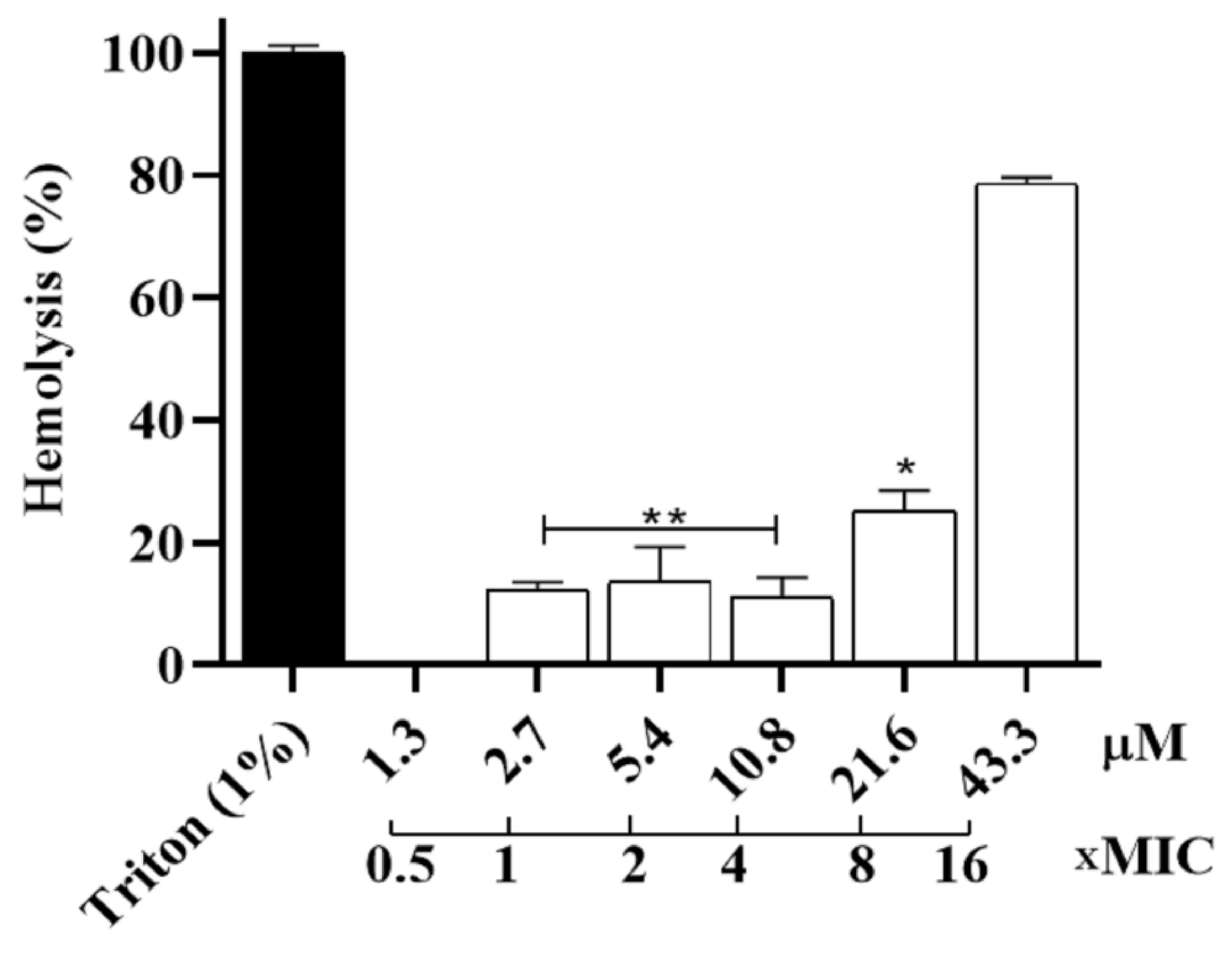
2.8. Hemolytic Assay
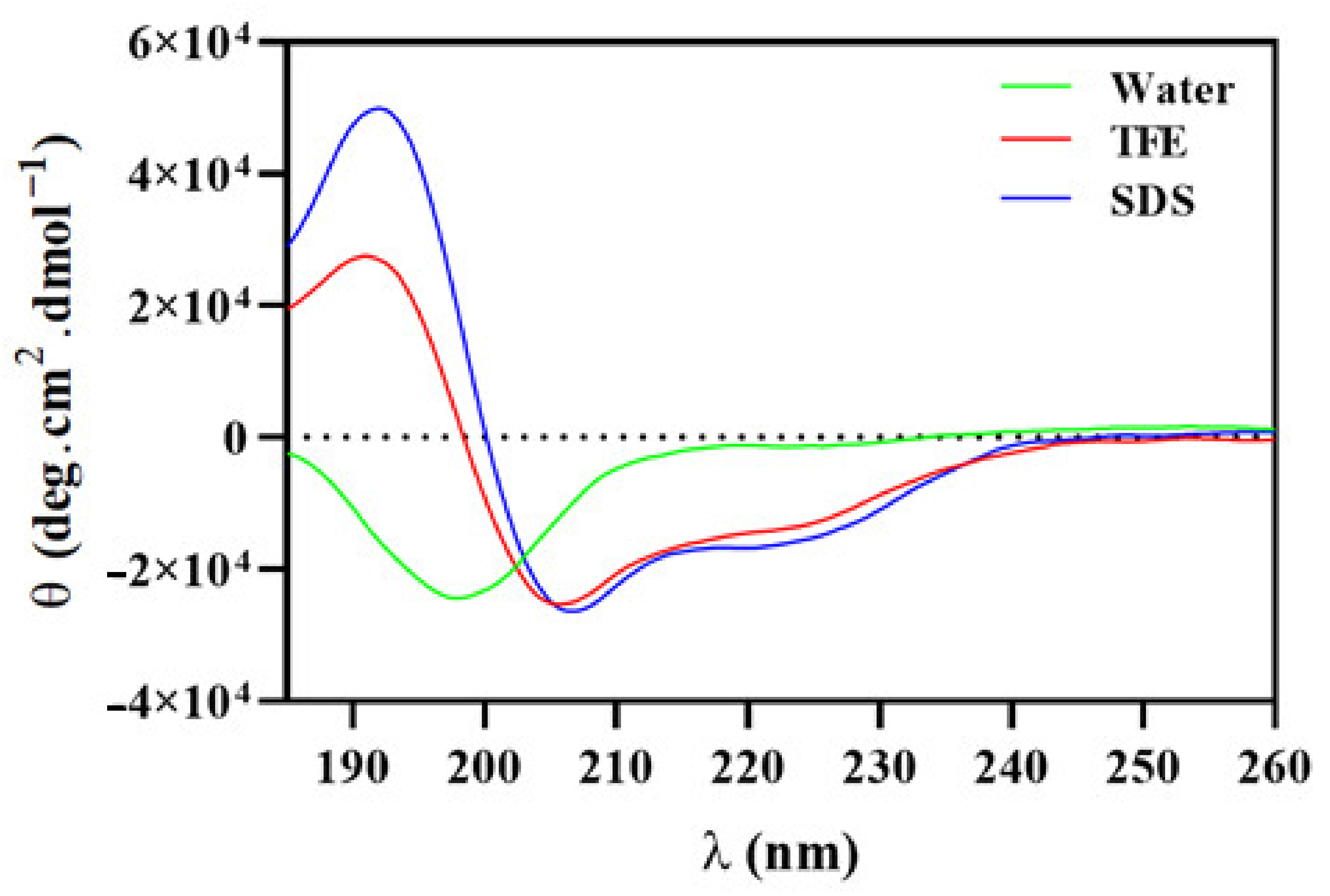
2.9. Secondary Structure of Hp-MAP3 in Different Environments
3. Discussion
4. Conclusions
5. Material and Methods
5.1. Rational Design and Synthesis
5.2. Minimal Inhibitory Concentration (MIC) and Minimal Bactericidal Concentration (MBC)
5.3. Membrane Permeabilization Using Sytox Green
5.4. Minimum Inhibitory Concentration of Biofilm (MICB)
5.5. Antibiofilm Activity
5.6. Yeast Strains and MIC Determinations
5.7. Cell Viability Test
5.8. Hemolytic Assay
5.9. Circular Dichroism Spectroscopy (CD)
5.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ventola, C.L. The Antibiotic Resistance Crisis: Part 1: Causes and Threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Bing, J.; Hu, T.; Ennis, C.L.; Nobile, C.J.; Huang, G. Candida Auris: Epidemiology, Biology, Antifungal Resistance, and Virulence. PLoS Pathog. 2020, 16, e1008921. [Google Scholar] [CrossRef] [PubMed]
- Calderone, R.A.; Fonzi, W.A. Virulence Factors of Candida Albicans. Trends Microbiol. 2001, 9, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Dadar, M.; Tiwari, R.; Karthik, K.; Chakraborty, S.; Shahali, Y.; Dhama, K. Candida Albicans—Biology, Molecular Characterization, Pathogenicity, and Advances in Diagnosis and Control—An Update. Microb. Pathog. 2018, 117, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Gunsalus, K.T.W.; Tornberg-Belanger, S.N.; Matthan, N.R.; Lichtenstein, A.H.; Kumamoto, C.A. Manipulation of Host Diet To Reduce Gastrointestinal Colonization by the Opportunistic Pathogen Candida Albicans. mSphere 2015, 1. [Google Scholar] [CrossRef]
- Andes, D.R.; Safdar, N.; Baddley, J.W.; Alexander, B.; Brumble, L.; Freifeld, A.; Hadley, S.; Herwaldt, L.; Kauffman, C.; Lyon, G.M.; et al. The Epidemiology and Outcomes of Invasive Candida Infections among Organ Transplant Recipients in the United States: Results of the Transplant-Associated Infection Surveillance Network (TRANSNET). Transpl. Infect. Dis. 2016, 18, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic Resistance of Bacterial Biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef]
- Batoni, G.; Maisetta, G.; Esin, S. Antimicrobial Peptides and Their Interaction with Biofilms of Medically Relevant Bacteria. Biochim. Biophys. Acta 2016, 1858, 1044–1060. [Google Scholar] [CrossRef]
- Cáncer. Available online: https://www.who.int/es/news-room/fact-sheets/detail/cancer (accessed on 21 August 2022).
- Yin, W.; Wang, J.; Jiang, L.; Kang, Y.J. Cancer and Stem Cells. Exp. Biol. Med. 2021, 246, 1791. [Google Scholar] [CrossRef]
- Nasim, F.; Sabath, B.F.; Eapen, G.A. Lung Cancer. Med. Clin. N. Am. 2019, 103, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Ferrall, L.; Lin, K.Y.; Roden, R.B.S.; Hung, C.F.; Wu, T.C. Cervical Cancer Immunotherapy: Facts and Hopes. Clin. Cancer Res. 2021, 27, 4953–4973. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.A.; Jhingran, A.; Oaknin, A.; Denny, L. Cervical Cancer. Lancet 2019, 393, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Corpus Uteri Source: Globocan 2020. Available online: https://gco.iarc.fr/today/data/factsheets/populations/900-world-fact-sheets.pdf (accessed on 21 August 2022).
- Kovachev, S.M. Cervical Cancer and Vaginal Microbiota Changes. Arch. Microbiol. 2020, 202, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Collignon, C.; Brisse, H.J.; Lemelle, L.; Cardoen, L.; Gauthier, A.; Pierron, G.; Roussel, A.; Dumont, B.; Alimi, A.; Cordero, C.; et al. Diagnostic Strategy in Pediatrics Soft Tissue Sarcomas. Bull. Cancer 2020, 107, 963–971. [Google Scholar] [CrossRef]
- Kapoor, G.; Das, K. Soft Tissue Sarcomas in Children. Indian J. Pediatr. 2012, 79, 936–942. [Google Scholar] [CrossRef]
- Skapek, S.X.; Ferrari, A.; Gupta, A.A.; Lupo, P.J.; Butler, E.; Shipley, J.; Barr, F.G.; Hawkins, D.S. Rhabdomyosarcoma. Nat. Rev. Dis. Prim. 2019, 5, 1. [Google Scholar] [CrossRef]
- Konai, M.M.; Barman, S.; Issa, R.; MacNeil, S.; Adhikary, U.; De, K.; Monk, P.N.; Haldar, J. Hydrophobicity-Modulated Small Antibacterial Molecule Eradicates Biofilm with Potent Efficacy against Skin Infections. ACS Infect. Dis. 2020, 6, 703–714. [Google Scholar] [CrossRef]
- Zakaryan, H.; Chilingaryan, G.; Arabyan, E.; Serobian, A.; Wang, G. Natural Antimicrobial Peptides as a Source of New Antiviral Agents. J. Gen. Virol. 2021, 102, 001661. [Google Scholar] [CrossRef]
- De Moura, G.A.; de Oliveira, J.R.; Rocha, Y.M.; de Oliveira Freitas, J.; Rodrigues, J.P.V.; Ferreira, V.P.G.; Nicolete, R. Antitumor and Antiparasitic Activity of Antimicrobial Peptides Derived from Snake Venom: A Systematic Review Approach. Curr. Med. Chem. 2022, 29, 5358–5368. [Google Scholar] [CrossRef]
- Guryanova, S.V.; Ovchinnikova, T.V. Immunomodulatory and Allergenic Properties of Antimicrobial Peptides. Int. J. Mol. Sci. 2022, 23, 2499. [Google Scholar] [CrossRef]
- Vanzolini, T.; Bruschi, M.; Rinaldi, A.C.; Magnani, M.; Fraternale, A. Multitalented Synthetic Antimicrobial Peptides and Their Antibacterial, Antifungal and Antiviral Mechanisms. Int. J. Mol. Sci. 2022, 23, 545. [Google Scholar] [CrossRef] [PubMed]
- Migliolo, L.; Silva, O.N.; Silva, P.A.; Costa, M.P.; Costa, C.R.; Nolasco, D.O.; Barbosa, J.A.R.G.; Silva, M.R.R.; Bemquerer, M.P.; Lima, L.M.P.; et al. Structural and Functional Characterization of a Multifunctional Alanine-Rich Peptide Analogue from Pleuronectes Americanus. PLoS ONE 2012, 7, e47047. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial Peptides of Multicellular Organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Bechinger, B. Insights into the Mechanisms of Action of Host Defence Peptides from Biophysical and Structural Investigations. J. Pept. Sci. 2011, 17, 306–314. [Google Scholar] [CrossRef]
- McMillan, K.A.M.; Coombs, M.R.P. Review: Examining the Natural Role of Amphibian Antimicrobial Peptide Magainin. Molecules 2020, 25, 5436. [Google Scholar] [CrossRef]
- Luisa Mangoni, M.; Di Grazia, A.; Cappiello, F.; Casciaro, B.; Luca, V. Naturally Occurring Peptides from Rana Temporaria: Antimicrobial Properties and More. Curr. Top. Med. Chem. 2015, 16, 54–64. [Google Scholar] [CrossRef]
- Srivastava, S.; Ghosh, J.K. Introduction of a Lysine Residue Promotes Aggregation of Temporin L in Lipopolysaccharides and Augmentation of Its Antiendotoxin Property. Antimicrob. Agents Chemother. 2013, 57, 2457. [Google Scholar] [CrossRef] [PubMed]
- Romero, S.M.; Cardillo, A.B.; Martínez Ceron, M.C.; Camperi, S.A.; Giudicessi, S.L. Temporins: An Approach of Potential Pharmaceutic Candidates. Surg. Infect. 2020, 21, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Souza e Silva, P.; Ferreira, M.A.; de Moraes, L.F.R.; de Barros, E.; Preza, S.L.E.; Cardoso, M.H.; Franco, O.L.; Migliolo, L. Synthetic Peptides Bioinspired in Temporin-PTa with Antibacterial and Antibiofilm Activity. Chem. Biol. Drug Des. 2022, 100, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Da Silva-Junio, A.G.; Frias, I.A.M.; Lima-Neto, R.G.; Migliolo, L.; e Silva, P.S.; Oliveira, M.D.L.; Andrade, C.A.S. Electrochemical Biosensor Based on Temporin-PTA Peptide for Detection of Microorganisms. J. Pharm. Biomed. Anal. 2022, 216, 114788. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Liu, C.J.; Zhang, X.Z. Multifunctional Peptides for Tumor Therapy. Adv. Drug Deliv. Rev. 2020, 160, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Diener, C.; Garza Ramos Martínez, G.; Moreno Blas, D.; Castillo González, D.A.; Corzo, G.; Castro-Obregon, S.; Del Rio, G. Effective Design of Multifunctional Peptides by Combining Compatible Functions. PLoS Comput. Biol. 2016, 12, e1004786. [Google Scholar] [CrossRef] [PubMed]
- Ang, Q.A.; Arfan, G.; Ong, C.Y.F.; Ng, F.M.; Ong, E.H.Q.; Chia, C.S.B. Designing a Leucine-Rich Antibacterial Nonapeptide with Potent Activity against Mupirocin-Resistant MRSA via a Structure-Activity Relationship Study. Chem. Biol. Drug Des. 2021, 97, 1185–1193. [Google Scholar] [CrossRef]
- Wang, C.; Dong, S.; Zhang, L.; Zhao, Y.; Huang, L.; Gong, X.; Wang, H.; Shang, D. Cell Surface Binding, Uptaking and Anticancer Activity of L-K6, a Lysine/Leucine-Rich Peptide, on Human Breast Cancer MCF-7 Cells. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Zelezetsky, I.; Tossi, A. Alpha-Helical Antimicrobial Peptides—Using a Sequence Template to Guide Structure-Activity Relationship Studies. Biochim. Biophys. Acta 2006, 1758, 1436–1449. [Google Scholar] [CrossRef] [PubMed]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of Antimicrobial Peptide Action and Resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef]
- Almeida, C.V.; de Oliveira, C.F.R.; dos Santos, E.L.; dos Santos, H.F.; Júnior, E.C.; Marchetto, R.; da Cruz, L.A.; Ferreira, A.M.T.; Gomes, V.M.; Taveira, G.B.; et al. Differential Interactions of the Antimicrobial Peptide, RQ18, with Phospholipids and Cholesterol Modulate Its Selectivity for Microorganism Membranes. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129937. [Google Scholar] [CrossRef]
- Sani, M.A.; Separovic, F. How Membrane-Active Peptides Get into Lipid Membranes. Acc. Chem. Res. 2016, 49, 1130–1138. [Google Scholar] [CrossRef]
- Domadia, P.N.; Bhunia, A.; Ramamoorthy, A.; Bhattacharjya, S. Structure, Interactions, and Antibacterial Activities of MSI-594 Derived Mutant Peptide MSI-594F5A in Lipopolysaccharide Micelles: Role of the Helical Hairpin Conformation in Outer-Membrane Permeabilization. J. Am. Chem. Soc. 2010, 132, 18417–18428. [Google Scholar] [CrossRef]
- Mishra, B.; Wang, G. Ab Initio Design of Potent Anti-MRSA Peptides Based on Database Filtering Technology. J. Am. Chem. Soc. 2012, 134, 12426–12429. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Kang, N.; Ko, S.J.; Park, J.; Park, E.; Shin, D.W.; Kim, S.H.; Lee, S.A.; Lee, J.I.; Lee, S.H.; et al. Antibacterial and Antibiofilm Activity and Mode of Action of Magainin 2 against Drug-Resistant Acinetobacter Baumannii. Int. J. Mol. Sci. 2018, 19, 3041. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.; Wang, X.; Lushnikova, T.; Zhang, Y.; Golla, R.M.; Narayana, J.L.; Wang, C.; McGuire, T.R.; Wang, G. Antibacterial, Antifungal, Anticancer Activities and Structural Bioinformatics Analysis of Six Naturally Occurring Temporins. Peptides 2018, 106, 9–20. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial Peptides: Pore Formers or Metabolic Inhibitors in Bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Melo, M.N.; Ferre, R.; Castanho, M.A.R.B. Antimicrobial Peptides: Linking Partition, Activity and High Membrane-Bound Concentrations. Nat. Rev. Microbiol. 2009, 7, 245–250. [Google Scholar] [CrossRef]
- Lebaron, P.; Catala, P.; Parthuisot, N. Effectiveness of SYTOX Green Stain for Bacterial Viability Assessment. Appl. Environ. Microbiol. 1998, 64, 2697–2700. [Google Scholar] [CrossRef]
- Mangoni, M.L.; Rinaldi, A.C.; Di Giulio, A.; Mignogna, G.; Bozzi, A.; Barra, D.; Simmaco, M. Structure-Function Relationships of Temporins, Small Antimicrobial Peptides from Amphibian Skin. Eur. J. Biochem. 2000, 267, 1447–1454. [Google Scholar] [CrossRef]
- Saviello, M.R.; Malfi, S.; Campiglia, P.; Cavalli, A.; Grieco, P.; Novellino, E.; Carotenuto, A. New Insight into the Mechanism of Action of the Temporin Antimicrobial Peptides. Biochemistry 2010, 49, 1477–1485. [Google Scholar] [CrossRef]
- Ciofu, O.; Tolker-Nielsen, T.; Jensen, P.Ø.; Wang, H.; Høiby, N. Antimicrobial Resistance, Respiratory Tract Infections and Role of Biofilms in Lung Infections in Cystic Fibrosis Patients. Adv. Drug Deliv. Rev. 2015, 85, 7–23. [Google Scholar] [CrossRef]
- Babapour, E.; Haddadi, A.; Mirnejad, R.; Angaji, S.A.; Amirmozafari, N. Biofilm Formation in Clinical Isolates of Nosocomial Acinetobacter Baumannii and Its Relationship with Multidrug Resistance. Asian Pac. J. Trop. Biomed. 2016, 6, 528–533. [Google Scholar] [CrossRef]
- Gaddy, J.A.; Actis, L.A. Regulation of Acinetobacter Baumannii Biofilm Formation. Future Microbiol. 2009, 4, 273–278. [Google Scholar] [CrossRef]
- Longo, F.; Vuotto, C.; Donelli, G. Biofilm Formation in Acinetobacter Baumannii. New Microbiol. 2014, 119–127. [Google Scholar]
- Grassi, L.; Maisetta, G.; Maccari, G.; Esin, S.; Batoni, G. Analogs of the Frog-Skin Antimicrobial Peptide Temporin 1Tb Exhibit a Wider Spectrum of Activity and a Stronger Antibiofilm Potential as Compared to the Parental Peptide. Front. Chem. 2017, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Segev-Zarko, L.; Saar-Dover, R.; Brumfeld, V.; Mangoni, M.L.; Shai, Y. Mechanisms of Biofilm Inhibition and Degradation by Antimicrobial Peptides. Biochem. J. 2015, 468, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Baindara, P.; Ghosh, A.K.; Mandal, S.M. Coevolution of Resistance Against Antimicrobial Peptides. Microb. Drug Resist. 2020, 26, 880–899. [Google Scholar] [CrossRef] [PubMed]
- Porto, W.F.; Irazazabal, L.N.; Humblot, V.; Haney, E.F.; Ribeiro, S.M.; Hancock, R.E.W.; Ladram, A.; Franco, O.L. EcDBS1R6: A Novel Cationic Antimicrobial Peptide Derived from a Signal Peptide Sequence. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129633. [Google Scholar] [CrossRef]
- Ma, L.; Ye, X.; Sun, P.; Xu, P.; Wang, L.; Liu, Z.; Huang, X.; Bai, Z.; Zhou, C. Antimicrobial and Antibiofilm Activity of the EeCentrocin 1 Derived Peptide EC1-17KV via Membrane Disruption. EBioMedicine 2020, 55, 102775. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden Killers: Human Fungal Infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef]
- Fisher, M.C.; Gurr, S.J.; Cuomo, C.A.; Blehert, D.S.; Jin, H.; Stukenbrock, E.H.; Stajich, J.E.; Kahmann, R.; Boone, C.; Denning, D.W.; et al. Threats Posed by the Fungal Kingdom to Humans, Wildlife, and Agriculture. MBio 2020, 11, e00449-20. [Google Scholar] [CrossRef]
- Mason, A.J.; Marquette, A.; Bechinger, B. Zwitterionic Phospholipids and Sterols Modulate Antimicrobial Peptide-Induced Membrane Destabilization. Biophys. J. 2007, 93, 4289–4299. [Google Scholar] [CrossRef]
- Bloch, K.E. Sterol Structure and Membrane Function. CRC Crit. Rev. Biochem. 1983, 14, 47–92. [Google Scholar] [CrossRef]
- Cirillo, S.; Tomeh, M.A.; Wilkinson, R.N.; Hill, C.; Brown, S.; Zhao, X. Designed Antitumor Peptide for Targeted SiRNA Delivery into Cancer Spheroids. ACS Appl. Mater. Interfaces 2021, 13, 49713–49728. [Google Scholar] [CrossRef]
- Nooranian, S.; Kazemi Oskuee, R.; Jalili, A. Characterization and Evaluation of Cell-Penetrating Activity of Brevinin-2R: An Amphibian Skin Antimicrobial Peptide. Mol. Biotechnol. 2022, 64, 546–559. [Google Scholar] [CrossRef]
- Rozek, T.; Wegener, K.L.; Bowie, J.H.; Olver, I.N.; Carver, J.A.; Wallace, J.C.; Tyler, M.J. The Antibiotic and Anticancer Active Aurein Peptides from the Australian Bell Frogs Litoria Aurea and Litoria Raniformis. Eur. J. Biochem. 2000, 267, 5330–5341. [Google Scholar] [CrossRef]
- Frihling, B.E.F.; Boleti, A.P.A.d; Oliveira, C.F.R.d; Sanches, S.C.; Cardoso, P.H.O.d.; Verbisck, N.; Macedo, M.L.R.; Rita, P.H.S.; Carvalho, C.M.E.; Migliolo, L. Purification, Characterization and Evaluation of the Antitumoral Activity of a Phospholipase A2 from the Snake Bothrops Moojeni. Pharmaceuticals 2022, 15, 724. [Google Scholar] [CrossRef] [PubMed]
- D’Abramo, M.; Rinaldi, A.C.; Bozzi, A.; Amadei, A.; Mignogna, G.; Di Nola, A.; Aschi, M. Conformational Behavior of Temporin A and Temporin L in Aqueous Solution: A Computational/Experimental Study. Biopolymers 2006, 81, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, M.H.; Orozco, R.Q.; Rezende, S.B.; Rodrigues, G.; Oshiro, K.G.N.; Cândido, E.S.; Franco, O.L. Computer-Aided Design of Antimicrobial Peptides: Are We Generating Effective Drug Candidates? Front. Microbiol. 2020, 10, 3097. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Rukmini, R.; Chattopadhyay, A. Modulation of Tryptophan Environment in Membrane-Bound Melittin by Negatively Charged Phospholipids: Implications in Membrane Organization and Function. Biochemistry 1997, 36, 14291–14305. [Google Scholar] [CrossRef]
- Chan, D.I.; Prenner, E.J.; Vogel, H.J. Tryptophan- and Arginine-Rich Antimicrobial Peptides: Structures and Mechanisms of Action. Biochim. Biophys. Acta 2006, 1758, 1184–1202. [Google Scholar] [CrossRef]
- Rinaldi, A.C.; Mangoni, M.L.; Rufo, A.; Luzi, C.; Barra, D.; Zhao, H.; Kinnunen, P.K.J.; Bozzi, A.; Di Giulio, A.; Simmaco, M. Temporin L: Antimicrobial, Haemolytic and Cytotoxic Activities, and Effects on Membrane Permeabilization in Lipid Vesicles. Biochem. J. 2002, 368, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, A.C. Antimicrobial Peptides from Amphibian Skin: An Expanding Scenario. Curr. Opin. Chem. Biol. 2002, 6, 799–804. [Google Scholar] [CrossRef] [PubMed]
- Domanov, Y.A.; Kinnunen, P.K.J. Antimicrobial Peptides Temporins B and L Induce Formation of Tubular Lipid Protrusions from Supported Phospholipid Bilayers. Biophys. J. 2006, 91, 4427. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fan, Y.; Zhou, Z.; Tu, H.; Ren, Q.; Wang, X.; Ding, L.; Zhou, X.; Zhang, L. De Novo Synthetic Short Antimicrobial Peptides against Cariogenic Bacteria. Arch. Oral Biol. 2017, 80, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Migliolo, L.; Felício, M.R.; Cardoso, M.H.; Silva, O.N.; Xavier, M.A.E.; Nolasco, D.O.; De Oliveira, A.S.; Roca-Subira, I.; Vila Estape, J.; Teixeira, L.D.; et al. Structural and Functional Evaluation of the Palindromic Alanine-Rich Antimicrobial Peptide Pa-MAP2. Biochim. Biophys. Acta Biomembr. 2016, 1858, 1488–1498. [Google Scholar] [CrossRef]
- Conlon, J.M. Reflections on a Systematic Nomenclature for Antimicrobial Peptides from the Skins of Frogs of the Family Ranidae. Peptides 2008, 29, 1815–1819. [Google Scholar] [CrossRef]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A Web Server to Screen Sequences with Specific α-Helical Properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-Web: Interactive Web Service for the Recognition of Errors in Three-Dimensional Structures of Proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Mohanram, H.; Bhattacharjya, S. Salt-Resistant Short Antimicrobial Peptides. Biopolymers 2016, 106, 345–356. [Google Scholar] [CrossRef]
- Calvo, B.; Melo, A.S.A.; Perozo-Mena, A.; Hernandez, M.; Francisco, E.C.; Hagen, F.; Meis, J.F.; Colombo, A.L. First Report of Candida Auris in America: Clinical and Microbiological Aspects of 18 Episodes of Candidemia. J. Infect. 2016, 73, 369–374. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Park, S.C.; Kim, M.H.; Lim, H.T.; Park, Y.; Hahm, K.S. Antimicrobial Activity Studies on a Trypsin–Chymotrypsin Protease Inhibitor Obtained from Potato. Biochem. Biophys. Res. Commun. 2005, 330, 921–927. [Google Scholar] [CrossRef] [PubMed]

| Microorganism | Hp-MAP3 (µM) | Ciprofloxacin (µM) | ||
|---|---|---|---|---|
| Gram-Negative | MIC a | MBC b | MIC a | MBC b |
| Acinetobacter baumannii (clinical isolated) | 2.7 | 2.7 | 54.4 | >100.8 |
| Escherichia coli (clinical isolated) | 5.4 | 21.6 | 54.4 | >100.8 |
| Escherichia coli (KPC + 34) | 10.8 | 21.6 | 100.8 | 100.8 |
| Klebsiella pneumoniae (ATCC 36) | 5.4 | 5.4 | 100.8 | 100.8 |
| Klebsiella pneumoniae (KPC + 39) | 10.8 | 21.6 | 54.4 | 54.4 |
| Pseudomonas aeruginosa (ATCC) | 43.3 | 43.3 | 6.3 | 6.3 |
| Gram-positive | ||||
| Staphylococcus aureus (clinical isolated) | 43.3 | 43.3 | 6.3 | 6.3 |
| Microorganism | Hp-MAP3 (µM) |
|---|---|
| Candida albicans ATCC 90029 | 21.6 |
| C. auris CBS 10913 | 10.8 |
| C. auris 2015/466 | 5.4 |
| C. auris 2015/467 | 10.8 |
| C. auris 2015/468 | 10.8 |
| C. auris 2015/470 | 10.8 |
| C. glabrata ATCC 9030 | 86.6 |
| C. krusei ATCC 6258 | 43.3 |
| C. parapsilosis ATCC 22019 | 43.3 |
| C. tropicalis ATCC 750 | 21.6 |
| Cell Line | Hp-MAP3 | |
|---|---|---|
| IC50 * | IS ** | |
| MRC-5 | 19.83 | - |
| RD | 24.01 | 0.82 |
| NCI-H292 | 15.44 | 1.28 |
| HeLa | 10.46 | 1.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, P.S.e.; Guindo, A.S.; Oliveira, P.H.C.; de Moraes, L.F.R.N.; Boleti, A.P.d.A.; Ferreira, M.A.; de Oliveira, C.F.R.; Macedo, M.L.R.; Rossato, L.; Simionatto, S.; et al. Evaluation of the Synthetic Multifunctional Peptide Hp-MAP3 Derivative of Temporin-PTa. Toxins 2023, 15, 42. https://doi.org/10.3390/toxins15010042
Silva PSe, Guindo AS, Oliveira PHC, de Moraes LFRN, Boleti APdA, Ferreira MA, de Oliveira CFR, Macedo MLR, Rossato L, Simionatto S, et al. Evaluation of the Synthetic Multifunctional Peptide Hp-MAP3 Derivative of Temporin-PTa. Toxins. 2023; 15(1):42. https://doi.org/10.3390/toxins15010042
Chicago/Turabian StyleSilva, Patrícia Souza e, Alexya Sandim Guindo, Pedro Henrique Cardoso Oliveira, Luiz Filipe Ramalho Nunes de Moraes, Ana Paula de Araújo Boleti, Marcos Antonio Ferreira, Caio Fernando Ramalho de Oliveira, Maria Ligia Rodrigues Macedo, Luana Rossato, Simone Simionatto, and et al. 2023. "Evaluation of the Synthetic Multifunctional Peptide Hp-MAP3 Derivative of Temporin-PTa" Toxins 15, no. 1: 42. https://doi.org/10.3390/toxins15010042
APA StyleSilva, P. S. e., Guindo, A. S., Oliveira, P. H. C., de Moraes, L. F. R. N., Boleti, A. P. d. A., Ferreira, M. A., de Oliveira, C. F. R., Macedo, M. L. R., Rossato, L., Simionatto, S., & Migliolo, L. (2023). Evaluation of the Synthetic Multifunctional Peptide Hp-MAP3 Derivative of Temporin-PTa. Toxins, 15(1), 42. https://doi.org/10.3390/toxins15010042






